Quantitative Structure–Activity Relationship Analysis of Isosteviol-Related Compounds as Activated Coagulation Factor X (FXa) Inhibitors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isosteviol Analogues
2.2. Geometry Optimization and Structural Descriptors
2.3. Statistical Analysis
2.4. MARSplines Analysis
2.5. Model Validation
3. Results
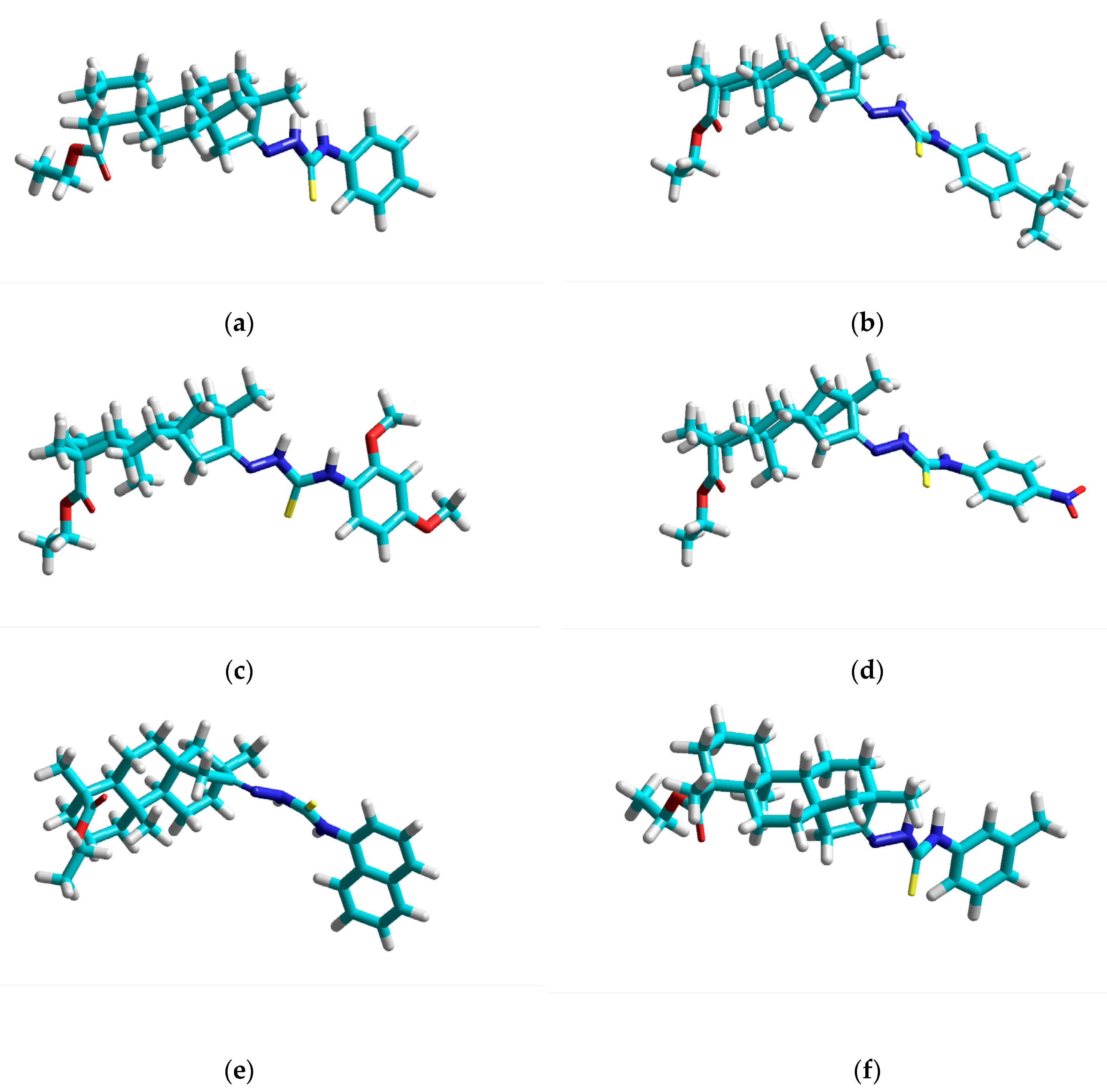
3.1. Geometry Optimization
3.2. Statistical Analysis
3.2.1. Model Building and Prediction of pIC50 Values
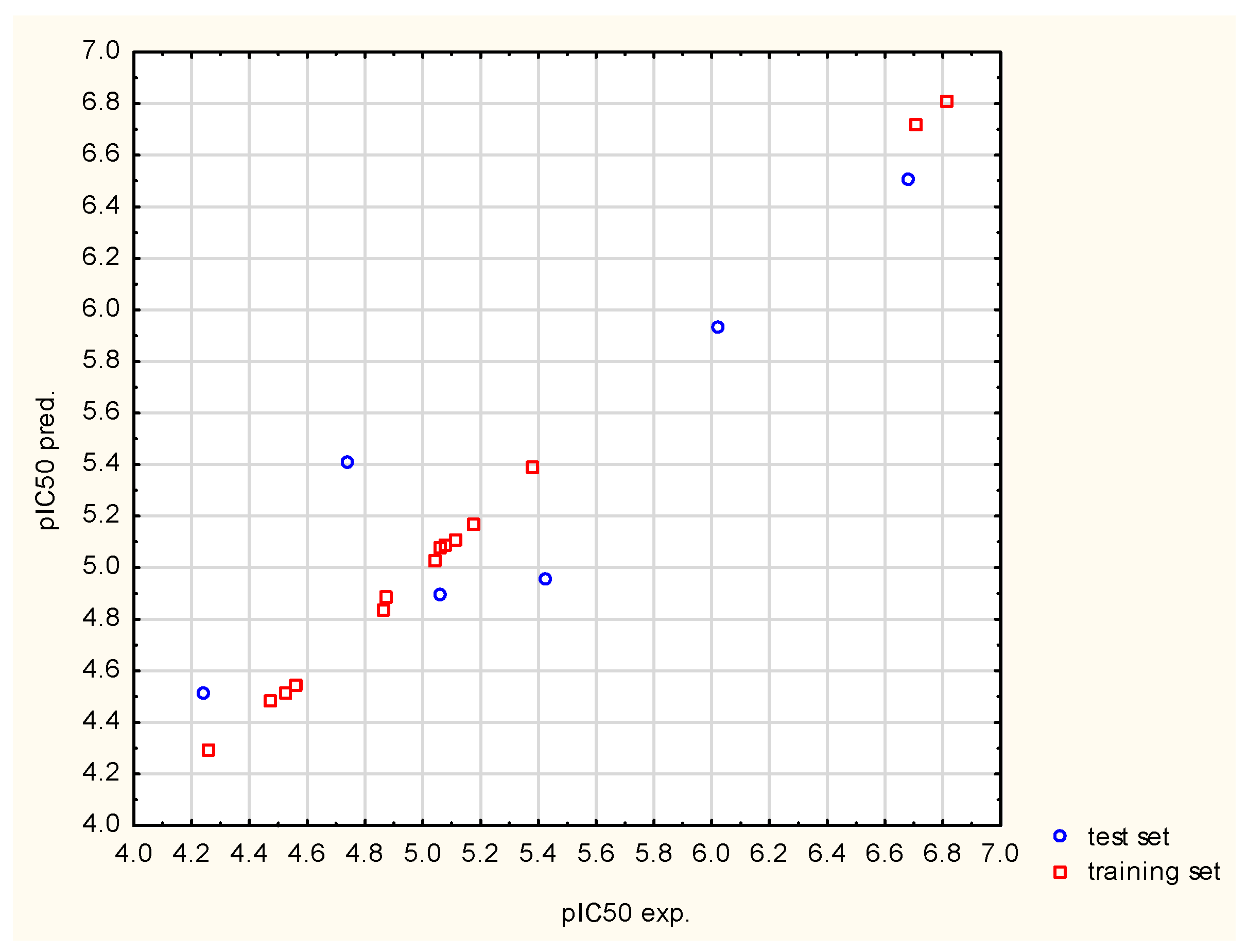
3.2.2. Validation and Selection of the Predictive Submodel
3.3. Values of Predicted Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ozsvár, D.; Nagy, V.; Zupkó, I.; Szakonyi, Z. Synthesis and Biological Application of Isosteviol-based 1,3-aminoalcohols. Int. J. Mol. Sci. 2021, 22, 11232. [Google Scholar] [CrossRef] [PubMed]
- Chatsudthipong, V.; Muanprasat, C. Stevioside and Related Compounds: Therapeutic Benefits beyond Sweetness. Pharmacol. Ther. 2009, 121, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.R.; De Oliveira, B.H. Stevioside and Related Sweet Diterpenoid Glycosides. Nat. Prod. Rep. 1993, 10, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Moons, N.; De Borggraeve, W.; Dehaen, W. Stevioside and Steviol as Starting Materials in Organic Synthesis. Curr. Org. Chem. 2012, 16, 1986–1995. [Google Scholar] [CrossRef]
- Corporate Authors. EC Regulation No. 231/2012, C.R. Commission Regulation (EU) No 231/2012. Off. J. Eur. Union 2012, 55, 1–295. [Google Scholar]
- Wang, M.; Li, H.; Xu, F.; Gao, X.; Li, J.; Xu, S.; Zhang, D.; Wu, X.; Xu, J.; Hua, H.; et al. Diterpenoid Lead Stevioside and Its Hydrolysis Products Steviol and Isosteviol: Biological Activity and Structural Modification. Eur. J. Med. Chem. 2018, 156, 885–906. [Google Scholar] [CrossRef]
- Mizushina, Y.; Akihisa, T.; Ukiya, M.; Hamasaki, Y.; Murakami-Nakai, C.; Kuriyama, I.; Takeuchi, T.; Sugawara, F.; Yoshida, H. Structural Analysis of Isosteviol and Related Compounds as DNA Polymerase and DNA Topoisomerase Inhibitors. Life Sci. 2005, 77, 2127–2140. [Google Scholar] [CrossRef]
- Takasaki, M.; Konoshima, T.; Kozuka, M.; Tokuda, H.; Takayasu, J.; Nishino, H.; Miyakoshi, M.; Mizutani, K.; Lee, K.H. Cancer Preventive Agents. Part 8: Chemopreventive Effects of Stevioside and Related Compounds. Bioorg. Med. Chem. 2009, 17, 600–605. [Google Scholar] [CrossRef]
- Abdullah Al-Dhabi, N.; Valan Arasu, M.; Rejiniemon, T.S. In Vitro Antibacterial, Antifungal, Antibiofilm, Antioxidant, and Anticancer Properties of Isosteviol Isolated from Endangered Medicinal Plant Pittosporum Tetraspermum. Evid.-Based Complement. Altern. Med. 2015, 2015, 164261. [Google Scholar] [CrossRef]
- Nordentoft, I.; Jeppesen, P.B.; Hong, J.; Abudula, R.; Hermansen, K. Isosteviol Increases Insulin Sensitivity and Changes Gene Expression of Key Insulin Regulatory Genes and Transcription Factors in Islets of the Diabetic KKAy Mouse. Diabetes Obes. Metab. 2008, 10, 939–949. [Google Scholar] [CrossRef]
- Yumuk, V.; Tsigos, C.; Fried, M.; Schindler, K.; Busetto, L.; Micic, D.; Toplak, H. European Guidelines for Obesity Management in Adults. Obes. Facts 2015, 8, 402–424. [Google Scholar] [CrossRef] [PubMed]
- Carretero Gómez, J.; Ena, J.; Arévalo Lorido, J.C.; Seguí Ripoll, J.M.; Carrasco-Sánchez, F.J.; Gómez-Huelgas, R.; Pérez Soto, M.I.; Delgado Lista, J.; Pérez Martínez, P. Obesity Is a Chronic Disease. Positioning Statement of the Diabetes, Obesity and Nutrition Workgroup of the Spanish Society of Internal Medicine (SEMI) for an Approach Centred on Individuals with Obesity. Rev. Clínica Española 2021, 221, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, E.; Bays, H.E. Cancer and Obesity: An Obesity Medicine Association (OMA) Clinical Practice Statement (CPS). Obes. Pillars 2022, 2022, 100026. [Google Scholar] [CrossRef]
- Shi, Y.; Pan, B.W.; Li, W.C.; Wang, Q.; Wu, Q.; Pan, M.; Fu, H.Z. Synthesis and Biological Evaluation of Isosteviol Derivatives as FXa Inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 126585. [Google Scholar] [CrossRef]
- Hayashi, Y.; Shimizu, I.; Yoshida, Y.; Ikegami, R.; Suda, M.; Katsuumi, G.; Fujiki, S.; Ozaki, K.; Abe, M.; Sakimura, K.; et al. Coagulation Factors Promote Brown Adipose Tissue Dysfunction and Abnormal Systemic Metabolism in Obesity. iScience 2022, 25, 104547. [Google Scholar] [CrossRef]
- Talete SRL List of Molecular Descriptors Calculated by Dragon. Available online: http://www.talete.mi.it/products/dragon_molecular_descriptor_list.pdf (accessed on 4 July 2022).
- Tropsha, A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef]
- Friedman, J.H. Multivariate Adaptive Regression Splines. Ann. Stat. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Gackowski, M.; Szewczyk-Golec, K.; Pluskota, R.; Koba, M.; Madra-Gackowska, K.; Woźniak, A. Application of Multivariate Adaptive Regression Splines (MARSplines) for Predicting Antitumor Activity of Anthrapyrazole Derivatives. Int. J. Mol. Sci. 2022, 23, 5132. [Google Scholar] [CrossRef]
- Roy, K.; Ambure, P.; Kar, S.; Ojha, P.K. Is It Possible to Improve the Quality of Predictions from an “Intelligent” Use of Multiple QSAR/QSPR/QSTR Models? J. Chemom. 2018, 32, e2992. [Google Scholar] [CrossRef]
- Todeschini, R.; Gramatica, P. The Whim Theory: New 3D Molecular Descriptors for Qsar in Environmental Modelling. SAR QSAR Environ. Res. 1997, 7, 89–115. [Google Scholar] [CrossRef]
- Kaur, D.; Thati, M.; Ruf, W. Factor X—Protease Activated Receptor-2 Signaling in the Regulation of Diet-Induced Obesity. Available online: https://abstracts.isth.org/abstract/factor-x-protease-activated-receptor-2-signaling-in-the-regulation-of-diet-induced-obesity (accessed on 21 July 2022).
- Cao, D.S.; Liang, Y.Z.; Xu, Q. Molecular Descriptors Guide Description of the Molecular Descriptors Appearing in the ChemoPy Software Package © 2012 China Computational Biology Drug Design Group Table of Contents. Man. Chemopy 2012. Available online: https://www.researchgate.net/profile/Dong-Sheng-Cao/publication/235919348_manual_for_chemopy/links/0912f5142d38a4eeb9000000/manual-for-chemopy.pdf?origin=publication_list (accessed on 9 August 2022).
- Devinyak, O.; Havrylyuk, D.; Lesyk, R. 3D-MoRSE Descriptors Explained. J. Mol. Graph. Model. 2014, 54, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; Wiley-VCH: Weinheim, Germany; New York, NY, USA; Chichester, UK; Brisbane, Australia; Sigapore; Toronto, ON, Canada, 2000; Volume 11, ISBN 3527299130. [Google Scholar]
- Suleiman, M.; Klenina, O.V.; Ogurtsov, V. Synthesis, Biological Activity Evaluation and QSAR Studies of Novel 3- Synthesis, Biological Activity Evaluation and QSAR Studies of Novel. J. Chem. Pharm. Res. 2014, 6, 1219–1235. [Google Scholar]
- Consonni, V.; Todeschini, R.; Pavan, M.; Gramatica, P. Structure/Response Correlations and Similarity/Diversity Analysis by GETAWAY Descriptors. 2 Application of the Novel 3D Molecular Descriptors to QSAR/QSPR Studies. J. Chem. Inf. Comput. Sci. 2002, 42, 693–705. [Google Scholar] [CrossRef] [PubMed]
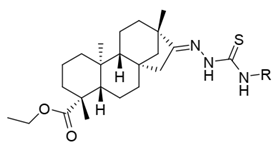 | |||
| Compound | Set | R * | Inhibition Activity Against FXa **: IC50 ***, M **** |
| i20 | training | 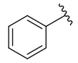 | 13,382.4 ± 183.85 |
| i21 | test | 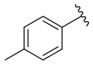 | 57,733.6 ± 315.07 |
| i22 | training | 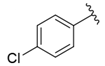 | 152.78 ± 3.18 |
| i23 | training |  | 27,546.6 ± 391.27 |
| i24 | training | 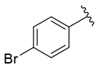 | 8772.8 ± 25.43 |
| i25 | training | 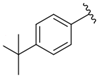 | 9034.4 ± 16.58 |
| i26 | training | 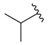 | 54,893.0 ± 588.77 |
| i27 | test | 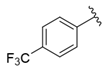 | 8658.8 ± 40.20 |
| i28 | training | 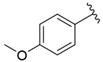 | 6651.8 ± 40.00 |
| i29 | training | 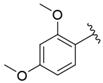 | 33,578.2 ± 275.65 |
| i30 | test | 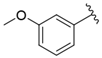 | 18,409.0 ± 435.88 |
| i31 | training |  | 29,616.6 ± 349.80 |
| i32 | training | 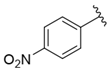 | 8463.4 ± 21.64 |
| i33 | training | 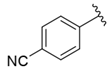 | 13,591.6 ± 410.15 |
| i34 | test | 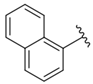 | 955.12 ± 18.74 |
| i35 | training | 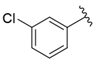 | 196.34 ± 5.37 |
| i36 | training | 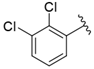 | 7727.8 ± 11.63 |
| i37 | training | 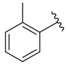 | 4154.2 ± 50.22 |
| i38 | test | 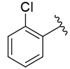 | 209.38 ± 4.76 |
| i39 | test | 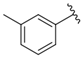 | 3759.2 ± 28.12 |
| Options | Values |
|---|---|
| Maximum number of basis functions | 40 |
| Degree of interactions | 3 |
| Penalty | 2 |
| Threshold | 0.0005 |
| Apply pruning | YES |
| Compound | Set | Descriptors | |||||
|---|---|---|---|---|---|---|---|
| B01[C-Cl] * | E2m ** | L3v *** | Mor06i **** | RDF070i ***** | HATS7s ****** | ||
| i20 | training | 0 | 0.166 | 1.551 | −4.428 | 50.027 | 0.614 |
| i21 | test | 0 | 0.164 | 1.477 | −4.302 | 55.231 | 0.592 |
| i22 | training | 1 | 0.139 | 1.525 | −3.421 | 50.427 | 0.677 |
| i23 | training | 0 | 0.153 | 1.518 | −3.909 | 51.834 | 0.757 |
| i24 | training | 0 | 0.114 | 1.519 | −3.619 | 50.99 | 0.650 |
| i25 | training | 0 | 0.155 | 1.54 | −4.578 | 54.079 | 0.477 |
| i26 | training | 0 | 0.183 | 1.468 | −4.294 | 54.897 | 0.585 |
| i27 | test | 0 | 0.133 | 1.485 | −4.321 | 53.827 | 0.645 |
| i28 | training | 0 | 0.151 | 1.62 | −4.445 | 52.251 | 0.577 |
| i29 | training | 0 | 0.194 | 1.444 | −4.451 | 47.955 | 0.559 |
| i30 | test | 0 | 0.152 | 1.685 | −4.568 | 49.918 | 0.600 |
| i31 | training | 0 | 0.157 | 1.469 | −4.434 | 59.622 | 0.525 |
| i32 | training | 0 | 0.134 | 1.577 | −3.925 | 50.96 | 0.633 |
| i33 | training | 0 | 0.148 | 1.495 | −4.196 | 50.649 | 0.646 |
| i34 | test | 0 | 0.178 | 2.086 | −5.578 | 52.789 | 0.566 |
| i35 | training | 1 | 0.147 | 1.574 | −3.515 | 54.923 | 0.629 |
| i36 | training | 1 | 0.281 | 1.716 | −1.044 | 44.579 | 0.597 |
| i37 | training | 0 | 0.161 | 1.824 | −4.695 | 57.552 | 0.546 |
| i38 | test | 1 | 0.191 | 1.296 | −4.037 | 45.968 | 0.597 |
| i39 | test | 0 | 0.165 | 1.648 | −4.638 | 55.429 | 0.609 |
| Symbol | Definition | Block | Dimensionality | Number in the Basis Function |
|---|---|---|---|---|
| B01[C-Cl] | Presence/absence of C-Cl at topological distance 1 | 2D Atom Pairs | 2D | 1 |
| E2m | 2nd component accessibility directional WHIM index/weighted by mass | WHIM * descriptors | 3D | 1 |
| L3v | 3rd component size directional WHIM index/weighted by van der Waals volume | WHIM descriptors | 3D | 1 |
| Mor06i | signal 06/weighted by ionization potential | 3D-MoRSE ** descriptors | 3D | 1 |
| RDF070i | Radial Distribution Function—070/weighted by ionization potential | RDF *** descriptors | 3D | 1 |
| HATS7s | leverage-weighted autocorrelation of lag 7/weighted by I-state | GETAWAY **** descriptors | 3D | 1 |
| Bm * | Definition | am ** |
|---|---|---|
| B0 | 1 | 5.74300 |
| B1 | (B01[C-Cl])+ | 2.08922 |
| B2 | (E2m-0.11400)+ | −1.17409 |
| B3 | (L3v-1.44400)+ | 2.16641 |
| B4 | (Mor06i+4.69500)+ | −3.32023 |
| B5 | (RDF070i-44.57900)+ | −4.22030 |
| B6 | (HATS7s-0.47700)+ | −1.17824 |
| Degree of Interaction | Number of Basis Functions | R2 | Q2 | MAE |
|---|---|---|---|---|
| 1 | 2 | 0.99272 | 0.15208 | 0.2973 |
| 6 | 0.99846 | 0.79223 | 0.1017 | |
| 2 | 1 | 0.98984 | 0.17375 | 0.4235 |
| 5 | 0.99745 | 0.67195 | 0.1635 | |
| 12 | 0.99631 | 0.50117 | 0.1476 | |
| 3 | 1 | 0.98984 | 0.17375 | 0.4235 |
| 5 | 0.99751 | 0.67195 | 0.1635 | |
| 12 | 0.99747 | 0.65840 | 0.1313 |
| Parameter | Value | Threshold | Meaning [20] |
|---|---|---|---|
| 0.9985 | ~1 | a measure of the variation of observed with the predicted data | |
| 0.7922 | ≥0.5 | cross-validated R2 (Q2) tested for internal validation | |
| 0.9874 | ≥0.5 | it measures the correlation between the observed and predicted data of the test set | |
| 0.7927 | ≥0.5 | almost equal or closer values of Q2(F2) and Q2(F1) infer that the training set mean lies in the close propinquity to that of the test set | |
| 0.9706 | ≥0.5 | it is a measure of the model predictability | |
| 0.9635 | ~1 | concordance correlation coefficient (CCC) measures both precision and accuracy, detecting the distance of the observations from the fitting line and the degree of deviation of the regression line from that passing through the origin, respectively | |
The terms k and k′ are explained as follows: | 0.0196 and 0.9216 | 2 > 0.5 | they reflect the overall predictability of the model for the whole data set |
| 0.8154 | assesses the model using the predicted residual sum of squares | ||
| 0.2020 | standard deviation of error of prediction (SDEP) is calculated from PRESS | ||
| 0.1017 | index of errors in the context of predictive modeling studies |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gackowski, M.; Szewczyk-Golec, K.; Mądra-Gackowska, K.; Pluskota, R.; Koba, M. Quantitative Structure–Activity Relationship Analysis of Isosteviol-Related Compounds as Activated Coagulation Factor X (FXa) Inhibitors. Nutrients 2022, 14, 3521. https://doi.org/10.3390/nu14173521
Gackowski M, Szewczyk-Golec K, Mądra-Gackowska K, Pluskota R, Koba M. Quantitative Structure–Activity Relationship Analysis of Isosteviol-Related Compounds as Activated Coagulation Factor X (FXa) Inhibitors. Nutrients. 2022; 14(17):3521. https://doi.org/10.3390/nu14173521
Chicago/Turabian StyleGackowski, Marcin, Karolina Szewczyk-Golec, Katarzyna Mądra-Gackowska, Robert Pluskota, and Marcin Koba. 2022. "Quantitative Structure–Activity Relationship Analysis of Isosteviol-Related Compounds as Activated Coagulation Factor X (FXa) Inhibitors" Nutrients 14, no. 17: 3521. https://doi.org/10.3390/nu14173521
APA StyleGackowski, M., Szewczyk-Golec, K., Mądra-Gackowska, K., Pluskota, R., & Koba, M. (2022). Quantitative Structure–Activity Relationship Analysis of Isosteviol-Related Compounds as Activated Coagulation Factor X (FXa) Inhibitors. Nutrients, 14(17), 3521. https://doi.org/10.3390/nu14173521










