Genetic Causal Association between Iron Status and Osteoarthritis: A Two-Sample Mendelian Randomization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Instrumental Variable Selection
2.3. Mendelian Randomization Analysis
2.4. Sensitivity Analysis
2.5. Further Validation of MR Results
3. Results
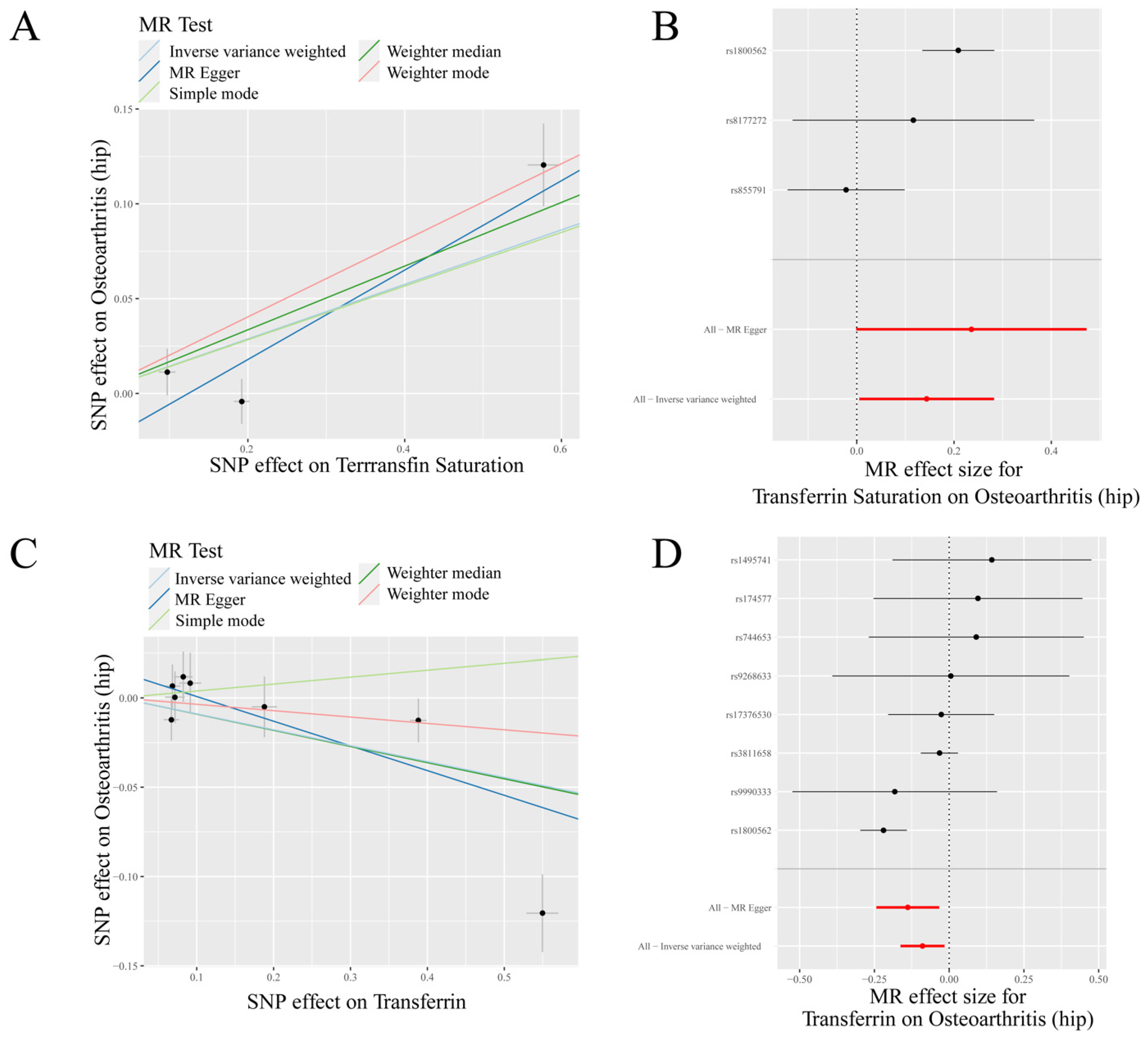
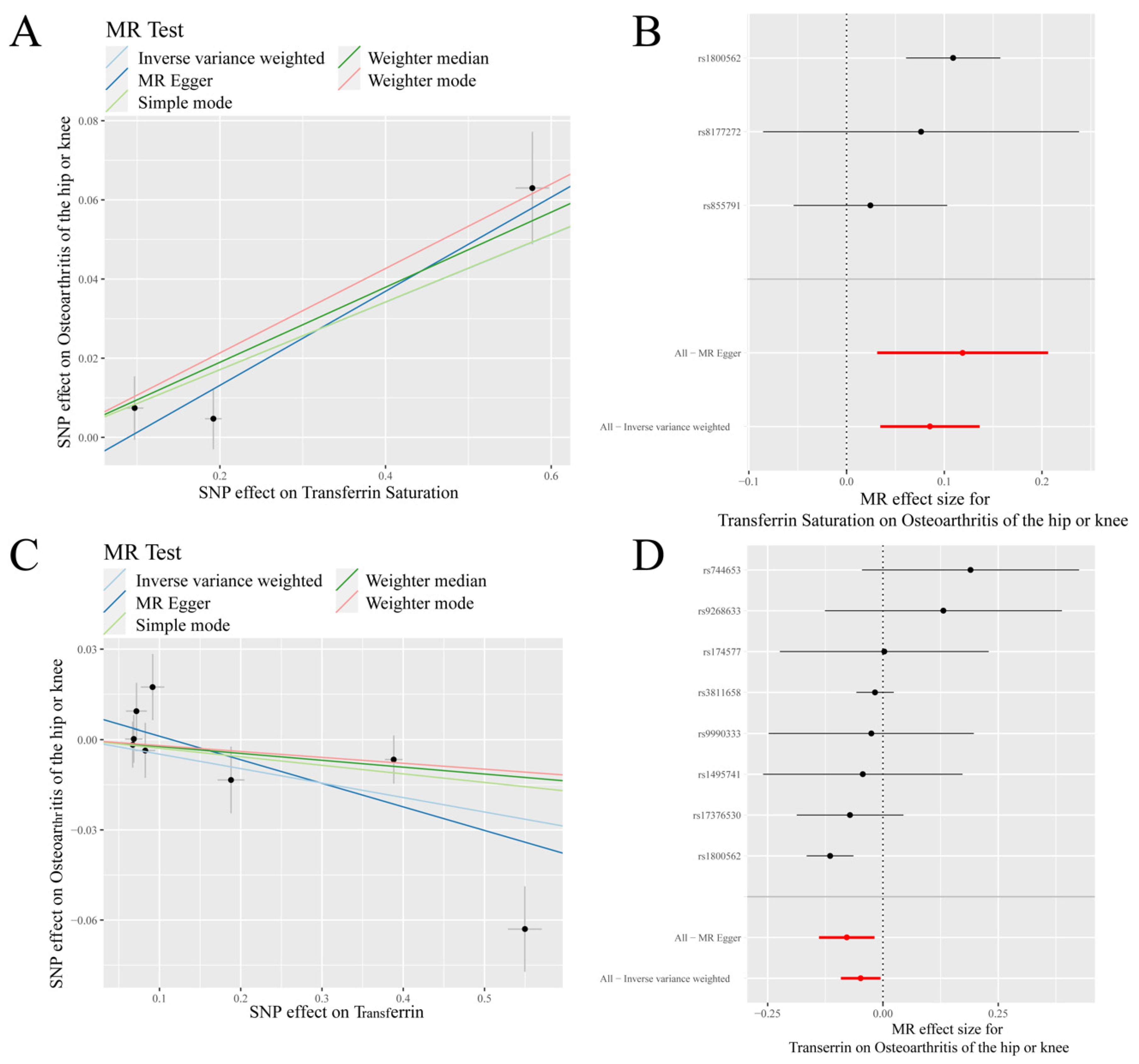
3.1. Mendelian Randomization Analysis
3.2. Sensitivity Analysis
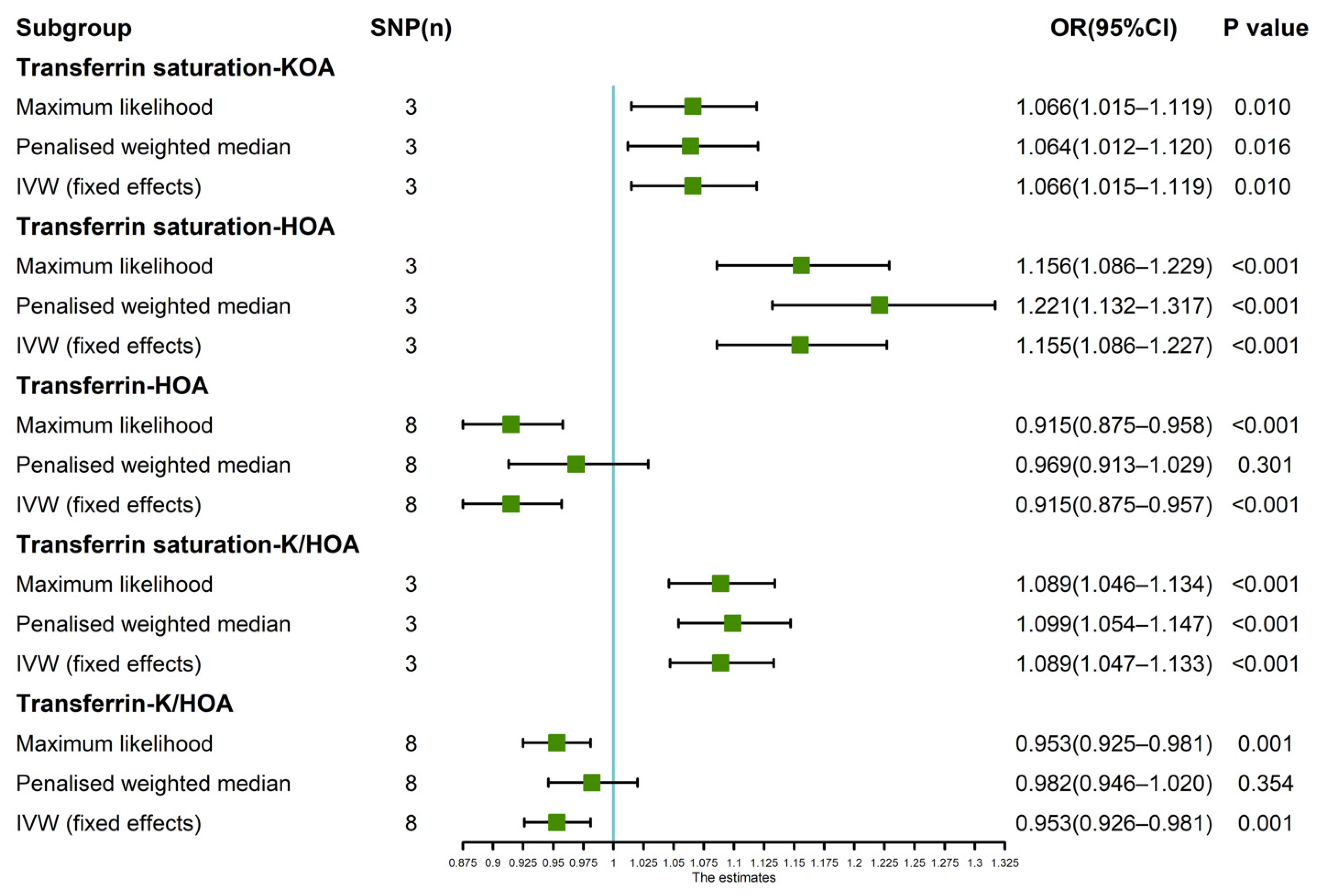
3.3. Further Validation of MR Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hunter, D.J.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef]
- Hunter, D.J.; March, L.; Chew, M. Osteoarthritis in 2020 and beyond: A Lancet Commission. Lancet 2020, 396, 1711–1712. [Google Scholar] [CrossRef]
- Mandl, L. Osteoarthritis year in review 2018: Clinical. Osteoarthr. Cartil. 2019, 27, 359–364. [Google Scholar] [CrossRef]
- Liu, L.; Wang, S.; Wen, Y.; Li, P.; Cheng, S.; Ma, M.; Zhang, L.; Cheng, B.; Qi, X.; Liang, C.; et al. Assessing the genetic relationships between osteoarthritis and human plasma proteins: A large scale genetic correlation scan. Ann. Transl. Med. 2020, 8, 677. [Google Scholar] [CrossRef]
- Meng, H.; Jiang, L.; Song, Z.; Wang, F. Causal Associations of Circulating Lipids with Osteoarthritis: A Bidirectional Mendelian Randomization Study. Nutrients 2022, 14, 1327. [Google Scholar] [CrossRef]
- Cai, C.; Hu, W.; Chu, T. Interplay Between Iron Overload and Osteoarthritis: Clinical Significance and Cellular Mechanisms. Front. Cell Dev. Biol. 2021, 9, 817104. [Google Scholar] [CrossRef]
- García-Ibarbia, C.; Neila, S.; Garcés, C.; Alonso, M.A.; Zarrabeitia, M.T.; Valero, C.; Ortiz, F.; Riancho, J.A. Non-synonymous WNT16 polymorphisms alleles are associated with different osteoarthritis phenotypes. Rheumatol. Int. 2017, 37, 1667–1672. [Google Scholar] [CrossRef]
- Loughlin, J. Translating osteoarthritis genetics research: Challenging times ahead. Trends Mol. Med. 2022, 28, 176–182. [Google Scholar] [CrossRef]
- Zhang, F.; Rao, S.; Baranova, A. Shared genetic liability between major depressive disorder and osteoarthritis. Bone Jt. Res. 2022, 11, 12–22. [Google Scholar] [CrossRef]
- Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends Biochem. Sci. 2016, 41, 274–286. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.; Guo, Z.; Hou, L.; Xu, J.; Du, T.; Xu, T.; Guo, F. Iron homeostasis in arthropathies: From pathogenesis to therapeutic potential. Ageing Res. Rev. 2021, 72, 101481. [Google Scholar] [CrossRef]
- Jing, X.; Lin, J.; Du, T.; Jiang, Z.; Li, T.; Wang, G.; Liu, X.; Cui, X.; Sun, K. Iron Overload Is Associated With Accelerated Progression of Osteoarthritis: The Role of DMT1 Mediated Iron Homeostasis. Front Cell Dev. Biol. 2020, 8, 594509. [Google Scholar] [CrossRef]
- Burton, L.H.; Radakovich, L.B.; Marolf, A.J.; Santangelo, K.S. Systemic iron overload exacerbates osteoarthritis in the strain 13 guinea pig. Osteoarthr. Cartil. 2020, 28, 1265–1275. [Google Scholar] [CrossRef]
- Holmes, M.V.; Ala-Korpela, M.; Smith, G.D. Mendelian randomization in cardiometabolic disease: Challenges in evaluating causality. Nat. Rev. Cardiol. 2017, 14, 577–590. [Google Scholar] [CrossRef]
- Zheng, J.; Baird, D.; Borges, M.-C.; Bowden, J.; Hemani, G.; Haycock, P.; Evans, D.M.; Smith, G.D. Recent Developments in Mendelian Randomization Studies. Curr. Epidemiol. Rep. 2017, 4, 330–345. [Google Scholar] [CrossRef]
- Yuan, S.; Larsson, S. Causal associations of iron status with gout and rheumatoid arthritis, but not with inflammatory bowel disease. Clin. Nutr. 2020, 39, 3119–3124. [Google Scholar] [CrossRef]
- Funck-Brentano, T.; Nethander, M.; Movérare-Skrtic, S.; Richette, P.; Ohlsson, C. Causal Factors for Knee, Hip, and Hand Osteoarthritis: A Mendelian Randomization Study in the UK Biobank. Arthritis Rheumatol. 2019, 71, 1634–1641. [Google Scholar] [CrossRef]
- Benyamin, B.; Esko, T.; Ried, J.S.; Radhakrishnan, A.; Vermeulen, S.H.; Traglia, M.; Gogele, M.; Anderson, D.; Broer, L.; Podmore, C.; et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 2014, 5, 4926. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Hatzikotoulas, K.; Southam, L.; Esparza-Gordillo, J.; Haberland, V.; Zheng, J.; Johnson, T.; Koprulu, M.; Zengini, E.; Steinberg, J.; et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat. Genet. 2019, 51, 230–236. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genetic Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Milne, R.L.; Kuchenbaecker, K.B.; Michailidou, K.; Beesley, J.; Kar, S.; Lindstrom, S.; Hui, S.; Lemacon, A.; Soucy, P.; Dennis, J.; et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017, 49, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, F.P.; Smith, G.D.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.J.; Li, J.R.; Zhu, Y.C.; Shen, H. Migraine and Ischemic Stroke: A Mendelian Randomization Study. Neurol. Ther. 2022, 11, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Mendelian Randomization Study Implies Causal Linkage Between Telomere Length and Juvenile Idiopathic Arthritis in a European Population. J. Inflamm. Res. 2022, 15, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, X.; Xie, Y.; Sun, Y.; Wang, X.; Hesketh, T. Irritable bowel syndrome and migraine: Evidence from Mendelian randomization analysis in the UK Biobank. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 1233–1239. [Google Scholar] [CrossRef]
- Xue, H.; Shen, X.; Pan, W. Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am. J. Hum. Genet. 2021, 108, 1251–1269. [Google Scholar] [CrossRef]
- Sandnes, M.; Vorland, M.; Ulvik, R.J.; Reikvam, H. HFE Genotype, Ferritin Levels and Transferrin Saturation in Patients with Suspected Hereditary Hemochromatosis. Genes 2021, 12, 1162. [Google Scholar] [CrossRef]
- Crownover, B.K.; Covey, C.J. Hereditary hemochromatosis. Am. Fam. Physician 2013, 87, 183–190. [Google Scholar]
- Bardou-Jacquet, E.; Lainé, F.; Guggenbuhl, P.; Morcet, J.; Jézéquel, C.; Guyader, D.; Moirand, R.; Deugnier, Y. Worse Outcomes of Patients With HFE Hemochromatosis With Persistent Increases in Transferrin Saturation During Maintenance Therapy. Clin. Gastroenterol. Hepatol. 2017, 15, 1620–1627. [Google Scholar] [CrossRef]
- Camacho, A.; Funck-Brentano, T.; Simão, M.; Cancela, L.; Ottaviani, S.; Cohen-Solal, M.; Richette, P. Effect of C282Y genotype on self-reported musculoskeletal complications in hereditary hemochromatosis. PLoS ONE 2015, 10, e0122817. [Google Scholar] [CrossRef]
- Oppl, B.; Husar-Memmer, E.; Pfefferkorn, S.; Blank, M.; Zenz, P.; Gollob, E.; Wurnig, C.; Engel, A.; Stadlmayr, A.; Uyanik, G.; et al. HFE hemochromatosis screening in patients with severe hip osteoarthritis: A prospective cross-sectional study. PLoS ONE 2018, 13, e0207415. [Google Scholar] [CrossRef] [PubMed]
- Simão, M.; Gavaia, P.J.; Camacho, A.; Porto, G.; Pinto, I.J.; Ea, H.-K.; Cancela, M.L. Intracellular iron uptake is favored in Hfe-KO mouse primary chondrocytes mimicking an osteoarthritis-related phenotype. Biofactors 2019, 45, 583–597. [Google Scholar] [CrossRef]
- Jing, X.; Du, T.; Li, T.; Yang, X.; Wang, G.; Liu, X.; Jiang, Z.; Cui, X. The detrimental effect of iron on OA chondrocytes: Importance of pro-inflammatory cytokines induced iron influx and oxidative stress. J. Cell. Mol. Med. 2021, 25, 5671–5680. [Google Scholar] [CrossRef] [PubMed]
- Boshuizen, M.; van der Ploeg, K.; von Bonsdorff, L.; Biemond, B.J.; Zeerleder, S.S.; van Bruggen, R.; Juffermans, N.P. Therapeutic use of transferrin to modulate anemia and conditions of iron toxicity. Blood Rev. 2017, 31, 400–405. [Google Scholar] [CrossRef]
- de Figueroa, P.L.; Lotz, M.K.; Blanco, F.J.; Caramés, B. Autophagy activation and protection from mitochondrial dysfunction in human chondrocytes. Arthritis Rheumatol. 2015, 67, 966–976. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, T.; Chen, J.; Shao, Z.; Wang, K.; Yan, Y.; Wu, C.; Lin, J.; Wang, H.; Gao, W.; et al. Parkin-mediated mitophagy as a potential therapeutic target for intervertebral disc degeneration. Cell Death Dis. 2018, 9, 980. [Google Scholar] [CrossRef]
- Qu, Z.; Yang, F.; Hong, J.; Wang, W.; Li, S.; Jiang, G.; Yan, S. Causal relationship of serum nutritional factors with osteoarthritis: A Mendelian randomization study. Rheumatology 2021, 60, 2383–2390. [Google Scholar] [CrossRef]
- Kennish, L.; Attur, M.; Oh, C.; Krasnokutsky, S.; Samuels, J.; Greenberg, J.D.; Huang, X.; Abramson, S.B. Age-dependent ferritin elevations and HFE C282Y mutation as risk factors for symptomatic knee osteoarthritis in males: A longitudinal cohort study. BMC Musculoskelet. Disord. 2014, 15, 8. [Google Scholar] [CrossRef]
- Nugzar, O.; Zandman-Goddard, G.; Oz, H.; Lakstein, D.; Feldbrin, Z.; Shargorodsky, M. The role of ferritin and adiponectin as predictors of cartilage damage assessed by arthroscopy in patients with symptomatic knee osteoarthritis. Best Pr. Res. Clin. Rheumatol. 2018, 32, 662–668. [Google Scholar] [CrossRef] [PubMed]
- de Boer, T.N.; van Spil, W.E.; Huisman, A.M.; Polak, A.A.; Bijlsma, J.W.; Lafeber, F.P.; Mastbergen, S.C. Serum adipokines in osteoarthritis; comparison with controls and relationship with local parameters of synovial inflammation and cartilage damage. Osteoarthr. Cartil. 2012, 20, 846–853. [Google Scholar] [CrossRef] [Green Version]
- Carroll, G.J.; Sharma, G.; Upadhyay, A.; Jazayeri, J.A. Ferritin concentrations in synovial fluid are higher in osteoarthritis patients with HFE gene mutations (C282Y or H63D). Scand. J. Rheumatol. 2010, 39, 413–420. [Google Scholar] [CrossRef]
- Brissot, P.; Ropert, M.; Le Lan, C.; Loréal, O. Non-transferrin bound iron: A key role in iron overload and iron toxicity. Biochim. Biophys. Acta 2012, 1820, 403–410. [Google Scholar] [CrossRef]
- Heiland, G.R.; Aigner, E.; Dallos, T.; Sahinbegovic, E.; Krenn, V.; Thaler, C.; Weiss, G.; Distler, J.H.; Datz, C.; Schett, G.; et al. Synovial immunopathology in haemochromatosis arthropathy. Ann. Rheum. Dis. 2010, 69, 1214–1219. [Google Scholar] [CrossRef]
- Elsayed, M.E.; Sharif, M.U.; Stack, A.G. Transferrin Saturation: A Body Iron Biomarker. Adv. Clin. Chem. 2016, 75, 71–97. [Google Scholar] [CrossRef]
- Lugrin, J.; Rosenblatt-Velin, N.; Parapanov, R.; Liaudet, L. The role of oxidative stress during inflammatory processes. Biol. Chem. 2014, 395, 203–230. [Google Scholar] [CrossRef]
- Ueda, N.; Takasawa, K. Impact of Inflammation on Ferritin, Hepcidin and the Management of Iron Deficiency Anemia in Chronic Kidney Disease. Nutrients 2018, 10, 1173. [Google Scholar] [CrossRef] [Green Version]

| Method | Serum Iron | Ferritin | Transferrin Saturation | Transferrin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP (n) | OR (95%CI) | p Value | SNP (n) | OR (95%CI) | p Value | SNP (n) | OR (95%CI) | p Value | SNP (n) | OR (95%CI) | p Value | ||
| KOA | MR Egger | 3 | 1.176 (1.035–1.337) | 0.244 | 4 | 1.194 (0.782–1.822) | 0.498 | 3 | 1.061 (0.978–1.151) | 0.389 | 8 | 0.942 (0.878–1.010) | 0.146 |
| Weighted median | 3 | 1.078 (1.008–1.153) | 0.028 | 4 | 1.185 (1.020–1.375) | 0.026 | 3 | 1.064 (1.013–1.119) | 0.014 | 8 | 0.959 (0.920–1.000) | 0.051 | |
| IVW | 3 | 1.068 (0.986–1.156) | 0.106 | 4 | 1.054 (0.860–1.292) | 0.610 | 3 | 1.066 (1.015–1.119) | 0.010 | 8 | 0.964 (0.920–1.011) | 0.128 | |
| Simple mode | 3 | 1.080 (0.995–1.172) | 0.207 | 4 | 1.163 (0.798–1.695) | 0.491 | 3 | 1.061 (0.992–1.135) | 0.228 | 8 | 0.918 (0.851–0.991) | 0.065 | |
| Weighted mode | 3 | 1.082 (1.003–1.168) | 0.178 | 4 | 1.186 (1.007–1.398) | 0.134 | 3 | 1.064 (1.009–1.123) | 0.149 | 8 | 0.958 (0.922–0.995) | 0.063 | |
| HOA | MR Egger | 3 | 1.482 (0.970–2.264) | 0.320 | 4 | 2.122 (1.589–2.835) | 0.036 | 3 | 1.266 (0.999–1.604) | 0.302 | 8 | 0.871 (0.784–0.967) | 0.042 |
| Weighted median | 3 | 1.142 (1.004–1.299) | 0.043 | 4 | 1.265 (1.004–1.593) | 0.047 | 3 | 1.183 (1.095–1.277) | <0.001 | 8 | 0.913 (0.852–0.979) | 0.010 | |
| IVW | 3 | 1.155 (0.896–1.489) | 0.266 | 4 | 1.358 (0.966–1.910) | 0.079 | 3 | 1.155 (1.005–1.326) | 0.042 | 8 | 0.915 (0.850–0.985) | 0.018 | |
| Simple mode | 3 | 0.920 (0.772–1.097) | 0.452 | 4 | 0.997 (0.718–1.384) | 0.986 | 3 | 1.152 (0.905–1.467) | 0.369 | 8 | 1.040 (0.893–1.211) | 0.633 | |
| Weighted mode | 3 | 0.983 (0.732–1.319) | 0.917 | 4 | 1.767 (1.377–2.267) | 0.021 | 3 | 1.224 (1.130–1.325) | 0.038 | 8 | 0.965 (0.896–1.039) | 0.377 | |
| K/HOA | MR Egger | 3 | 1.246 (1.080–1.438) | 0.204 | 4 | 1.434 (1.020–2.014) | 0.174 | 3 | 1.126 (1.032–1.229) | 0.229 | 8 | 0.925 (0.871–0.982) | 0.043 |
| Weighted median | 3 | 1.095 (1.028–1.167) | 0.005 | 4 | 1.164 (1.002–1.353) | 0.047 | 3 | 1.099 (1.051–1.150) | <0.001 | 8 | 0.977 (0.935–1.022) | 0.316 | |
| IVW | 3 | 1.094 (0.973–1.231) | 0.134 | 4 | 1.146 (0.922–1.424) | 0.220 | 3 | 1.089 (1.035–1.146) | <0.001 | 8 | 0.953 (0.913–0.995) | 0.028 | |
| Simple mode | 3 | 1.007 (0.878–1.155) | 0.932 | 4 | 0.917 (0.660–1.274) | 0.641 | 3 | 1.089 (0.996–1.192) | 0.203 | 8 | 0.972 (0.895–1.056) | 0.523 | |
| Weighted mode | 3 | 1.106 (1.025–1.193) | 0.139 | 4 | 1.310 (1.138–1.509) | 0.033 | 3 | 1.113 (1.058–1.170) | 0.054 | 8 | 0.981 (0.937–1.027) | 0.429 | |
| Exposure | Transferrin Saturation | Transferrin Saturation | Transferrin | Transferrin Saturation | Transferrin | |
|---|---|---|---|---|---|---|
| Outcome | KOA | HOA | HOA | K/HOA | K/HOA | |
| IVW (heterogeneity) | p value | 0.898 | 0.006 | 0.009 | 0.196 | 0.035 |
| Q | 0.214 | 10.279 | 18.860 | 3.255 | 15.098 | |
| MR Egger (heterogeneity) | p value | 0.659 | 0.020 | 0.020 | 0.188 | 0.071 |
| Q | 0.195 | 5.402 | 15.001 | 1.735 | 11.637 | |
| MR Egger (pleiotropy) | p value | 0.912 | 0.516 | 0.260 | 0.521 | 0.230 |
| intercept | 0.001 | −0.029 | 0.015 | −0.011 | 0.009 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Zhang, S.; Tian, Y.; Si, H.; Zeng, Y.; Wu, Y.; Liu, Y.; Li, M.; Sun, K.; Wu, L.; et al. Genetic Causal Association between Iron Status and Osteoarthritis: A Two-Sample Mendelian Randomization. Nutrients 2022, 14, 3683. https://doi.org/10.3390/nu14183683
Xu J, Zhang S, Tian Y, Si H, Zeng Y, Wu Y, Liu Y, Li M, Sun K, Wu L, et al. Genetic Causal Association between Iron Status and Osteoarthritis: A Two-Sample Mendelian Randomization. Nutrients. 2022; 14(18):3683. https://doi.org/10.3390/nu14183683
Chicago/Turabian StyleXu, Jiawen, Shaoyun Zhang, Ye Tian, Haibo Si, Yi Zeng, Yuangang Wu, Yuan Liu, Mingyang Li, Kaibo Sun, Limin Wu, and et al. 2022. "Genetic Causal Association between Iron Status and Osteoarthritis: A Two-Sample Mendelian Randomization" Nutrients 14, no. 18: 3683. https://doi.org/10.3390/nu14183683
APA StyleXu, J., Zhang, S., Tian, Y., Si, H., Zeng, Y., Wu, Y., Liu, Y., Li, M., Sun, K., Wu, L., & Shen, B. (2022). Genetic Causal Association between Iron Status and Osteoarthritis: A Two-Sample Mendelian Randomization. Nutrients, 14(18), 3683. https://doi.org/10.3390/nu14183683




