Diet and SIRT1 Genotype Interact to Modulate Aging-Related Processes in Patients with Coronary Heart Disease: From the CORDIOPREV Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Population
2.2. Diet, Dietary Assessment and Follow-Up Visits
2.3. Laboratory Measurements
2.4. OxS- and Inflammation-Related Parameters
2.5. DNA Isolation from Blood Samples
2.6. Genotyping
2.7. Quantitative PCR Analysis of Leucocyte Telomere Length (LTL)
2.8. Statistics
3. Results
3.1. Baseline Characteristics according to the SIRT1 SNPs Genotype
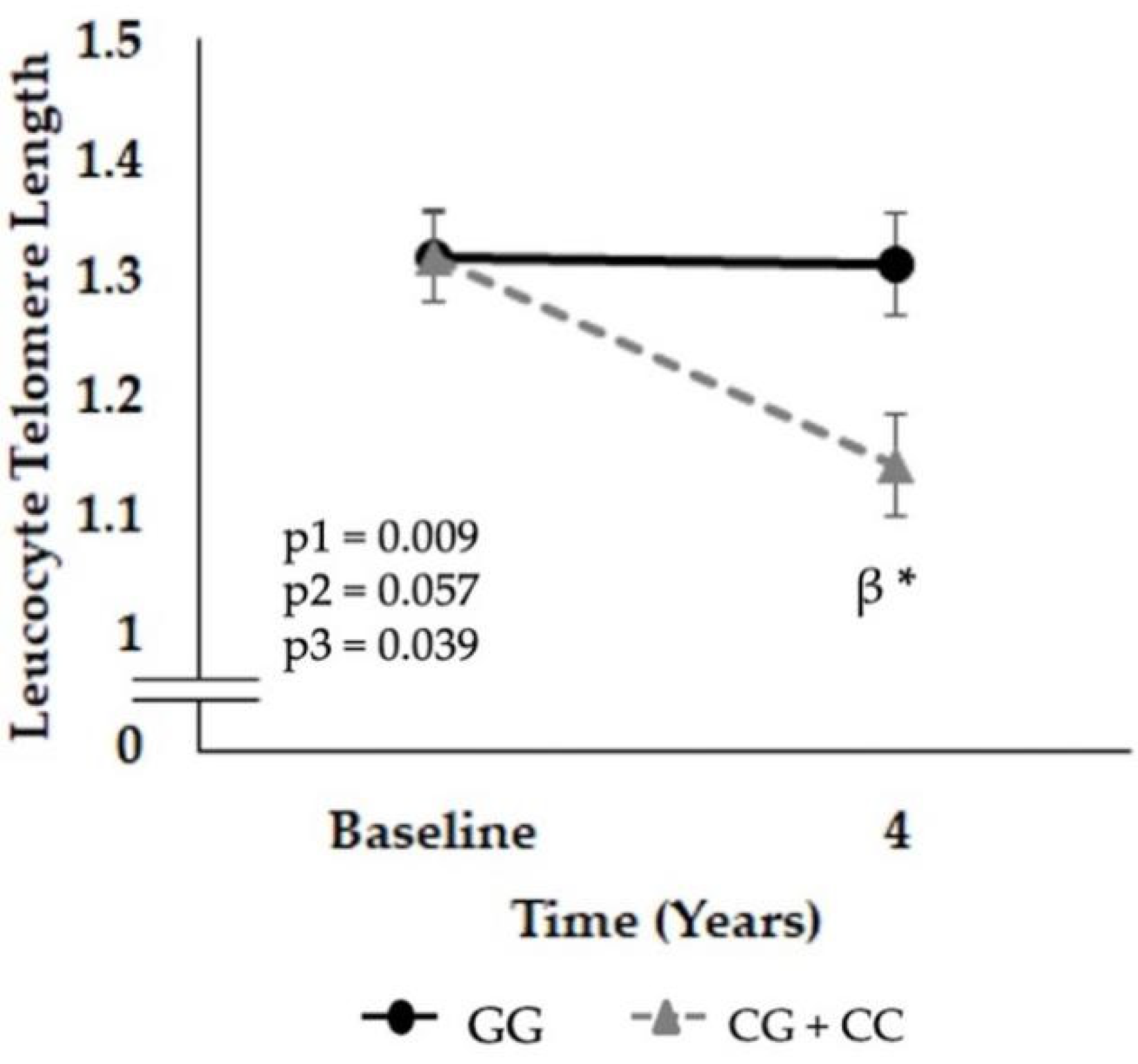
3.2. Relationship between the SNP rs7069102 and LTL
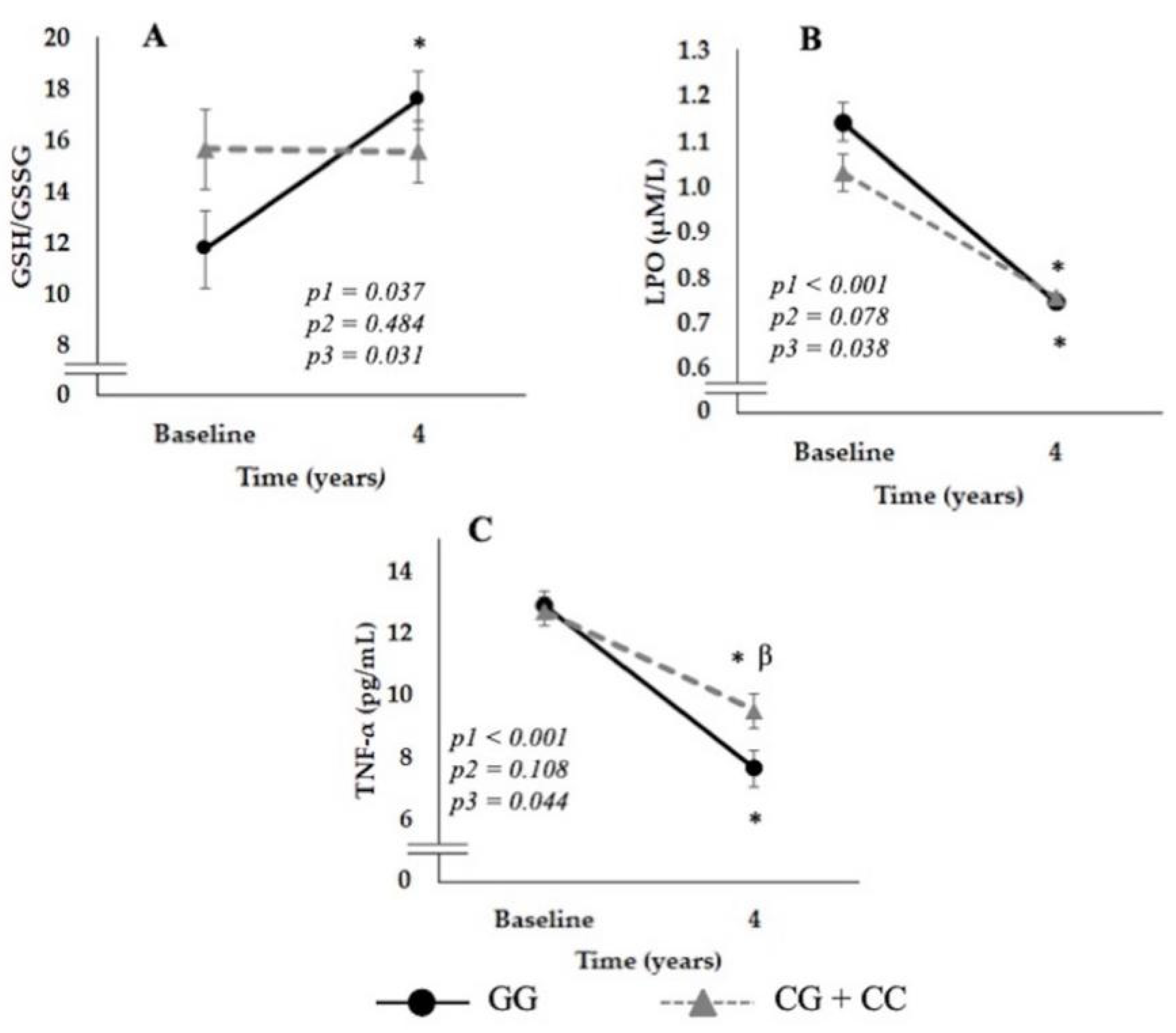
3.3. Relationship between the SNP rs7069102 and Inflammation and OxS-Related Parameters
3.4. Effect of Diet on LTL and Inflammation-Related Parameters According to SIRT1 Gene Variants
3.4.1. Patients Randomized to the LF Diet
3.4.2. Patients Randomized to Med Diet
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ebeling, M.; Rau, R.; Malmstrom, H.; Ahlbom, A.; Modig, K. The rate by which mortality increases with age is the same for those who experienced chronic disease as for the general population. Age Ageing 2021, 50, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ageing and Health. Available online: https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (accessed on 25 May 2022).
- Corina, A.; Rangel-Zuniga, O.A.; Jimenez-Lucena, R.; Alcala-Diaz, J.F.; Quintana-Navarro, G.; Yubero-Serrano, E.M.; Lopez-Moreno, J.; Delgado-Lista, J.; Tinahones, F.; Ordovas, J.M.; et al. Low Intake of Vitamin E Accelerates Cellular Aging in Patients With Established Cardiovascular Disease: The CORDIOPREV Study. J. Gerontol. A Biol. Sci. Med. Sci. 2019, 74, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Demissie, S.; Levy, D.; Benjamin, E.J.; Cupples, L.A.; Gardner, J.P.; Herbert, A.; Kimura, M.; Larson, M.G.; Meigs, J.B.; Keaney, J.F.; et al. Insulin resistance, oxidative stress, hypertension, and leukocyte telomere length in men from the Framingham Heart Study. Aging Cell 2006, 5, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Lista, J.; Alcala-Diaz, J.F.; Torres-Pena, J.D.; Quintana-Navarro, G.M.; Fuentes, F.; Garcia-Rios, A.; Ortiz-Morales, A.M.; Gonzalez-Requero, A.I.; Perez-Caballero, A.I.; Yubero-Serrano, E.M.; et al. Long-term secondary prevention of cardiovascular disease with a Mediterranean diet and a low-fat diet (CORDIOPREV): A randomised controlled trial. Lancet 2022, 399, 1876–1885. [Google Scholar] [CrossRef]
- Estruch, R.; Ros, E.; Salas-Salvado, J.; Covas, M.I.; Corella, D.; Aros, F.; Gomez-Gracia, E.; Ruiz-Gutierrez, V.; Fiol, M.; Lapetra, J.; et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef]
- Galie, S.; Canudas, S.; Muralidharan, J.; Garcia-Gavilan, J.; Bullo, M.; Salas-Salvado, J. Impact of Nutrition on Telomere Health: Systematic Review of Observational Cohort Studies and Randomized Clinical Trials. Adv. Nutr. 2020, 11, 576–601. [Google Scholar] [CrossRef]
- Dolinsky, V.W.; Dyck, J.R. Calorie restriction and resveratrol in cardiovascular health and disease. Biochim. Biophys. Acta 2011, 1812, 1477–1489. [Google Scholar] [CrossRef]
- Eyre, H.; Kahn, R.; Robertson, R.M.; Clark, N.G.; Doyle, C.; Hong, Y.; Gansler, T.; Glynn, T.; Smith, R.A.; Taubert, K.; et al. Preventing cancer, cardiovascular disease, and diabetes: A common agenda for the American Cancer Society, the American Diabetes Association, and the American Heart Association. Circulation 2004, 109, 3244–3255. [Google Scholar] [CrossRef]
- Weiss, E.P.; Fontana, L. Caloric restriction: Powerful protection for the aging heart and vasculature. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1205–H1219. [Google Scholar] [CrossRef]
- Johnson, S.C.; Dong, X.; Vijg, J.; Suh, Y. Genetic evidence for common pathways in human age-related diseases. Aging Cell 2015, 14, 809–817. [Google Scholar] [CrossRef]
- Richardson, A.G.; Schadt, E.E. The role of macromolecular damage in aging and age-related disease. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. 1), S28–S32. [Google Scholar] [CrossRef] [PubMed]
- Angelakopoulou, A.; Shah, T.; Sofat, R.; Shah, S.; Berry, D.J.; Cooper, J.; Palmen, J.; Tzoulaki, I.; Wong, A.; Jefferis, B.J.; et al. Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: Cardiovascular Biomarker Genetics Collaboration. Eur. Heart J. 2012, 33, 393–407. [Google Scholar] [CrossRef]
- Corsetti, J.P.; Gansevoort, R.T.; Navis, G.; Sparks, C.E.; Dullaart, R.P. LPL polymorphism (D9N) predicts cardiovascular disease risk directly and through interaction with CETP polymorphism (TaqIB) in women with high HDL cholesterol and CRP. Atherosclerosis 2011, 214, 373–376. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vasto, S.; Candore, G.; Duro, G.; Lio, D.; Grimaldi, M.P.; Caruso, C. Alzheimer’s disease and genetics of inflammation: A pharmacogenomic vision. Pharmacogenomics 2007, 8, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.; Li, T.; Liu, M.; Wang, Y.; Ma, H.; Mu, N.; Wang, H. SIRT6 in Senescence and Aging-Related Cardiovascular Diseases. Front. Cell Dev. Biol. 2021, 9, 641315. [Google Scholar] [CrossRef]
- Singh, V.; Ubaid, S. Role of Silent Information Regulator 1 (SIRT1) in Regulating Oxidative Stress and Inflammation. Inflammation 2020, 43, 1589–1598. [Google Scholar] [CrossRef]
- Chalkiadaki, A.; Guarente, L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012, 16, 180–188. [Google Scholar] [CrossRef]
- Russo, M.A.; Sansone, L.; Polletta, L.; Runci, A.; Rashid, M.M.; De Santis, E.; Vernucci, E.; Carnevale, I.; Tafani, M. Sirtuins and resveratrol-derived compounds: A model for understanding the beneficial effects of the Mediterranean diet. Endocr. Metab. Immune Disord. Drug Targets 2014, 14, 300–308. [Google Scholar] [CrossRef]
- Chang, H.C.; Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. TEM 2014, 25, 138–145. [Google Scholar] [CrossRef]
- Wenzel, U. Nutrition, sirtuins and aging. Genes Nutr. 2006, 1, 85–93. [Google Scholar] [CrossRef]
- Yamamoto, H.; Schoonjans, K.; Auwerx, J. Sirtuin functions in health and disease. Mol. Endocrinol. 2007, 21, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Pang, J.; Han, Y.; Dai, Y.; Dai, D.; Cai, J.; Zhang, T.M. Age-dependent tissue expression patterns of Sirt1 in senescence-accelerated mice. Mol. Med. Rep. 2014, 10, 3296–3302. [Google Scholar] [CrossRef]
- Kilic, U.; Gok, O.; Bacaksiz, A.; Izmirli, M.; Elibol-Can, B.; Uysal, O. SIRT1 gene polymorphisms affect the protein expression in cardiovascular diseases. PLoS ONE 2014, 9, e90428. [Google Scholar] [CrossRef]
- Quintana-Navarro, G.M.; Alcala-Diaz, J.F.; Lopez-Moreno, J.; Perez-Corral, I.; Leon-Acuna, A.; Torres-Pena, J.D.; Rangel-Zuniga, O.A.; Arenas de Larriva, A.P.; Corina, A.; Camargo, A.; et al. Long-term dietary adherence and changes in dietary intake in coronary patients after intervention with a Mediterranean diet or a low-fat diet: The CORDIOPREV randomized trial. Eur. J. Nutr. 2020, 59, 2099–2110. [Google Scholar] [CrossRef]
- Delgado-Lista, J.; Perez-Martinez, P.; Garcia-Rios, A.; Alcala-Diaz, J.F.; Perez-Caballero, A.I.; Gomez-Delgado, F.; Fuentes, F.; Quintana-Navarro, G.; Lopez-Segura, F.; Ortiz-Morales, A.M.; et al. CORonary Diet Intervention with Olive oil and cardiovascular PREVention study (the CORDIOPREV study): Rationale, methods, and baseline characteristics: A clinical trial comparing the efficacy of a Mediterranean diet rich in olive oil versus a low-fat diet on cardiovascular disease in coronary patients. Am. Heart J. 2016, 177, 42–50. [Google Scholar] [CrossRef]
- Rangel-Zuniga, O.A.; Corina, A.; Lucena-Porras, B.; Cruz-Teno, C.; Gomez-Delgado, F.; Jimenez-Lucena, R.; Alcala-Diaz, J.F.; Haro-Mariscal, C.; Yubero-Serrano, E.M.; Delgado-Lista, J.; et al. TNFA gene variants related to the inflammatory status and its association with cellular aging: From the CORDIOPREV study. Exp. Gerontol. 2016, 83, 56–62. [Google Scholar] [CrossRef]
- Ortiz-Morales, A.M.; Alcala-Diaz, J.F.; Rangel-Zuniga, O.A.; Corina, A.; Quintana-Navarro, G.; Cardelo, M.P.; Yubero-Serrano, E.; Malagon, M.M.; Delgado-Lista, J.; Ordovas, J.M.; et al. Biological senescence risk score. A practical tool to predict biological senescence status. Eur. J. Clin. Investig. 2020, 50, e13305. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Bińkowski, J.; Miks, S. Gene-Calc [Computer Software]. September 2018. Available online: www.gene-calc.pl (accessed on 11 September 2022).
- Cawthon, R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002, 30, e47. [Google Scholar] [CrossRef]
- Gomez-Delgado, F.; Delgado-Lista, J.; Lopez-Moreno, J.; Rangel-Zuniga, O.A.; Alcala-Diaz, J.F.; Leon-Acuna, A.; Corina, A.; Yubero-Serrano, E.; Torres-Pena, J.D.; Camargo, A.; et al. Telomerase RNA Component Genetic Variants Interact With the Mediterranean Diet Modifying the Inflammatory Status and its Relationship with Aging: CORDIOPREV Study. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 327–332. [Google Scholar] [CrossRef]
- Gonzalez-Guardia, L.; Yubero-Serrano, E.M.; Rangel-Zuniga, O.; Marin, C.; Camargo, A.; Perez-Martinez, P.; Delgado-Lista, J.; Gomez-Delgado, F.; Garcia-Rios, A.; Tinahones, F.J.; et al. Influence of endothelial dysfunction on telomere length in subjects with metabolic syndrome: LIPGENE study. Age 2014, 36, 9681. [Google Scholar] [CrossRef] [PubMed]
- Passos, J.F.; Saretzki, G.; von Zglinicki, T. DNA damage in telomeres and mitochondria during cellular senescence: Is there a connection? Nucleic Acids Res. 2007, 35, 7505–7513. [Google Scholar] [CrossRef] [PubMed]
- Salpea, K.D.; Talmud, P.J.; Cooper, J.A.; Maubaret, C.G.; Stephens, J.W.; Abelak, K.; Humphries, S.E. Association of telomere length with type 2 diabetes, oxidative stress and UCP2 gene variation. Atherosclerosis 2010, 209, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, N.; Hastings, J.L.; Bhella, P.S.; Shibata, S.; Gandhi, N.K.; Carrick-Ranson, G.; Palmer, D.; Levine, B.D. Effect of ageing on left ventricular compliance and distensibility in healthy sedentary humans. J. Physiol. 2012, 590, 1871–1880. [Google Scholar] [CrossRef]
- Inamori, T.; Goda, T.; Kasezawa, N.; Yamakawa-Kobayashi, K. The combined effects of genetic variation in the SIRT1 gene and dietary intake of n-3 and n-6 polyunsaturated fatty acids on serum LDL-C and HDL-C levels: A population-based study. Lipids Health Dis. 2013, 12, 4. [Google Scholar] [CrossRef]

| Parameters | LF Diet | Med Diet | ||||
|---|---|---|---|---|---|---|
| GG | CG + CC | p Value | GG | CG + CC | p Value | |
| N | 162 | 171 | 189 | 194 | ||
| Men/Women (n) | 134/28 | 145/26 | 0.607 | 164/25 | 151/43 | 0.022 * |
| Age (years) | 58.7 ± 8.7 | 59.6 ± 8.1 | 0.329 | 59.3 ± 8.9 | 59.8 ± 9.2 | 0.629 |
| Weight (kg) | 85.3 ± 12.8 | 84.5 ± 12.2 | 0.547 | 86.6 ± 14.2 | 83.7 ± 14 | 0.043 * |
| Waist circumference (cm) | 104.5 ± 10.4 | 105.1 ± 10.2 | 0.628 | 105.2 ± 11.5 | 104.1 ± 11 | 0.311 |
| BMI (kg/m2) | 31.2 ± 4.3 | 30.7 ± 4 | 0.289 | 31.4 ± 4.4 | 30.9 ± 4.4 | 0.315 |
| Total cholesterol (mg/dL) | 157.2± 29.8 | 160.1 ± 28.2 | 0.359 | 158.5 ± 33.1 | 159.6 ± 31.5 | 0.723 |
| HDL-C (mg/dL) | 42.1 ± 10 | 42.9 ± 10.1 | 0.452 | 42.2 ± 10.3 | 42.1 ± 10.3 | 0.931 |
| LDL-C (mg/dL) | 86.8 ± 23.9 | 89.7± 23.5 | 0.281 | 88.2 ± 26.6 | 90.1 ± 25.4 | 0.486 |
| TG (mg/dL) | 128 ± 64.4 | 142.2 ± 73.4 | 0.063 | 137.1 ± 71 | 130.1 ± 64.5 | 0.315 |
| Glucose (mg/dL) | 108.8 ± 29.5 | 113.4 ± 38.3 | 0.219 | 115.4 ± 40.8 | 111.4 ± 36.3 | 0.316 |
| CRP (mg/dL) | 3.6 ± 4.3 | 2.8 ± 3.2 | 0.073 | 2.7 ± 3.7 | 2.8 ± 3.1 | 0.945 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hidalgo-Moyano, C.; Rangel-Zuñiga, O.A.; Gomez-Delgado, F.; Alcala-Diaz, J.F.; Rodriguez-Cantalejo, F.; Yubero-Serrano, E.M.; Torres-Peña, J.D.; Arenas-de Larriva, A.P.; Camargo, A.; Perez-Martinez, P.; et al. Diet and SIRT1 Genotype Interact to Modulate Aging-Related Processes in Patients with Coronary Heart Disease: From the CORDIOPREV Study. Nutrients 2022, 14, 3789. https://doi.org/10.3390/nu14183789
Hidalgo-Moyano C, Rangel-Zuñiga OA, Gomez-Delgado F, Alcala-Diaz JF, Rodriguez-Cantalejo F, Yubero-Serrano EM, Torres-Peña JD, Arenas-de Larriva AP, Camargo A, Perez-Martinez P, et al. Diet and SIRT1 Genotype Interact to Modulate Aging-Related Processes in Patients with Coronary Heart Disease: From the CORDIOPREV Study. Nutrients. 2022; 14(18):3789. https://doi.org/10.3390/nu14183789
Chicago/Turabian StyleHidalgo-Moyano, Cristina, Oriol Alberto Rangel-Zuñiga, Francisco Gomez-Delgado, Juan F. Alcala-Diaz, Fernando Rodriguez-Cantalejo, Elena M. Yubero-Serrano, Jose D. Torres-Peña, Antonio P. Arenas-de Larriva, Antonio Camargo, Pablo Perez-Martinez, and et al. 2022. "Diet and SIRT1 Genotype Interact to Modulate Aging-Related Processes in Patients with Coronary Heart Disease: From the CORDIOPREV Study" Nutrients 14, no. 18: 3789. https://doi.org/10.3390/nu14183789
APA StyleHidalgo-Moyano, C., Rangel-Zuñiga, O. A., Gomez-Delgado, F., Alcala-Diaz, J. F., Rodriguez-Cantalejo, F., Yubero-Serrano, E. M., Torres-Peña, J. D., Arenas-de Larriva, A. P., Camargo, A., Perez-Martinez, P., Lopez-Miranda, J., & Delgado-Lista, J. (2022). Diet and SIRT1 Genotype Interact to Modulate Aging-Related Processes in Patients with Coronary Heart Disease: From the CORDIOPREV Study. Nutrients, 14(18), 3789. https://doi.org/10.3390/nu14183789






