Gene-Environment Interactions in Vitamin D Status and Sun Exposure: A Systematic Review with Recommendations for Future Research
Abstract
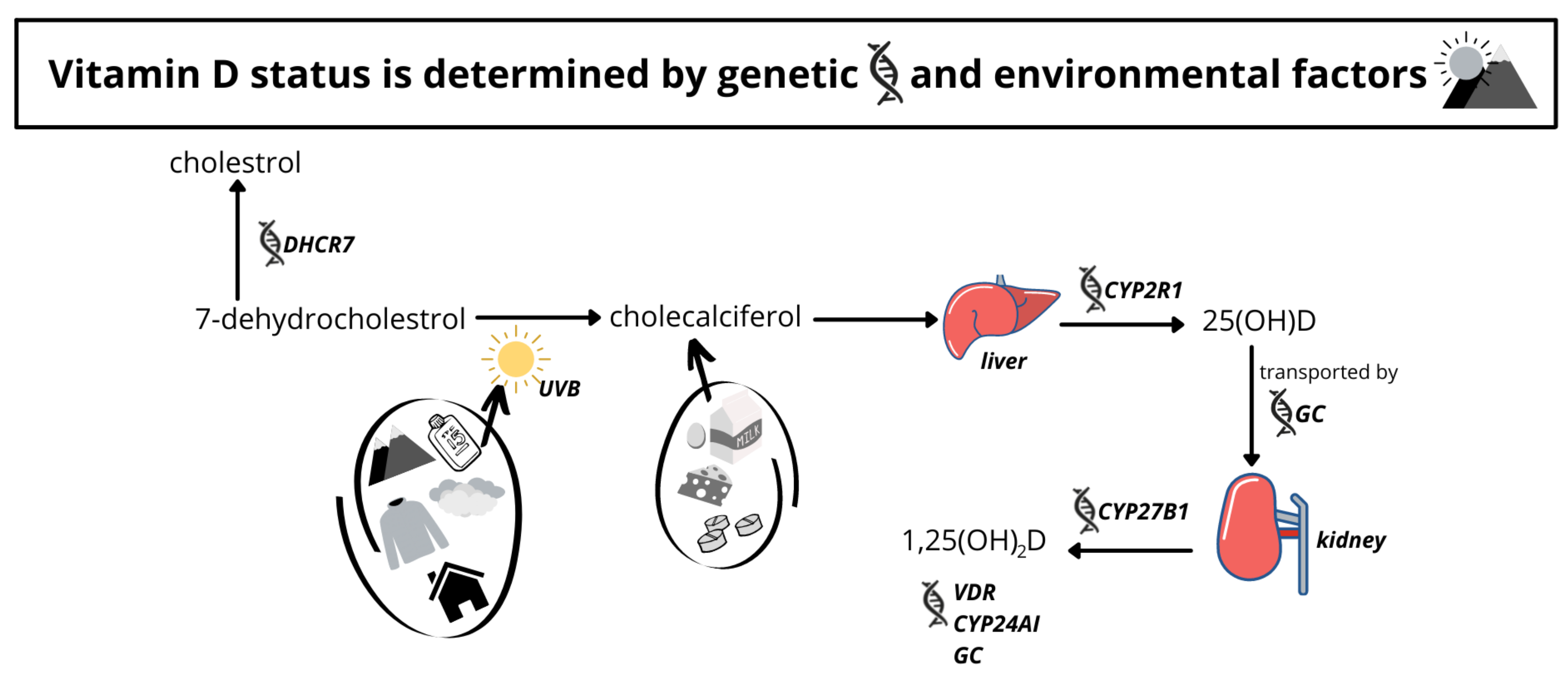
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Study Selection
2.4. Data Extraction and Quality Assessment
3. Results
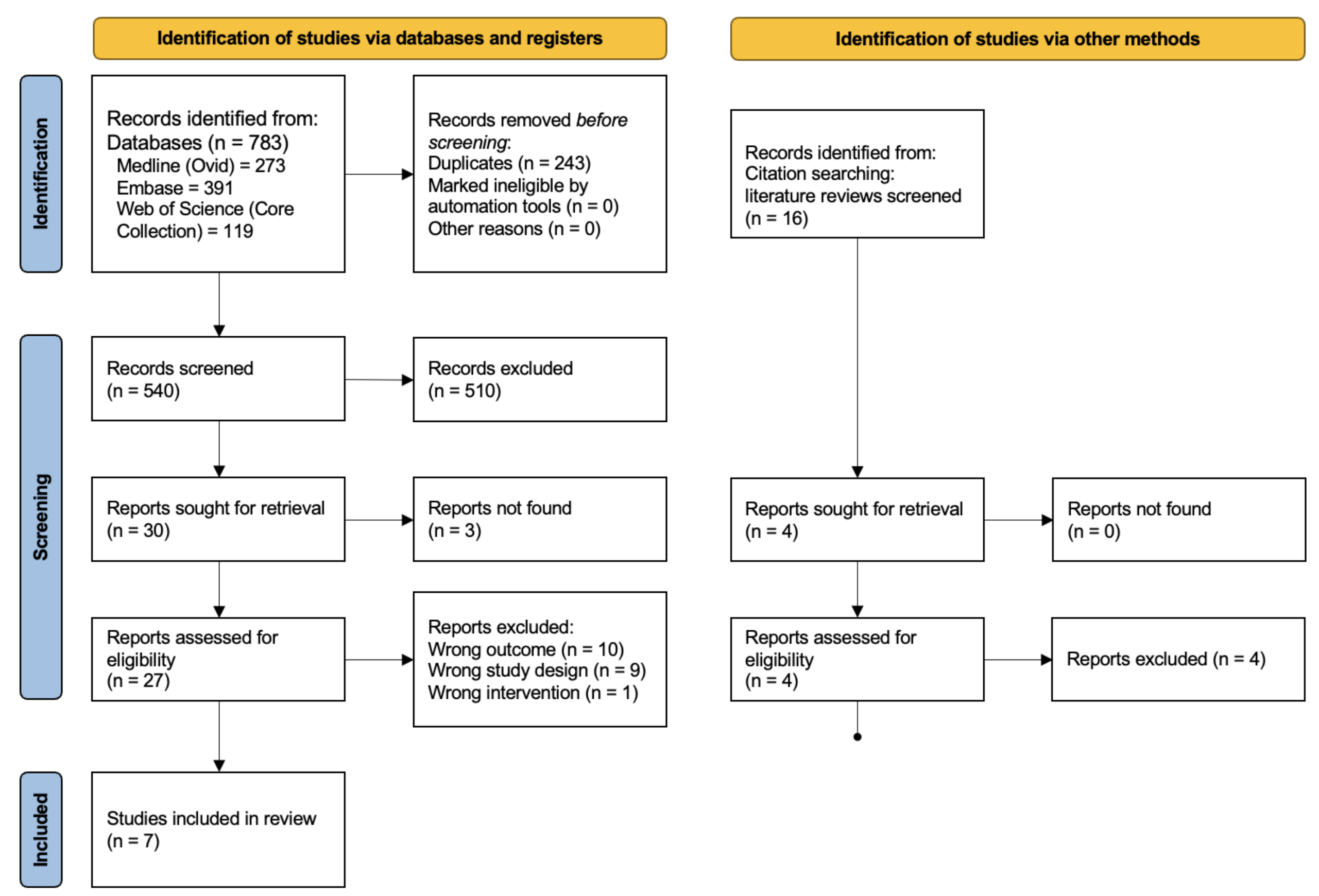
3.1. Study Selection and Characteristics
3.2. Risk of Bias in Studies
3.3. Research Design and Study Samples
3.4. Environmental Exposure
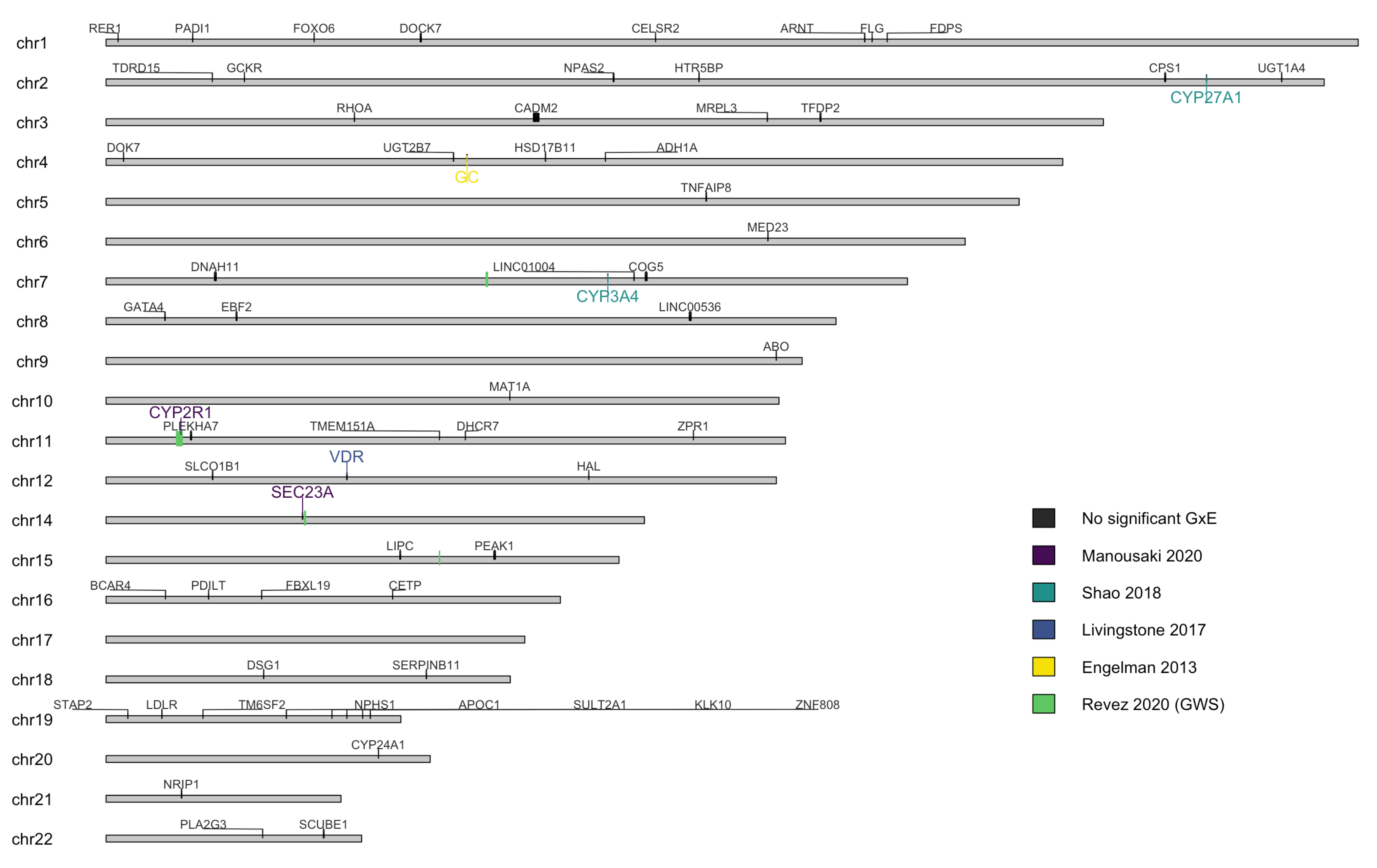
3.5. Genetic Factors
3.6. Covariates
3.7. Interaction Findings
3.8. Interaction in GC
3.9. Interaction in VDR
3.10. Interaction in CYP2R1
3.11. Interaction with Other Genetic Variants
4. Discussion
4.1. Sample Size and Power
4.2. Variability in Exposures
4.3. Choice of Environmental Exposure
4.4. Significance Thresholds
4.5. Methodological Implications
4.6. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chowdhury, R.; Kunutsor, S.; Vitezova, A.; Oliver-Williams, C.; Chowdhury, S.; Kiefte-de-Jong, J.; Khan, H.; Baena, C.; Prabhakaran, D.; Hoshen, M. Vitamin D and risk of cause specific death: Systematic review and meta-analysis of observational cohort and randomised intervention studies. BMJ 2014, 348, g1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodoratou, E.; Tzoulaki, I.; Zgaga, L.; Ioannidis, J. Vitamin D and multiple health outcomes: Umbrella review of systematic reviews and meta-analyses of observational studies and randomised trials. BMJ 2014, 348, g2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Wang, J.; Song, M.; Giovannucci, E.; Ma, H.; Jin, G.; Hu, Z.; Shen, H.; Hang, D. Vitamin D Status and Risk of All-Cause and Cause-Specific Mortality in a Large Cohort: Results From the UK Biobank. J. Clin. Endocrinol. Metab. 2020, 105, e3606–e3619. [Google Scholar] [CrossRef] [PubMed]
- Palacios, C.; Gonzalez, L. Is vitamin D deficiency a major global public health problem? J. Steroid Biochem. Mol. Biol. 2014, 144, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraff, V.; Shaw, N. Sunshine and vitamin D. Arch. Dis. Child. 2016, 101, 190–192. [Google Scholar] [CrossRef]
- Neville, J.; Palmieri, T.; Young, A. Physical Determinants of Vitamin D Photosynthesis: A Review. JBMR Plus 2021, 5, e10460. [Google Scholar] [CrossRef]
- Leal, A.; Correa, M.; Holick, M.; Melo, E.; Lazaretti-Castro, M. Sun-induced production of vitamin D3 throughout 1 year in tropical and subtropical regions: Relationship with latitude, cloudiness, UV-B exposure and solar zenith angle. Photochem. Photobiol. Sci. 2021, 20, 265–274. [Google Scholar] [CrossRef]
- McCullough, M.; Weinstein, S.; Freedman, D.; Helzlsouer, K.; Flanders, W.; Koenig, K.; Kolonel, L.; Laden, F.; Le Marchand, L.; Purdue, M. Correlates of circulating 25-hydroxyvitamin D: Cohort Consortium Vitamin D Pooling Project of Rarer Cancers. Am. J. Epidemiol. 2010, 172, 21–35. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zhang, F.; Richards, J.; Kestenbaum, B.; Meurs, J.; Berry, D.; Kiel, D.; Streeten, E.; Ohlsson, C.; Koller, D. Common genetic determinants of vitamin D insufficiency: A genome-wide association study. Lancet 2010, 376, 180–188. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D. Gene–environment-wide association studies: Emerging approaches. Nat. Rev. Genet. 2010, 11, 259–272. [Google Scholar] [CrossRef]
- Lagunova, Z.; Porojnicu, A.; Lindberg, F.; Hexeberg, S.; Moan, J. The dependency of vitamin D status on body mass index, gender, age and season. Anticancer Res. 2009, 29, 3713–3720. [Google Scholar] [CrossRef] [PubMed]
- Manousaki, D.; Mitchell, R.; Dudding, T.; Haworth, S.; Harroud, A.; Forgetta, V.; Shah, R.; Luan, J.; Langenberg, C.; Timpson, N. Genome-wide Association Study for Vitamin D Levels Reveals 69 Independent Loci. Am. J. Hum. Genet. 2020, 106, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Karohl, C.; Su, S.; Kumari, M.; Tangpricha, V.; Veledar, E.; Vaccarino, V.; Raggi, P. Heritability and seasonal variability of vitamin D concentrations in male twins. Am. J. Clin. Nutr. 2010, 92, 1393–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snellman, G.; Melhus, H.; Gedeborg, R.; Olofsson, S.; Wolk, A.; Pedersen, N.; Michaelsson, K. Seasonal genetic influence on serum 25-hydroxyvitamin D levels: A twin study. PLoS ONE 2009, 4, e7747. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Kiel, D.; Kraft, P. The genetics of vitamin D. Bone 2019, 126, 59–77. [Google Scholar] [CrossRef]
- Revez, J.; Lin, T.; Qiao, Z.; Xue, A.; Holtz, Y.; Zhu, Z.; Zeng, J.; Wang, H.; Sidorenko, J.; Kemper, K. Genome-wide association study identifies 143 loci associated with 25 hydroxyvitamin D concentration. Nat. Commun. 2020, 11, 1647. [Google Scholar] [CrossRef] [Green Version]
- Mills, N.; Wright, M.; Henders, A.; Eyles, D.; Baune, B.; McGrath, J.; Byrne, E.; Hansell, N.; Birosova, E.; Scott, J. Heritability of Transforming Growth Factor-beta1 and Tumor Necrosis Factor-Receptor Type 1 Expression and Vitamin D Levels in Healthy Adolescent Twins. Twin Res. Hum. Genet. 2015, 18, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; O’Reilly, P.; Aschard, H.; Hsu, Y.; Richards, J.; Dupuis, J.; Ingelsson, E.; Karasik, D.; Pilz, S.; Berry, D. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat. Commun. 2018, 9, 260. [Google Scholar] [CrossRef]
- Hunter, D. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005, 6, 287–298. [Google Scholar] [CrossRef]
- Khoury, M.; Wacholder, S. Invited commentary: From genome-wide association studies to gene-environment-wide interaction studies–challenges and opportunities. Am. J. Epidemiol. 2009, 169, 227–230. [Google Scholar] [CrossRef]
- Picot, J.; Hartwell, D.; Harris, P.; Mendes, D.; Clegg, A.; Takeda, A. The preferred reporting items for systematic reviews and meta-analyses checklist. In The Effectiveness of Interventions to Treat Severe Acute Malnutrition in Young Children: A Systematic Review; NIHR Journals Library: Southampton, UK, 2012. [Google Scholar]
- Covidence Systematic Review Software, Veritas Health Innovation, Melbourne, Australia. Available online: www.covidence.org (accessed on 5 May 2022).
- Wells, G.A.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Available online: https://www.ohri.ca//programs/clinical_epidemiology/oxford.Asp (accessed on 5 May 2022).
- Balduzzi, S.; Rücker, G.; Schwarzer, G. How to perform a meta-analysis with R: A practical tutorial. Evid.-Based Ment. Health 2019, 22, 153–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viechtbauer, W. Bias and Efficiency of Meta-Analytic Variance Estimators in the Random-Effects Model. J. Educ. Behav. Stat. 2005, 3, 261–293. [Google Scholar] [CrossRef]
- Robien, K.; Butler, L.; Wang, R.; Beckman, K.; Walek, D.; Koh, W.; Yuan, J. Genetic and environmental predictors of serum 25-hydroxyvitamin D concentrations among middle-aged and elderly Chinese in Singapore. Br. J. Nutr. 2013, 109, 493–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, B.; Jiang, S.; Muyiduli, X.; Wang, S.; Mo, M.; Li, M.; Wang, Z.; Yu, Y. Vitamin D pathway gene polymorphisms influenced vitamin D level among pregnant women. Clin. Nutr. 2018, 37, 2230–2237. [Google Scholar] [CrossRef]
- Engelman, C.; Meyers, K.; Iyengar, S.; Liu, Z.; Karki, C.; Igo, R., Jr.; Truitt, B.; Robinson, J.; Sarto, G.; Wallace, R. Vitamin D intake and season modify the effects of the GC and CYP2R1 genes on 25-hydroxyvitamin D concentrations. J. Nutr. 2013, 143, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Livingstone, K.; Celis-Morales, C.; Hoeller, U.; Lambrinou, C.; Moschonis, G.; Macready, A.; Fallaize, R.; Baur, M.; Roos, F.; Bendik, I. Weekday sunlight exposure, but not vitamin D intake, influences the association between vitamin D receptor genotype and circulating concentration 25-hydroxyvitamin D in a pan-European population: The Food4Me study. Mol. Nutr. Food Res. 2017, 61, 1600476. [Google Scholar] [CrossRef] [Green Version]
- Hatchell, K.; Lu, Q.; Mares, J.; Michos, E.; Wood, A.; Engelman, C. Multi-ethnic analysis shows genetic risk and environmental predictors interact to influence 25(OH)D concentration and optimal vitamin D intake. Genet. Epidemiol. 2020, 44, 208–217. [Google Scholar] [CrossRef]
- GeneCards®: The Human Gene Database. Available online: https://www.genecards.org/ (accessed on 5 May 2022).
- Machiela, M.; Chanock, S. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- Hatchell, K.; Lu, Q.; Hebbring, S.; Michos, E.; Wood, A.; Engelman, C. Ancestry-specific polygenic scores and SNP heritability of 25(OH)D in African- and European-ancestry populations. Hum. Genet. 2019, 138, 1155–1169. [Google Scholar] [CrossRef]
- Hong, J.; Hatchell, K.; Bradfield, J.; Bjonnes, A.; Chesi, A.; Lai, C.; Langefeld, C.; Lu, L.; Lu, Y.; Lutsey, P. Transethnic Evaluation Identifies Low-Frequency Loci Associated With 25-Hydroxyvitamin D Concentrations. J. Clin. Endocrinol. Metab. 2018, 103, 1380–1392. [Google Scholar] [CrossRef]
- Pearce, N. Epidemiology in a changing world: Variation, causation and ubiquitous risk factors. Int. J. Epidemiol. 2011, 40, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, F. The Role of Vitamin D and UVB on the Risk and Survival of Oesophageal and Gastric Cancer. Ph.D. Thesis, Trinity College Dublin, Dublin, Ireland, 2018. [Google Scholar]
- Bjork, A.; Andersson, A.; Johansson, G.; Bjorkegren, K.; Bardel, A.; Kristiansson, P. Evaluation of sun holiday, diet habits, origin and other factors as determinants of vitamin D status in Swedish primary health care patients: A cross-sectional study with regression analysis of ethnic Swedish and immigrant women. BMC Fam. Pract. 2013, 14, 129. [Google Scholar] [CrossRef]
- Brouwer-Brolsma, E.; Vaes, A.; Zwaluw, N.; Wijngaarden, J.; Swart, K.; Ham, A.; Dijk, S.; Enneman, A.; Sohl, E.; Schoor, N. Relative importance of summer sun exposure, vitamin D intake, and genes to vitamin D status in Dutch older adults: The B-PROOF study. J. Steroid Biochem. Mol. Biol. 2016, 164, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Dunn, E.; Uddin, M.; Subramanian, S.; Smoller, J.; Galea, S.; Koenen, K. Research review: Gene-environment interaction research in youth depression—A systematic review with recommendations for future research. J. Child Psychol. Psychiatry 2011, 52, 1223–1238. [Google Scholar] [CrossRef] [PubMed]
- Vandenbrouckel, J.P.; Elm, E.; Altman, D.G.; Gotzsche, P.C.; Mulrow, C.D.; Pocock, S.J.; Poole, C.; Schlesselman, J.J.; Egger, M. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and elaboration. PLoS Med. 2007, 4, 1628–1655. [Google Scholar]
- Little, J.; Higgins, J.P.; Ioannidis, J.P.; Moher, D.; Gagnon, F.; Elm, E.V.; Khoury, M.J. STrengthening the REporting of Genetic Association Studies (STREGA)—An extension of the STROBE statement. Genet. Epidemiol. Off. Publ. Int. Genet. Epidemiol. Soc. 2009, 33, 581–598. [Google Scholar]
- Kechichian, E.; Ezzedine, K. Vitamin D and the Skin: An Update for Dermatologists. Am. J. Clin. Dermatol. 2018, 19, 223–235. [Google Scholar] [CrossRef]
- Chang, S.; Lee, H. Vitamin D and health—The missing vitamin in humans. Pediatr. Neonatol. 2019, 60, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C. Nutrigenomics of Vitamin D. Nutrients 2019, 11, 676. [Google Scholar] [CrossRef] [Green Version]

| Study | Sample Characteristics (GxE) | Mean 25(OH)D Concentration | Gene (G) (Quality Control) | Environment (E) | Other Covariates | GxE Findings |
|---|---|---|---|---|---|---|
| Engelman 2013 [28] | 1204 postmenopausal European women age 50–79 recruited in the US 1993–1998 (sampled from CAREDS eye disease cohort) | Dec–May: 50.1 (SD 22.1), Jun–Nov: 63.3 (SD 22.7) nmol/L (chemiluminescence or radioimmunoassay) | SNPs in GC, DHCR7, CYP2R1, and CYP24A1 (HWE, MAF, call rate, heterozygosity, concordance rate) | season of blood draw: winter/spring (Dec–May) and summer/fall (June–Nov); individual sun exposure: weekly duration of total recreational physical activity and yard work; time spent in direct sunglight at baseline: a sunlight exposure questionnaire administered at baseline (2001–2004) | vitamin D intake, waist circumference, season of blood draw, total cholesterol, and hours in sunlight | SNP × season: interaction was significant for only one gene-environment pair (rs7041- season; p = 0.01), where the b-coefficient for the high-exposure group was much more than twice that in the low-exposure group (−0.33 vs. −0.02, respectively); interaction between genetic risk score and external source of vitamin D was significant for season of blood draw (p = 0.04) but not for vitamin D intake (p = 0.26) |
| Robien 2013 [26] | 504 government-built housing estate residents with Hokkien or Cantonese dialect age 55.7 (7.8), 56% F, recruited in Singapore 1993–1998 | 68.6 nmol/L (SD 18.3) (chemiluminescence immunoassay) | GC haplotype (HWE, MAF, call rate) | average number of hours spent sitting at work and hours spent doing vigorous work, taken as surrogates for time spent indoors and outdoors, respectively | Dialect group, education level, menopausal status (women), BMI, height, weight, body surface area, physical activity, smoking status, hours spent sitting at work, season of blood draw, use of cod liver oil supplements and dietary intake of vitamin D, Ca, fish, dairy products and alcohol | GC haplotype × hours spent sitting at work: p-interaction = 0.24 (not significant) |
| Livingstone 2017 [29] | 1312 healthy university students age 40.2 (13.0), 97% Caucasian, recruited in Ireland, the Netherlands, Greece, the UK, Poland, and Germany 2012–2013 | 60.6 nmol/L (SD 26.4) (chromatography) | SNPs from VDR, GC and PGS from the minor alleles of VDR and GC (HWE, LD) | Weekend and weekday sunlight exposure (during day light on a typical week day and on a weekend day during the sunny months of the year (i.e., April to September) collapsed into <20 min‚ 20 min–2 h, and >2 h (dietary intake of vitamin D: Online food frequency questionnaire (FFQ) of foods and supplements) | age, sex, BMI, ethnicity, country, season, vitamin D intake (food only) and vitamin D supplementation | SNP × sunlight exposure (and SNP × diet): The relationship between VDR rs2228570 genotype and 25(OH)D concentration was modulated by time spent in the sunlight during the week (p-interaction = 0.009). When total sunlight exposure (week- days plus weekend days) was considered, the interaction with VDR rs2228570 remained significant but evidence for the interaction was weaker (p = 0.045). No significant interactions were observed between genotype and dietary vitamin D |
| Shao 2018 [27] | 759 healthy pregnant Chinese women age 28 (3), recruited in China 2011–2014 (no history of chronic or acute disease or mental disorders) | 39 (SD 16.25) nmol/L (chromatography) | DHCR7, GC, CYP24A1, CYP27A1, CYP27B1, CYP2R1, CYP3A4, LRP2, NADSYN1, VDR (HWE, MAF, ) | season, merged into summer/fall (June-November) and winter/spring (December–May) | Age, pre-pregnancy BMI, sampling season, vitamin D supplements, physical activity | SNP × season: interactions were observed between season and CYP27A1 rs933994 (p = 0.02), CYP3A4 rs2246709 (p = 0.004); similar trends were also found in the logistic analysis of interactions between CYP27A1 rs933994 (p = 0.05), CYP3A4 rs2246709 (p = 0.03) and seasons on vitamin D deficiency (see [27] Table 8) |
| Hatchell 2020 [30] | 9688 European and African ancestry individuals age 45–84, 59% F, 11% African, recruited in the USA 1990–2002 (sampled from Atherosclerosis cohorts and population-based cohorts) | range of 18.9 to 30.1 ng/ml (see [30] Table 1) (chemiluminescence and chromatography) | PGS, (HWE, MAF, imputation quality score, sample and SNP call rate) | continuous UV radiation based on month of blood draw and location using UV data from the National Weather Service Climate Prediction Center historical database (range: 0.7–9.5 UV index units) | age, sex, BMI, cohort, vitamin D intake, and available UV radiation (physical activity where available) | PGS × season (and PGS × vitamin D intake): in European and PGS*UV model, beta (SE) = 0.017 (0.0073) (p-value <0.021) (see [30] Table 2). The 2-DF PGS*intake, 1-DF PGS*UV, and 2-DF PGS*UV results were statistically significant in participants of European ancestry p = , p = , and p = , respectively). No significant interactions in African ancestry sample (limited power). |
| Manousaki 2020 [12] | 193,809 white British individuals age 56.8 (8.0), 54.1% F recruited in the UK 2006–2010 (population-based cohort UKBB) | 70.0 (SD 34.7) (chemiluminescence) | 138 conditionally independent SNPs (HWE, MAF, imputation quality score) | season of measurement, winter (Jan–March), summer (July–Sept) | age, sex, season of measurement, and vitamin D supplementation (BMI excluded to avoid introducing collider bias) | SNP × season of measurement: significant interaction with season in 11 independent SNPs in the CYP2R1 locus on chromosome 11 and in a single variant in the SEC23A locus on chromosome 14 (all p < ), strongest interaction was found for rs117913124 in CYP2R1 (p -interaction ) |
| Revez 2020 [16] | 318,851 white British individuals age 40–69 recruited in the UK 2006–2010 (population-based cohort UKBB) | median, mean and interquartile range of 47.9, 49.6, 33.5–63.2 nmol/L (chemiluminescence) | 1127 genome-wide significant variants, (MAF, genome-wide significance) | season of blood draw, winter (Dec–April) and summer (June–Oct) | age, sex, (with and without BMI), genotyping batch, assessment centre, month of testing, supplement intake and thefirst four ancestry PCs | variant × season of blood draw: Of 6,098,063 variants tested (MAF > 0.05), 1127 had a GWS (p < ) interaction with season, and 1120 (99%) were also GWS in the vQTL analysis. Of the 20 vQTL loci without significant GxE with season, at least half showed no evidence at all for GxE with season, so these variants are candidates for GxE with other environmental factors |
| Study | Main Analysis | G in GxE | GxE Significance |
|---|---|---|---|
| Robien 2013 [26] | 55 SNPs in VDR, CYP2R1, CYP3A4, CYP27B1, CYP24A1, and GC | GC haplotype | p < 0.05 |
| Engelman 2013 [28] | 29 SNPs in GC, DHCR7, CYP2R1, and CYP24A1 | GC (rs4588, rs7401) and CYPR21 (rs2060793, rs10500804, rs11023380, rs11023374) | p < 0.05 |
| Livingstone 2017 [29] | 5 SNPs from VDR and GC | VDR (rs2228570) | p < 0.05 |
| Shao 2018 [27] | 51 SNPs in NADSYN1/DHCR7, GC, CYP3A4, CYP2R1, CYP27A1, CYP27B1, VDR, CYP24A1, and LRP2 | CYP27A1 (rs933994) and CYP3A4 (rs2246709) (not clear if any other snps were tested) | p < 0.05 |
| Hatchell 2020 [30] | PGS | PGS | p < 0.05 |
| Manousaki 2020 [12] | genome-wide (20,370,874 variants) | 138 conditionally independent lead SNPs | p < , Bonferroni-corrected threshold (0.05/number of SNPs) |
| Revez 2020 [16] | genome-wide (8,806,780 SNPs GWAS, MAF > 0.01) | 6,098,063 variants (MAF > 0.05) | p < , genome-wide significance |
| Vitamin D | Specify which vitamin D measure was used (e.g., 25(OH)D) and details of the measurement method. Include descriptive statistics of vitamin D levels in the sample. Report whether this outcome was defined as continuous or categorical (e.g., very deficient, deficient, adequate). Standardise the distribution to enable comparison across populations, which may differ significantly in mean or range of vitamin D. |
| Genetics | Report clearly on chosen genetic factor. Researchers are also encouraged to aim to replicate previous findings where possible. |
| Environment | Use independent UV radiation data from sources such as NASA or Google Earth alongside personal sun exposure habits. Quantitative sun exposure data allows comparison across studies. |
| Interaction | Report clearly on the model parameters and interaction term(s) as well as the effect estimates and statistical significance of G, E, and GxE. Include the reasoning for choosing the model and assumptions made. Report GxE results even if not significant. |
| Sample | Include descriptive statistics of the sample such as age and sex. While the field broadly would benefit from larger and more ethnically and geographically diverse samples, this may not be possible for individual studies. Where possible, researchers should consider sampling underrepresented populations to broaden ancestry coverage within vitamin D research. Report on the ethnicity and geography of the sampled population and any analysis of population structure. |
| Covariates | Evaluate known covariates associated with vitamin D—age, sex, and BMI. Consider other covariates such as season of blood draw, ethnicity, skin colour, and vitamin D supplement intake. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shraim, R.; MacDonnchadha, C.; Vrbanic, L.; McManus, R.; Zgaga, L. Gene-Environment Interactions in Vitamin D Status and Sun Exposure: A Systematic Review with Recommendations for Future Research. Nutrients 2022, 14, 2735. https://doi.org/10.3390/nu14132735
Shraim R, MacDonnchadha C, Vrbanic L, McManus R, Zgaga L. Gene-Environment Interactions in Vitamin D Status and Sun Exposure: A Systematic Review with Recommendations for Future Research. Nutrients. 2022; 14(13):2735. https://doi.org/10.3390/nu14132735
Chicago/Turabian StyleShraim, Rasha, Conor MacDonnchadha, Lauren Vrbanic, Ross McManus, and Lina Zgaga. 2022. "Gene-Environment Interactions in Vitamin D Status and Sun Exposure: A Systematic Review with Recommendations for Future Research" Nutrients 14, no. 13: 2735. https://doi.org/10.3390/nu14132735
APA StyleShraim, R., MacDonnchadha, C., Vrbanic, L., McManus, R., & Zgaga, L. (2022). Gene-Environment Interactions in Vitamin D Status and Sun Exposure: A Systematic Review with Recommendations for Future Research. Nutrients, 14(13), 2735. https://doi.org/10.3390/nu14132735







