Molecular Alterations Caused by Alcohol Consumption in the UK Biobank: A Mendelian Randomisation Study
Abstract
:1. Introduction
2. Methods
2.1. Beverage Specific Agnostic Association Analyses
2.2. Two Sample MR
2.2.1. Instrument Selection (Multiple Instrument MR)
2.2.2. Single-Instrument MR Analysis
2.3. Phenome-Wide Association Analysis
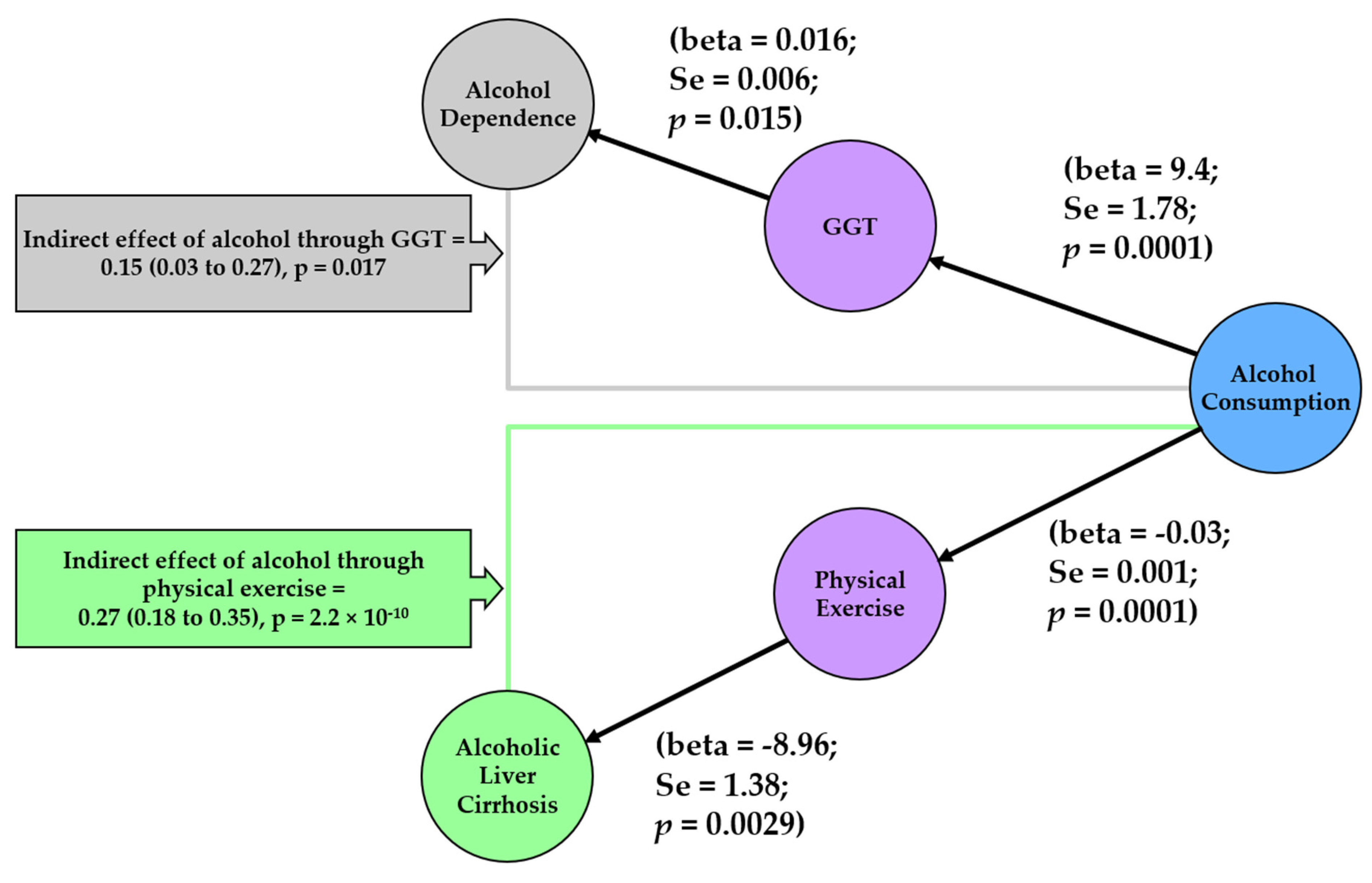
2.4. Mediation Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Status Report on Alcohol and Health 2018: Executive Summary; World Health Organization: Geneva, Switzerland, 2018.
- Poli, A.; Marangoni, F.; Avogaro, A.; Barba, G.; Bellentani, S.; Bucci, M.; Cambieri, R.; Catapano, A.L.; Costanzo, S.; Cricelli, C.; et al. Moderate alcohol use and health: A consensus document. Nutr. Metab. Cardiovasc. Dis. 2013, 23, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Lea, W. UK Chief Medical Officers’ Low Risk Drinking Guidelines; Department of Health and Aged Care, Ed.; The UK Government: London, UK, 2016; pp. 1–11. [Google Scholar]
- Holmes, J.; Beard, E.; Brown, J.; Brennan, A.; Meier, P.S.; Michie, S.; Stevely, A.K.; Webster, L.; Buykx, P.F. Effects on alcohol consumption of announcing and implementing revised UK low-risk drinking guidelines: Findings from an interrupted time series analysis. J. Epidemiol. Community Health 2020, 74, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Chiva-Blanch, G.; Arranz, S.; Lamuela-Raventos, R.M.; Estruch, R. Effects of wine, alcohol and polyphenols on cardiovascular disease risk factors: Evidences from human studies. Alcohol Alcohol. 2013, 48, 270–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jani, B.D.; McQueenie, R.; Nicholl, B.I.; Field, R.; Hanlon, P.; Gallacher, K.I.; Mair, F.S.; Lewsey, J. Association between patterns of alcohol consumption (beverage type, frequency and consumption with food) and risk of adverse health outcomes: A prospective cohort study. BMC Med. 2021, 19, 8. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Ritchie, M.D. Current Scope and Challenges in Phenome-Wide Association Studies. Curr. Epidemiol. Rep. 2017, 4, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.C.; Ritchie, M.D.; Basford, M.A.; Pulley, J.M.; Bastarache, L.; Brown-Gentry, K.; Wang, D.; Masys, D.R.; Roden, D.M.; Crawford, D.C. PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010, 26, 1205–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davey Smith, G.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Baird, D.; Borges, M.-C.; Bowden, J.; Hemani, G.; Haycock, P.; Evans, D.M.; Smith, G.D. Recent Developments in Mendelian Randomization Studies. Curr. Epidemiol. Rep. 2017, 4, 330–345. [Google Scholar] [CrossRef] [Green Version]
- Alfaro-Almagro, F.; McCarthy, P.; Afyouni, S.; Andersson, J.L.R.; Bastiani, M.; Miller, K.L.; Nichols, T.E.; Smith, S.M. Confound modelling in UK Biobank brain imaging. Neuroimage 2021, 224, 117002. [Google Scholar] [CrossRef]
- Kyle, S.D.; Sexton, C.E.; Feige, B.; Luik, A.I.; Lane, J.; Saxena, R.; Anderson, S.G.; Bechtold, D.A.; Dixon, W.; Little, M.A.; et al. Sleep and cognitive performance: Cross-sectional associations in the UK Biobank. Sleep Med. 2017, 38, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peakman, T.C.; Elliott, P. The UK Biobank sample handling and storage validation studies. Int. J. Epidemiol. 2008, 37 (Suppl. S1), i2–i6. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jiang, Y.; Wedow, R.; Li, Y.; Brazel, D.M.; Chen, F.; Datta, G.; Davila-Velderrain, J.; McGuire, D.; Tian, C.; et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019, 51, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Jorgenson, E.; Thai, K.K.; Hoffmann, T.J.; Sakoda, L.C.; Kvale, M.N.; Banda, Y.; Schaefer, C.; Risch, N.; Mertens, J.; Weisner, C.; et al. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol. Psychiatry 2017, 22, 1359–1367. [Google Scholar] [CrossRef] [Green Version]
- Olfson, E.; Bierut, L.J. Convergence of genome-wide association and candidate gene studies for alcoholism. Alcohol. Clin. Exp. Res. 2012, 36, 2086–2094. [Google Scholar] [CrossRef] [Green Version]
- Buch, S.; Stickel, F.; Trépo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.; Berg, T.; Ridinger, M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015, 47, 1443–1448. [Google Scholar] [CrossRef]
- Davison, A.C.; Hinkley, D.V. Bootstrap Methods and Their Application; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef]
- Clarke, L.; Zheng-Bradley, X.; Smith, R.; Kulesha, E.; Xiao, C.; Toneva, I.; Vaughan, B.; Preuss, D.; Leinonen, R.; Shumway, M.; et al. The 1000 Genomes Project: Data management and community access. Nat. Methods 2012, 9, 459–462. [Google Scholar] [CrossRef] [Green Version]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Duell, E.J.; Sala, N.; Travier, N.; Muñoz, X.; Boutron-Ruault, M.C.; Clavel-Chapelon, F.; Barricarte, A.; Arriola, L.; Navarro, C.; Sánchez-Cantalejo, E.; et al. Genetic variation in alcohol dehydrogenase (ADH1A, ADH1B, ADH1C, ADH7) and aldehyde dehydrogenase (ALDH2), alcohol consumption and gastric cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort. Carcinogenesis 2011, 33, 361–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edenberg, H.J.; McClintick, J.N. Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol. Clin. Exp. Res. 2018, 42, 2281–2297. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bierut, L.J.; Agrawal, A.; Bucholz, K.K.; Doheny, K.F.; Laurie, C.; Pugh, E.; Fisher, S.; Fox, L.; Howells, W.; Bertelsen, S.; et al. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. USA 2010, 107, 5082–5087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobel, M.E. Asymptotic Confidence Intervals for Indirect Effects in Structural Equation Models. In Sociological Methodology; Leinhart, S., Ed.; Jossey-Bass: San Francisco, CA, USA, 1982; pp. 290–312. [Google Scholar]
- Lee, D.-H.; Ha, M.-H.; Christiani, D.C. Body weight, alcohol consumption and liver enzyme activity—A 4-year follow-up study. Int. J. Epidemiol. 2001, 30, 766–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alatalo, P.I.; Koivisto, H.M.; Hietala, J.P.; Puukka, K.S.; Bloigu, R.; Niemelä, O.J. Effect of moderate alcohol consumption on liver enzymes increases with increasing body mass index. Am. J. Clin. Nutr. 2008, 88, 1097–1103. [Google Scholar] [CrossRef] [Green Version]
- Roerecke, M.; Vafaei, A.; Hasan, O.S.M.; Chrystoja, B.R.; Cruz, M.; Lee, R.; Rehm, J. Alcohol Consumption and Risk of Liver Cirrhosis: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2019, 114, 1574–1586. [Google Scholar] [CrossRef]
- Tedesco, I.; Russo, M.; Russo, P.; Iacomino, G.; Russo, G.L.; Carraturo, A.; Faruolo, C.; Moio, L.; Palumbo, R. Antioxidant effect of red wine polyphenols on red blood cells. J. Nutr. Biochem. 2000, 11, 114–119. [Google Scholar] [CrossRef]
- Galani-Nikolakaki, S.; Kallithrakas-Kontos, N.; Katsanos, A.A. Trace element analysis of Cretan wines and wine products. Sci. Total Environ. 2002, 285, 155–163. [Google Scholar] [CrossRef]
- Myrhed, M.; Berglund, L.; Böttiger, L.E. Alcohol consumption and hematology. Acta Med. Scand. 1977, 202, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Ballard, H.S. The hematological complications of alcoholism. Alcohol Health Res. World 1997, 21, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Toth, A.; Sandor, B.; Papp, J.; Rabai, M.; Botor, D.; Horvath, Z.; Kenyeres, P.; Juricskay, I.; Toth, K.; Czopf, L. Moderate red wine consumption improves hemorheological parameters in healthy volunteers. Clin. Hemorheol. Microcirc. 2014, 56, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Campion, E.W.; Glynn, R.J.; DeLabry, L.O. Asymptomatic hyperuricemia. Risks and consequences in the Normative Aging Study. Am. J. Med. 1987, 82, 421–426. [Google Scholar] [CrossRef]
- Jee, Y.H.; Jung, K.J.; Park, Y.B.; Spiller, W.; Jee, S.H. Causal effect of alcohol consumption on hyperuricemia using a Mendelian randomization design. Int. J. Rheum. Dis. 2019, 22, 1912–1919. [Google Scholar] [CrossRef]
- Gaffo, A.L.; Roseman, J.M.; Jacobs, D.R., Jr.; Lewis, C.E.; Shikany, J.M.; Mikuls, T.R.; Jolly, P.E.; Saag, K.G. Serum urate and its relationship with alcoholic beverage intake in men and women: Findings from the Coronary Artery Risk Development in Young Adults (CARDIA) cohort. Ann. Rheum. Dis. 2010, 69, 1965–1970. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.K.; Curhan, G. Beer, liquor, and wine consumption and serum uric acid level: The Third National Health and Nutrition Examination Survey. Arthritis Rheum. 2004, 51, 1023–1029. [Google Scholar] [CrossRef]
- Li, X.; Meng, X.; Spiliopoulou, A.; Timofeeva, M.; Wei, W.-Q.; Gifford, A.; Shen, X.; He, Y.; Varley, T.; McKeigue, P.; et al. MR-PheWAS: Exploring the causal effect of SUA level on multiple disease outcomes by using genetic instruments in UK Biobank. Ann. Rheum. Dis. 2018, 77, 1039–1047. [Google Scholar] [CrossRef] [Green Version]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Sanderson, E. Multivariable Mendelian Randomization and Mediation. Cold Spring Harb. Perspect. Med. 2021, 11, a038984. [Google Scholar] [CrossRef]
- Taylor, M.; Simpkin, A.J.; Haycock, P.C.; Dudbridge, F.; Zuccolo, L. Exploration of a Polygenic Risk Score for Alcohol Consumption: A Longitudinal Analysis from the ALSPAC Cohort. PLoS ONE 2016, 11, e0167360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
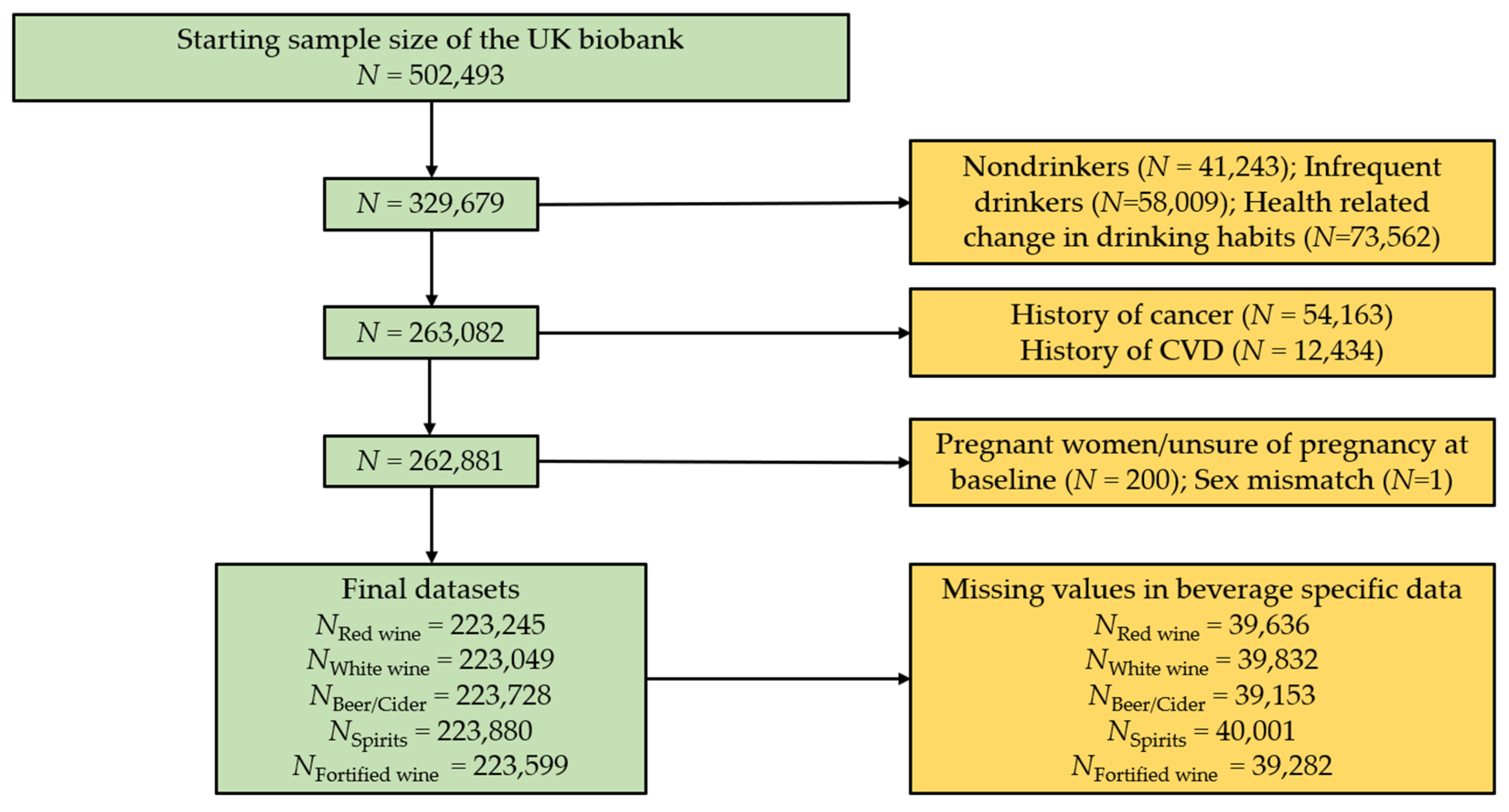
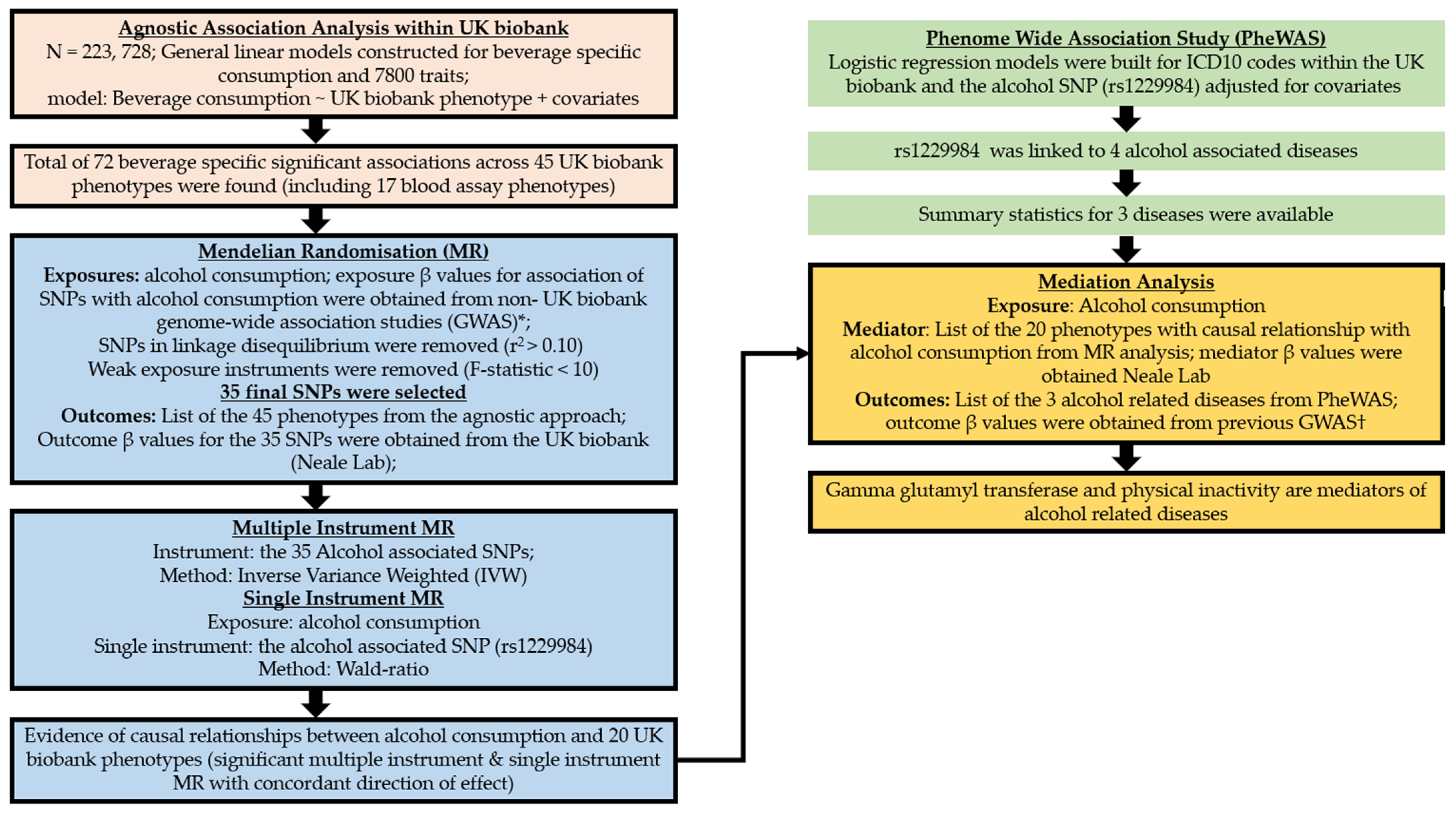

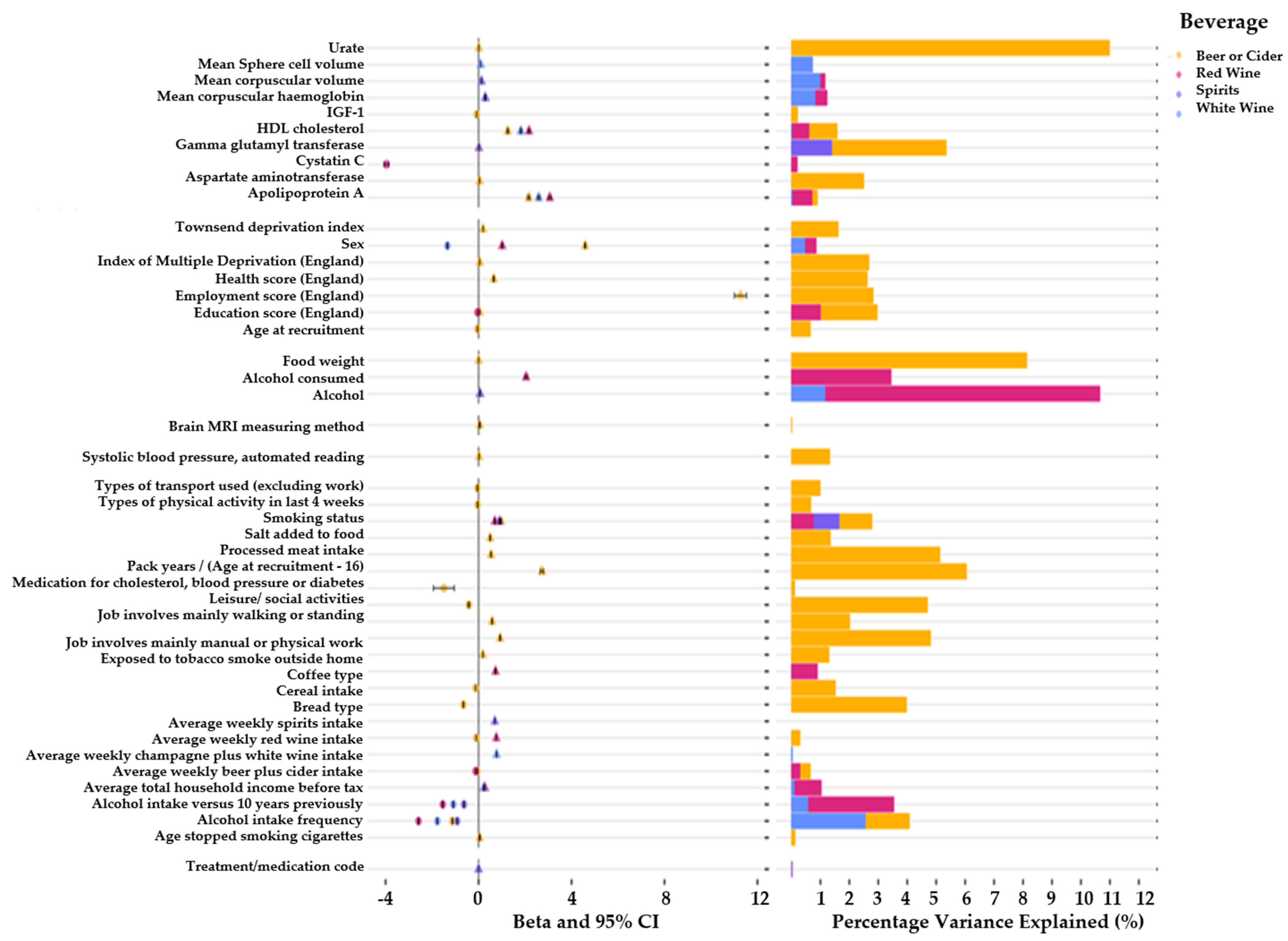
| Characteristics | Red Wine Dataset (N = 223,245) | White Wine/Sparkling White Wine Dataset (N = 223,049) | Beer or Cider Dataset (N = 223,728) | Spirits Dataset (N = 222,880) | Fortified Wine Dataset (N = 223,599) |
|---|---|---|---|---|---|
| Age-yr | 55.5 (±8.01) | 55.5 (±8.01) | 55.5 (±8.01) | 55.5 (±8.01) | 55.593 (±8.01) |
| Male sex-no. (%) | 108,467 (48.59%) | 108,509 (48.61%) | 108,579 (48.64%) | 108,529 (48.61%) | 108,458 (48.58%) |
| Lipid treatment-no./total no. (%) | 24,259 (10.87%) | 24,246 (10.86%) | 19,778 (8.86%) | 19,756 (8.85%) | 19,774 (8.86%) |
| Diabetes mellitus-no./total no. (%) | 5840 (2.62%) | 5829 (2.61%) | 5847 (2.62%) | 5830 (2.62%) | 5837 (2.61%) |
| Body mass index | 26.9 (±4.33) | 26.9 (±4.33) | 26.9 (±4.34) | 26.9 (±4.33) | 26.9 (±4.34) |
| MET Score | 2642.4 (±2664.35) | 2642.8 (±2665.44) | 2642.5 (±2665.28) | 2642.1 (±2664.77) | 2641.4 (±2663.52) |
| current smoking-no. (%) | 23,593 (10.57%) | 23,599 (10.57%) | 23,659 (10.60%) | 23,553 (10.55%) | 23,620 (10.58%) |
| past smoking-no. (%) | 77,860 (34.88%) | 77,884 (34.89%) | 77,875 (34.88%) | 77,881 (34.89%) | 77,875 (34.88%) |
| never smoking-no. (%) | 121,130 (54.26%) | 121,097 (54.24%) | 121,044 (54.22%) | 121,148 (54.27%) | 121,084 (54.24%) |
| Systolic blood pressure-mean (SD)-mmHg | 140.4 (±19.47) | 140.4 (±19.48) | 140.4 (±19.48) | 140.4 (±19.48) | 140.4 (±19.49) |
| Diastolic blood pressure-mean (SD)-mmHg | 83.4 (±10.82) | 83.4 (±10.81) | 83.4 (±10.81) | 83.4 (±10.81) | 83.4 (±10.82) |
| Red wine intake-mean (SD)-glass/week | 3.9 (±5.68) | 3.9 (±5.68) | 3.9 (±5.68) | 3.93 (±5.68) | 3.9 (±5.68) |
| White wine intake-mean (SD)-glass/week | 2.7 (±4.88) | 2.7 (±4.88) | 2.7 (±4.88) | 2.7 (±4.88) | 2.7 (±4.88) |
| Fortified wine intake- mean (SD)-glass/week | 0.2 (±1.21) | 0.2 (±1.21) | 0.2 (±1.22) | 0.2 (±1.22) | 0.2 (±1.22) |
| Beer intake-mean (SD)-pints/week | 2.9 (±5.59) | 2.9 (±5.59) | 2.9(±5.62) | 2.9 (±5.60) | 2.9 (±5.60) |
| Spirits intake-mean (SD)-measures/week | 1.8 (±5.29) | 1.8 (±5.29) | 1.8 (±5.32) | 1.8 (±5.36) | 1.8 (±5.30) |
| Single Instrument MR | Multiple Instrument MR | |||||
|---|---|---|---|---|---|---|
| Beta | 95% CI | Observed p-Value | Beta | 95% CI | Observed p-Value | |
| Gamma Glutamyl Transferase | 9.7 | 5.8, 13.6 | 0.0001 | 9.4 | 5.9, 12.9 | 0 |
| Mean Sphered Cell Volume | 0.2 | 0.15, 0.31 | 0 | 0.19 | 0.11, 027 | 0.0001 |
| Mean Corpuscular Haemoglobin | 0.3 | 0.18, 0.36 | 0 | 0.3 | 0.19, 0.35 | 0 |
| Mean Corpuscular Volume | 0.3 | 0.18, 0.36 | 0 | 0.3 | 0.19, 0.36 | 0 |
| Unplanned physical activity by method of transport | −0.04 | −0.07, −0.01 | 0.01 | −0.06 | −0.1, −0.03 | 0.0008 |
| Wholemeal/ wholegrain bread consumption | −0.05 | −0.09, −0.02 | 0.006 | −0.06 | −0.1, −0.02 | 0.008 |
| White bread consumption | 0.05 | 0.02, 0.09 | 0.002 | 0.05 | 0.02, 0.09 | 0.008 |
| Description | Effect Estimate | 95% CI | Odds Ratio | p-Value |
|---|---|---|---|---|
| Alcohol-related disorders | 0.24 | (0.16, 0.32) | 1.3 | 4.87 × 10−10 |
| Alcoholism | 0.26 | (0.16, 0.36) | 1.3 | 2.52 × 10−8 |
| Alcoholic liver damage | 0.27 | (0.15, 0.39) | 1.3 | 3.47 × 10−6 |
| Enthesopathy | −0.06 | (−0.08, −0.04) | 0.9 | 1.05 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Farrell, F.; Jiang, X.; Aljifri, S.; Pazoki, R. Molecular Alterations Caused by Alcohol Consumption in the UK Biobank: A Mendelian Randomisation Study. Nutrients 2022, 14, 2943. https://doi.org/10.3390/nu14142943
O’Farrell F, Jiang X, Aljifri S, Pazoki R. Molecular Alterations Caused by Alcohol Consumption in the UK Biobank: A Mendelian Randomisation Study. Nutrients. 2022; 14(14):2943. https://doi.org/10.3390/nu14142943
Chicago/Turabian StyleO’Farrell, Felix, Xiyun Jiang, Shahad Aljifri, and Raha Pazoki. 2022. "Molecular Alterations Caused by Alcohol Consumption in the UK Biobank: A Mendelian Randomisation Study" Nutrients 14, no. 14: 2943. https://doi.org/10.3390/nu14142943
APA StyleO’Farrell, F., Jiang, X., Aljifri, S., & Pazoki, R. (2022). Molecular Alterations Caused by Alcohol Consumption in the UK Biobank: A Mendelian Randomisation Study. Nutrients, 14(14), 2943. https://doi.org/10.3390/nu14142943






