The Response of the Human Umbilical Vein Endothelial Cell Transcriptome to Variation in Magnesium Concentration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Extraction
2.3. Microarray Analysis
2.4. Bioinformatics and Statistical Analysis
3. Results
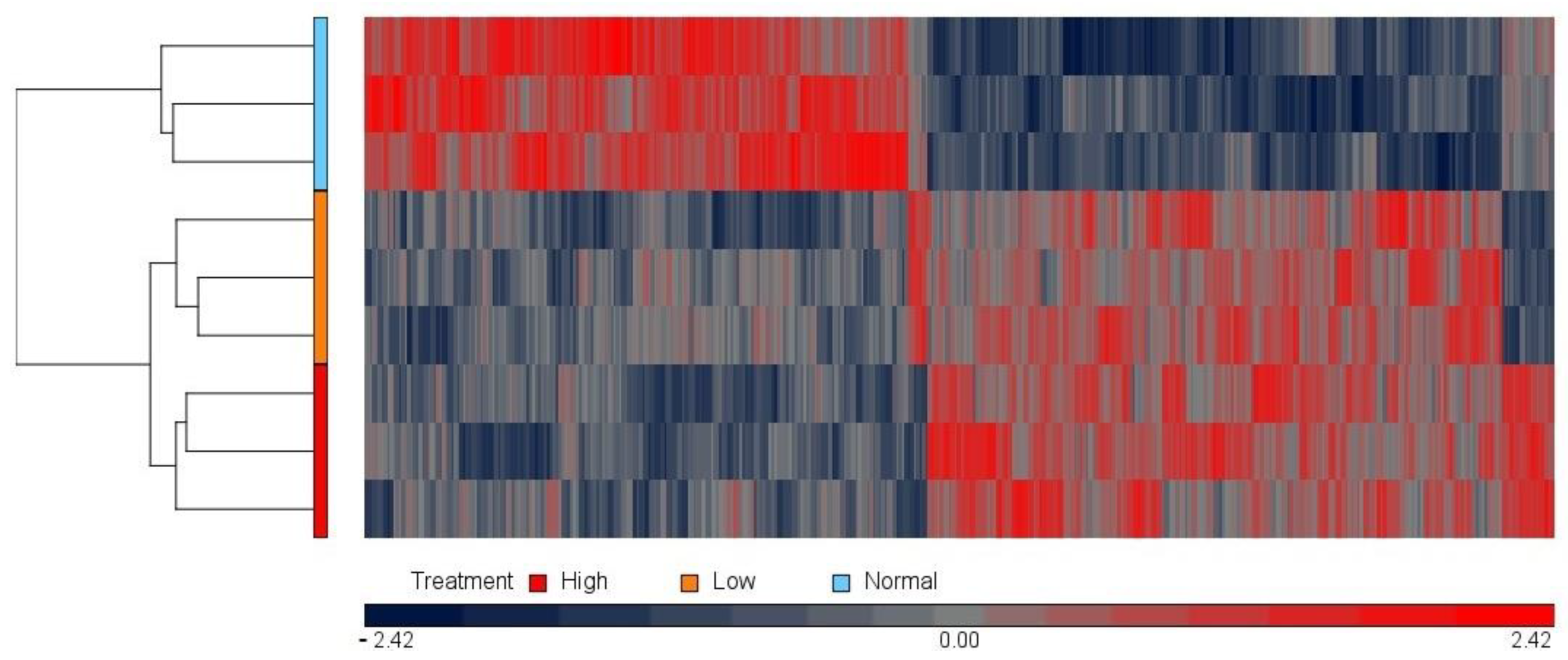
3.1. Microarray Approach: Effect of Magnesium Concentration on the HUVEC Transcriptome
3.2. Gene Ontology Analysis
3.3. Effects of Magnesium Concentration on Genes Involved in Inflammation, Mediated by the Chemokine and Cytokine Signalling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bouïs, D.; Hospers, G.; Meijer, C.; Molema, G.; Mulder, N. Endothelium In Vitro: A review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis 2001, 4, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Deanfield, J.E.; Halcox, J.P.; Rabelink, T.J. Endothelial function and dysfunction: Testing and clinical relevance. Circulation 2007, 115, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; DeFelice, A.; Hanig, J.; Colatsky, T. Biomarkers of endothelial cell activation serve as potential surrogate markers for drug-induced vascular injury. Toxicol. Pathol. 2010, 38, 856–871. [Google Scholar] [CrossRef]
- Bernardini, D.; Nasulewic, A.; Mazur, A.; Maier, J.A. Magnesium and microvascular endothelial cells: A role in inflammation and angiogenesis. Front. Biosci. J. Virtual Libr. 2005, 10, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, S.; Tucker, G.; Brameld, J. Effects of dietary polyphenols on gene expression in human vascular endothelial cells. Proc. Nutr. Soc. 2008, 67, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Koltsova, S.; Trushina, Y.; Haloui, M.; Akimova, O.A.; Tremblay, J.; Hamet, P.; Orlov, S.N. Ubiquitous [Na+] i/[K+] i-sensitive transcriptome in mammalian cells: Evidence for Ca2+ i-independent excitation-transcription coupling. PLoS ONE 2012, 7, e38032. [Google Scholar] [CrossRef]
- Ambra, R.; Manca, S.; Palumbo, M.; Leoni, G.; Natarelli, L.; De Marco, A.; Consoli, A.; Pandolfi, A.; Virgili, F. Transcriptome analysis of human primary endothelial cells (HUVEC) from umbilical cords of gestational diabetic mothers reveals candidate sites for an epigenetic modulation of specific gene expression. Genomics 2014, 103, 337–348. [Google Scholar] [CrossRef]
- Chakraborti, S.; Chakraborti, T.; Mandal, M.; Mandal, A.; Das, S.; Ghosh, S. Protective role of magnesium in cardiovascular diseases: A review. Mol. Cell. Biochem. 2002, 238, 163–179. [Google Scholar] [CrossRef]
- Whitton, C.; Nicholson, S.K.; Roberts, C.; Prynne, C.J.; Pot, G.; Olson, A.; Fitt, E.; Cole, D.; Teucher, B.; Bates, B. National Diet and Nutrition Survey: UK food consumption and nutrient intakes from the first year of the rolling programme and comparisons with previous surveys. Br. J. Nutr. 2011, 106, 1899. [Google Scholar] [CrossRef]
- Swaminathan, R. Magnesium metabolism and its disorders. Clin. Biochem. Rev. 2003, 24, 47. [Google Scholar]
- Jiang, L.; He, P.; Chen, J.; Liu, Y.; Liu, D.; Qin, G.; Tan, N. Magnesium levels in drinking water and coronary heart disease mortality risk: A meta-analysis. Nutrients 2016, 8, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, J. Low magnesium and atherosclerosis: An evidence-based link. Mol. Asp. Med. 2003, 24, 137–146. [Google Scholar] [CrossRef]
- De Baaij, J.; Hoenderop, J.; Bindels, R. Magnesium in man: Implications for health and disease. Physiol. Rev. 2015, 95, 1–46. [Google Scholar] [CrossRef]
- Almousa, L.; Salter, A.; Langley-Evans, S. Magnesium deficiency heightens lipopolysaccharide-induced inflammation and enhances monocyte adhesion in human umbilical vein endothelial cells. Magnes. Res. 2018, 31, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Banai, S.; Haggroth, L.; Epstein, S.; Casscells, W. Influence of extracellular magnesium on capillary endothelial cell proliferation and migration. Circ. Res. 1990, 67, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, S.; Baldoli, E.; Leidi, M.; Maier, J. Magnesium deficiency promotes a pro-atherogenic phenotype in cultured human endothelial cells via activation of NFkB. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2010, 1802, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Maier, J. Endothelial cells and magnesium: Implications in atherosclerosis. Clin. Sci. 2012, 122, 397–407. [Google Scholar] [CrossRef]
- Guo, F.; Tang, J.; Zhou, Z.; Dou, Y.; Van Lonkhuyzen, D.; Gao, C.; Huan, J. GEF-H1-RhoA signaling pathway mediates LPS-induced NF-κB transactivation and IL-8 synthesis in endothelial cells. Mol. Immunol. 2012, 50, 98–107. [Google Scholar] [CrossRef]
- Hoffmann, E.; Dittrich-Breiholz, O.; Holtmann, H.; Kracht, M. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 2002, 72, 847–855. [Google Scholar]
- Berg, A.; Baldwin, A. The Ikb proteins: Multifunctional regulators of rel/NFk-B transcription. Genes Dev. 1993, 7, 2064–2070. [Google Scholar]
- Li, A.; Varney, M.L.; Valasek, J.; Godfrey, M.; Dave, B.J.; Singh, R.K. Autocrine role of interleukin-8 in induction of endothelial cell proliferation, survival, migration and MMP-2 production and angiogenesis. Angiogenesis 2005, 8, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Baggiolini, M.; Loetscher, P.; Moser, B. Interleukin-8 and the chemokine family. Int. J. Immunopharmacol. 1995, 17, 103–108. [Google Scholar] [CrossRef]
- Lukacs, N.W.; Strieter, R.; Elner, V.; Evanoff, H.; Burdick, M.; Kunkel, S. Production of chemokines, interleukin-8 and monocyte chemoattractant protein-1, during monocyte: Endothelial cell interactions. Blood 1995, 86, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Sironi, M.; Toniatti, C.; Polentarutti, N.; Fruscella, P.; Ghezzi, P.; Faggioni, R.; Luini, W.; Van Hinsbergh, V.; Sozzani, S.; et al. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity 1997, 6, 315–325. [Google Scholar] [CrossRef]
- Ferrè, S.; Mazur, A.; Maier, J. Low-magnesium induces senescent features in cultured human endothelial cells. Magnes. Res. 2007, 20, 66–71. [Google Scholar]
- Maier, J.; MalpuechBrugère, C.; Zimowska, W.; Rayssiguier, Y.; Mazur, A. Low magnesium promotes endothelial cell dysfunction: Implications for atherosclerosis, inflammation and thrombosis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2004, 1689, 13–21. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.; Thomas, P. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551. [Google Scholar] [CrossRef]
- Mazur, A.; Maier, J.; Rock, E.; Gueux, E.; Nowacki, W.; Rayssiguier, Y. Magnesium and the inflammatory response: Potential physiopathological implications. Arch. Biochem. Biophys. 2007, 458, 48–56. [Google Scholar] [CrossRef]
- Shechter, M.; Sherer, Y. Endothelial dysfunction: A crystal ball prediction for enhanced cardiovascular risk? IMAJ 2003, 5, 736–738. [Google Scholar]
- Almousa, L.; Salter, A.; Langley-Evans, S. The effect of varying concentrations of magnesium on expression of human endothelia adhesion molecules. J. Nutr. Food Sci. 2016, 6, 53. [Google Scholar]
- Martin, H.; Staedtler, F.; Lamboley, C.; Adrian, M.; Schumacher, M.; Chibout, S.-D.; Laurant, P.; Richert, L.; Berthelot, A. Effects of long-term dietary intake of magnesium on rat liver transcriptome. Magnes. Res. 2007, 20, 259–265. [Google Scholar] [PubMed]
- Chen, C.; Coats, S.; Bumgarner, R.; Darveau, R. Hierarchical gene expression profiles of HUVEC stimulated by different lipid A structures obtained from Porphyromonas gingivalis and Escherichia coli. Cell. Microbiol. 2007, 9, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Bal, G.; Kamhieh-Milz, J.; Futschik, M.; Häupl, T.; Salama, A.; Moldenhauer, A. Transcriptional profiling of the hematopoietic support of interleukin-stimulated human umbilical vein endothelial cells (HUVECs). Cell Transplant. 2012, 21, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Wolf, F.; Trapani, V.; Cittadini, A. Magnesium and the control of cell proliferation: Looking for a needle in a haystack. Magnes. Res. 2008, 21, 83–91. [Google Scholar]
- Sugimoto, J.; Romani, A.; Valentin-Torres, A.; Luciano, A.; Kitchen, C.; Funderburg, N.; Mesiano, S.; Bernstein, H.B. Magnesium decreases inflammatory cytokine production: A novel innate immunomodulatory mechanism. J. Immunol. 2012, 188, 6338–6346. [Google Scholar] [CrossRef]
- Wolf, F.; Trapani, V. Cell (patho) physiology of magnesium. Clin. Sci. 2008, 114, 27–35. [Google Scholar] [CrossRef]
- Saris, N.-E.; Mervaala, E.; Karppanen, H.; Khawaja, J.; Lewenstam, A. Magnesium: An update on physiological, clinical and analytical aspects. Clin. Chim. Acta 2000, 294, 1–26. [Google Scholar] [CrossRef]
- Noronha, L.; Matuschak, G. Magnesium in critical illness: Metabolism, assessment, and treatment. Intensive Care Med. 2002, 28, 667–679. [Google Scholar] [CrossRef]
- Yee, N.; Kazi, A.; Yee, R. Cellular and developmental biology of TRPM7 channel-kinase: Implicated roles in cancer. Cells 2014, 3, 751–777. [Google Scholar] [CrossRef]
- Nielsen, F. Magnesium deficiency and increased inflammation: Current perspectives. J. Inflamm. Res. 2018, 18, 25–34. [Google Scholar] [CrossRef]
- Almousa, L.A.; Salter, A.M.; Langley-Evans, S.C. Varying magnesium concentration elicits changes in inflammatory response in human umbilical vein endothelial cells (HUVECs). Magnes. Res. 2018, 31, 99–109. [Google Scholar] [CrossRef] [PubMed]

| Prob Set Name | Entrez Gene | Symbol | Gene Title | p-Value | Log 2 Fold-Change |
|---|---|---|---|---|---|
| 225160_x_at | 4193 | MDM2 | Mdm2 p53 binding protein homolog (mouse) | 0.046 | 2.1 |
| 231108_at | 2521 | FUS | Fused in sarcoma | 0.031 | 1.7 |
| 232423_at | 414 | ARSD | Arylsulfatase D | 0.038 | 1.7 |
| 215123_at | 23,117 | NPIPL3 | Nuclear pore complex interacting protein-like 3 | 0.031 | 1.7 |
| 244872_at | 5928 | RBBP4 | retinoblastoma binding protein 4 | 0.023 | 1.6 |
| 213742_at | 9295 | SRSF11 | Serine/arginine-rich splicing factor 11 | 0.046 | 1.6 |
| 209936_at | 10,181 | RBM5 | RNA binding motif protein 5 | 0.021 | 1.5 |
| 232898_at | 1601 | DAB2 | Disabled homolog 2, mitogen-responsive phosphoprotein | 0.003 | 1.5 |
| 243599_at | 100,507,226 | LOC100507226 | Hypothetical LOC100507226 | 0.04 | 1.5 |
| 216983_s_at | 7767 | ZNF224 | Zinc finger protein 224 | 0.008 | 1.4 |
| 224576_at | 57,222 | ERGIC1 | Endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 | 0.045 | −1.7 |
| 211611_s_at | 1388///7148 | ATF6B | Activating transcription factor 6 beta | 0.045 | −1.7 |
| 1555561_a_at | 55,757 | UGGT2 | UDP-glucose glycoprotein glucosyltransferase 2 | 0.001 | −1.6 |
| 210932_s_at | 6049 | RNF6 | Ring finger protein (C3H2C3 type) 6 | 0.023 | −1.5 |
| 229667_s_at | 3218 | HOXB8 | Homeobox B8 | 0.024 | −1.5 |
| 242157_at | 80,205 | CHD9 | Chromodomain helicase DNA binding protein 9 | 0.019 | −1.5 |
| 1555106_a_at | 51,496 | CTDSPL2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like | 0.042 | −1.5 |
| 221440_s_at | 10,741 | RBBP9 | Retinoblastoma binding protein 9 | 0.001 | −1.4 |
| 213606_s_at | 396 | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 0.042 | −1.4 |
| 206288_at | 5229 | PGGT1B | Protein geranylgeranyltransferase type I, beta subunit | 0.001 | −1.4 |
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|
| 228656_at | 5629 | PROX1 | Prospero homeobox 1 | 0.001 | 1.6 |
| 205410_s_at | 493 | ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | 0.01 | 1.5 |
| 209401_s_at | 6560 | SLC12A4 | Solute carrier family 12 (potassium/chloride transporters), member 4 | 0.034 | 1.5 |
| 241985_at | 133,746 | JMY | Junction mediating and regulatory protein, p53 cofactor | 0.04 | 1.4 |
| 243323_s_at | 463 | ZFHX3 | Zinc finger homeobox 3 | 0.002 | 1.4 |
| 221085_at | 9966 | TNFSF15 | Tumor necrosis factor (ligand) superfamily, member 15 | 0.018 | 1.4 |
| 207332_s_at | 7037 | TFRC | Transferrin receptor (p90, CD71) | p < 0.001 | 1.4 |
| 219772_s_at | 23,676 | SMPX | Small muscle protein, X-linked | 0.004 | 1.4 |
| 216950_s_at | 100,132,417///2209 | FCGR1A | Fc fragment of IgG, high affinity Ia, receptor (CD64) | 0.018 | 1.4 |
| 210954_s_at | 9819 | TSC22D2 | TSC22 domain family, member 2 | 0.038 | 1.3 |
| 236361_at | 117,248 | GALNTL2 | UDP-N-acetyl-alpha-D-galactosamine | p < 0.001 | −2.0 |
| 1556325_at | 27,145 | FILIP1 | Filamin A interacting protein 1 | p < 0.001 | −1.9 |
| 206336_at | 6372 | CXCL6 | Chemokine (C-X-C motif) ligand 6 | p < 0.001 | −1.8 |
| 230543_at | 8239 | USP9X | Ubiquitin specific peptidase 9, X-linked | 0.039 | −1.7 |
| 240757_at | 23,332 | CLASP1 | Cytoplasmic linker associated protein 1 | 0.007 | −1.6 |
| 203889_at | 6447 | SCG5 | Secretogranin V | 0.004 | −1.6 |
| 1570515_a_at | 27,145 | FILIP1 | Filamin A interacting protein 1 | p < 0.001 | −1.6 |
| 219825_at | 56,603 | CYP26B1 | Cytochrome P450, family 26, subfamily B, polypeptide 1 | 0.004 | −1.5 |
| 207542_s_at | 358 | AQP1 | Aquaporin 1 (Colton blood group) | p < 0.001 | −1.5 |
| 211506_s_at | 3576 | IL8 | Interleukin 8 | 0.011 | −1.5 |
| GO Processes | Enrichment Score | Enrichment p Value | Number of Transcripts Differentially Regulated |
|---|---|---|---|
| Biological processes | |||
| Cellular process | 1.19 | 3.71 × 10−9 | 1052 |
| Sensory perception of chemical signals | 0.20 | 2.21 × 10−5 | 6 |
| Metabolic process | 1.19 | 2.64 × 10−5 | 748 |
| Cellular component organisation or biogenesis | 1.31 | 2.87 × 10−4 | 296 |
| Primary metabolic process | 1.18 | 4.31 × 10−4 | 604 |
| Cell adhesion | 1.78 | 4.94 × 10−4 | 68 |
| Phosphate containing compound metabolic process | 1.33 | 9.64 × 10−4 | 228 |
| Intracellular signal transduction | 1.39 | 2.22 × 10−3 | 160 |
| Cell death | 1.70 | 2.90 × 10−3 | 65 |
| Apoptotic process | 1.72 | 3.46 × 10−3 | 62 |
| G-protein coupled receptor signalling pathway | 0.49 | 3.46 × 10−3 | 24 |
| Nucleobase-containing compound metabolic processes | 1.21 | 3.69 × 10−3 | 364 |
| Molecular functions | |||
| Catalytic activity | 1.21 | 4.30 × 10−4 | 547 |
| Protein binding | 1.23 | 6.15 × 10−3 | 365 |
| Binding | 1.15 | 8.45 × 10−3 | 608 |
| Hydrolase activity | 1.27 | 3.14 × 10−4 | 251 |
| Cellular components | |||
| Intracellular | 1.23 | 1.91 × 10−7 | 694 |
| Cell part | 1.21 | 4.49 × 10−7 | 720 |
| Cytoplasm | 1.25 | 4.99 × 10−5 | 427 |
| Organelle | 1.21 | 1.71 × 10−4 | 507 |
| Nucleus | 1.27 | 1.91 × 10−3 | 265 |
| GO Processes | Enrichment Score | Enrichment p Value | Number of Transcripts Differentially Regulated |
|---|---|---|---|
| Biological processes | |||
| Metabolic process | 1.15 | 4.75 × 10−3 | 590 |
| Cellular component organisation or biogenesis | 1.33 | 1.02 × 10−3 | 243 |
| Cellular process | 1.14 | 3.73 × 10−4 | 819 |
| Cellular components | |||
| Ribosome macromolecular complex | 1.32 | 3.24 x10−4 | 247 |
| Cytoplasm | 1.29 | 3.08 × 10−5 | 355 |
| Intracellular | 1.27 | 2.39 × 10−8 | 583 |
| Nucleus | 1.32 | 6.89 × 10−4 | 223 |
| Organelle | 1.24 | 3.94 × 10−5 | 424 |
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 207445_s_at | 10,803 | CCR9 | Chemokine (C-C motif) receptor 9 | Chemotaxis/inferred from electronic annotation/chemotaxis | 0.042 | 1.3 |
| 211230_s_at | 5293 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | B cell homeostasis/inferred from electronic annotation/ | 0.004 | 1.3 |
| 216834_at | 5996 | RGS1 | Regulator of G-protein signaling 1 | Immune response/traceable author statement/signal transduce | 0.047 | 1.2 |
| 230202_at | 5970 | RELA | Transcription factor p65 | Liver development/inferred from electronic annotation | 0.047 | 1.2 |
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation/ | 0.022 | 1.2 |
| 224909_s_at | 57,580 | PREX1 | Phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | Superoxide metabolic process/traceable author statement | 0.019 | 1.1 |
| 210162_s_at | 4772 | NFATC1 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | G1/S transition of mitotic cell cycle/inferred from electronic annotation | 0.036 | 1.1 |
| 222912_at | 408 | ARRB1 | Arrestin, beta 1 | G-protein coupled receptor internalization/inferred from mutant phenotype | 0.015 | 1.1 |
| 203175_at | 391 | RHOG | Rho-related GTP-binding protein, member G (rho G) | Small GTPase mediated signal transduction/inferred from electronic annota | 0.035 | 1.1 |
| 212777_at | 6654 | SOS1 | Son of sevenless homolog 1 | Apoptosis/not recorded signal transduction | 0.017 | 1.1 |
| 211543_s_at | 2870 | GRK6 | G protein-coupled receptor kinase 6 | Protein phosphorylation/inferred from electronic annotation | 0.006 | 1.1 |
| 205884_at | 3676 | ITGA4 | Integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | Blood vessel remodeling inferred from electronic annotation | 0.029 | 1.1 |
| 205127_at | 5742 | PTGS1 | Prostaglandin-endoperoxide synthase 1 | Prostaglandin biosynthetic process/inferred from sequence or structural s | 0.017 | 1.1 |
| 33814_at | 10,298 | PAK4 | P21 protein (Cdc42/Rac)-activated kinase 4 | Protein phosphorylation/inferred from electronic annotation | 0.019 | 1.1 |
| 228388_at | 4793 | NFKBIB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | Transcription/traceable author statement/signal transducti | 0.01 | 1.1 |
| 200885_at | 389 | RHOC | Rho-related GTP binding protein, member C | Small GTPase mediated signal transduction/inferred from electronic annota | 0.018 | 1.1 |
| 208075_s_at | 6354 | CCL7 | Chemokine (C-C motif) ligand 7 | Cellular calcium ion homeostasis/traceable author statement | 0.042 | 1.1 |
| 201188_s_at | 3710 | ITPR3 | Inositol 1,4,5-triphosphate receptor, type 3 | Transport/inferred from electronic annotation/ion transport | 0.038 | 1.1 |
| 225363_at | 5728 | PTEN | Phosphatase and tensin homolog | Regulation of cyclin-dependent protein kinase activity/traceable authors | 0.03 | 1.1 |
| 205125_at | 5333 | PLCD1 | Phospholipase C, delta 1 | Lipid metabolic process/inferred from electronic annotation/ | 0.03 | 1.1 |
| 201895_at | 369 | ARAF | Serine/threonine protein kinase A raf | Protein modification process/traceable author statement | p < 0.001 | 1.1 |
| 212647_at | 6237 | RRAS | Related RAS viral (r-ras) oncogene homolog | Signal transduction/inferred from electronic annotation | 0.041 | 1.1 |
| 224994_at | 817 | CAMK2D | Calcium/calmodulin-dependent protein kinase II delta | G1/S transition of mitotic cell cycle//inferred from electronic annotation | 0.038 | 1.1 |
| 206336_at | 6372 | CXCL6 | Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | Chemotaxis/inferred from electronic annotation/chemotaxis | p < 0.001 | −1.8 |
| 211506_s_at | 3576 | IL8 | Interleukin 8 | Angiogenesis/traceable author statement/cellular component | 0.011 | −1.5 |
| 204470_at | 2919 | CXCL1 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | Chemotaxis/traceable author statement inflammatory response | 0.027 | −1.2 |
| 225139_at | 4775 | NFATC3 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | Transcription/inferred from electronic annotation | 0.033 | −1.0 |
| 204010_s_at | 3845 | KRAS | GTPase KRas | Apoptosis/inferred from electronic annotation/signal trans | 0.029 | −1.1 |
| 38290_at | 10,636 | RGS14 | Regulator of G-protein signaling 14 | Mitosis/inferred from electronic annotation/signal transduce | 0.001 | −1.1 |
| 204103_at | 6351 | CCL4 | Chemokine (C-C motif) ligand 4 | Cellular component movement/traceable author statement | 0.008 | −1.1 |
| 207952_at | 3567 | IL5 | Interleukin 5 (colony-stimulating factor, eosinophil) | Inflammatory response/inferred from electronic annotation | 0.024 | −1.1 |
| 213044_at | 6093 | ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | Cytokinesis/inferred from electronic annotation/protein ph | 0.002 | −1.1 |
| 209774_x_at | 2920 | CXCL2 | Chemokine (C-X-C motif) ligand 2 | Chemotaxis/inferred from electronic annotation/chemotaxis | 0.045 | −1.1 |
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation | 0.022 | 1.2 |
| 228388_at | 4793 | NF-κB IB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | Transcription/traceable author statement/signal transducti | 0.01 | 1.1 |
| 202284_s_at | 1026 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Regulation of cyclin-dependent protein kinase activity/traceable authors | 0.006 | 1.1 |
| 205170_at | 6773 | STAT2 | Signal transducer and activator of transcription 2 | Transcription/inferred from electronic annotation | 0.04 | 1.0 |
| 211506_s_at | 3576 | IL8 | Interleukin 8 | Angiogenesis/traceable author statement//cellular component | 0.011 | −1.5 |
| 234066_at | 9173 | IL1RL1 | Interleukin 1 receptor-like 1 | Immune response/non-traceable author statement/signal tran | p < 0.001 | −1.3 |
| 243541_at | 133,396 | IL31RA | Interleukin 31 receptor A | MAPKKK cascade/non-traceable author statement anti-apoptos | 0.028 | −1.2 |
| 207952_at | 3567 | IL5 | Interleukin 5 (colony-stimulating factor, eosinophil) | Inflammatory response/inferred from electronic annotation | 0.024 | −1.2 |
| 209821_at | 90,865 | IL33 | Interleukin 33 | Positive regulation of macrophage activation/inferred from direct assay | 0.014 | −1.2 |
| 226333_at | 3570 | IL6R | Interleukin 6 receptor | Hepatic immune response/traceable author statement | 0.044 | −1.1 |
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 211230_s_at | 5293 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | B cell homeostasis/inferred from electronic annotation | 0.006 | 1.3 |
| 205127_at | 5742 | PTGS1 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | Prostaglandin biosynthetic process/inferred from sequence or structural | 0.008 | 1.2 |
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation | 0.01 | 1.2 |
| 201188_s_at | 3710 | ITPR3 | Inositol 1,4,5-triphosphate receptor, type 3 | Transport/inferred from electronic annotation ion transpor | 0.03 | 1.2 |
| 233254_x_at | 5728 | PTEN | Phosphatase and tensin homolog | Regulation of cyclin-dependent protein kinase activity/traceable authors | 0.006 | 1.2 |
| 232043_at | 2788 | GNG7 | Guanine nucleotide binding protein (G protein), gamma 7 | Behavioral fear response/inferred from electronic annotation | 0.03 | 1.1 |
| 1568926_x_at | 91,807 | MYLK3 | Myosin light chain kinase 3 | Protein phosphorylation/inferred from electronic annotation | 0.018 | 1.1 |
| 216190_x_at | 3688 | ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, M | G1/S transition of mitotic cell cycle/inferred from electronic annotation | 0.014 | 1.1 |
| 224994_at | 817 | CAMK2D | Calcium/calmodulin-dependent protein kinase II delta | G1/S transition of mitotic cell cycle//inferred from electronic annotation | 0.01 | 1.1 |
| 205962_at | 5062 | PAK2 | P21 protein (Cdc42/Rac)-activated kinase 2 | Protein phosphorylation/inferred from direct assay/protein | 0.017 | 1.1 |
| 243829_at | 673 | BRAF | V-raf murine sarcoma viral oncogene homolog B1 | MAPKKK cascade/inferred from electronic annotation/protein | 0.003 | 1.1 |
| 210162_s_at | 4772 | NFATC1 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | G1/S transition of mitotic cell cycle/inferred from electronic annotation | 0.04 | 1.1 |
| 209201_x_at | 7852 | CXCR4 | Chemokine (C-X-C motif) receptor 4 | Activation of MAPK activity traceable author statement | 0.01 | −1.3 |
| 1560524_at | 400,581 | GRAP | GRB2-related adaptor protein | Ras protein signal transduction/traceable author statement | 0.03 | −1.2 |
| 224965_at | 54,331 | GNG2 | Guanine nucleotide binding protein (G protein), gamma 2 | Signal transduction/inferred from electronic annotation | 0.01 | −1.2 |
| 208641_s_at | 5879 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Endocytosis/inferred from electronic annotation/apoptosis | 0.01 | −1.1 |
| 204174_at | 241 | ALOX5AP | Arachidonate 5-lipoxygenase-activating protein | Leukotriene production involved in inflammatory response/inferred from el | 0.04 | −1.1 |
| 222912_at | 408 | ARRB1 | Arrestin, beta 1 | G-protein coupled receptor internalization/inferred from mutant phenotype | 0.04 | −1.1 |
| 201179_s_at | 2773 | GNAI3 | Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | Transport/non-traceable author statement/vesicle fusion/ | 0.007 | −1.1 |
| 212590_at | 22,800 | RRAS2 | Related RAS viral (r-ras) oncogene homolog 2 | Signal transduction/inferred from electronic annotation | 0.013432 | −1.1 |
| 204103_at | 6351 | CCL4 | Chemokine (C-C motif) ligand 4 | Cellular component movement/traceable author statement | 0.01 | −1.1 |
| 225141_at | 4775 | NFATC3 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | Transcription/inferred from electronic annotation | 0.036 | −1.1 |
| 202647_s_at | 4893 | NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | Apoptosis/inferred from electronic annotation/signal trans | 0.005 | −1.1 |
| 211434_s_at | 9034 | CCRL2 | Chemokine (C-C motif) receptor-like 2 | Chemotaxis/traceable author statement/signal transduction | 0.03 | −1.1 |
| 39402_at | 3553 | IL1B | Interleukin 1, beta | Activation of MAPK activity/inferred from direct assay | 0.02 | −1.1 |
| 203154_s_at | 10,298 | PAK4 | P21 protein (Cdc42/Rac)-activated kinase 4 | Protein phosphorylation/inferred from electronic annotation | 0.03 | −1.05 |
| 38290_at | 10,636 | RGS14 | Regulator of G-protein signaling 14 | Mitosis/inferred from electronic annotation/signal transdu | 0.004 | −1.0 |
| 207157_s_at | 2787 | GNG5 | Guanine nucleotide binding protein (G protein), gamma 5 | Signal transduction/non-traceable author statement/signal | 0.03 | −1.0 |
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 204906_at | 6196 | RPS6KA2 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | Mitotic metaphase/inferred from electronic annotation | 0.01 | 1.2 |
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation | 0.01 | 1.2 |
| 243829_at | 673 | BRAF | V-raf murine sarcoma viral oncogene homolog B1 | MAPKKK cascade/inferred from electronic annotation/protein | 0.003 | 1.1 |
| 202647_s_at | 4893 | NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | Apoptosis/inferred from electronic annotation/signal trans | 0.005 | −1.1 |
| 207538_at | 3565 | IL4 | Interleukin 4 | Chemotaxis/traceable author statement/immune response | 0.04 | −1.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almousa, L.A.; Salter, A.M.; Castellanos, M.; May, S.T.; Langley-Evans, S.C. The Response of the Human Umbilical Vein Endothelial Cell Transcriptome to Variation in Magnesium Concentration. Nutrients 2022, 14, 3586. https://doi.org/10.3390/nu14173586
Almousa LA, Salter AM, Castellanos M, May ST, Langley-Evans SC. The Response of the Human Umbilical Vein Endothelial Cell Transcriptome to Variation in Magnesium Concentration. Nutrients. 2022; 14(17):3586. https://doi.org/10.3390/nu14173586
Chicago/Turabian StyleAlmousa, Lujain A., Andrew M. Salter, Marcos Castellanos, Sean T. May, and Simon C. Langley-Evans. 2022. "The Response of the Human Umbilical Vein Endothelial Cell Transcriptome to Variation in Magnesium Concentration" Nutrients 14, no. 17: 3586. https://doi.org/10.3390/nu14173586
APA StyleAlmousa, L. A., Salter, A. M., Castellanos, M., May, S. T., & Langley-Evans, S. C. (2022). The Response of the Human Umbilical Vein Endothelial Cell Transcriptome to Variation in Magnesium Concentration. Nutrients, 14(17), 3586. https://doi.org/10.3390/nu14173586







