The Modulatory Effect of Cyclocarya paliurus Flavonoids on Intestinal Microbiota and Hypothalamus Clock Genes in a Circadian Rhythm Disorder Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of CPFs
2.3. CPF Dose Determination in Animal Experiments
2.4. Animals and Experimental Design
2.5. Intestinal Microbiota Analysis
2.6. Fecal and Cecum Sample Metabolite Extraction
2.7. Untargeted Metabolomics Analyses
2.8. Single Cell Preparation and scRNA-seq
2.9. Bioinformatic Analysis of scRNA-seq Data
2.10. Statistical Analysis
3. Results
3.1. The Contents of CPF in the Extract
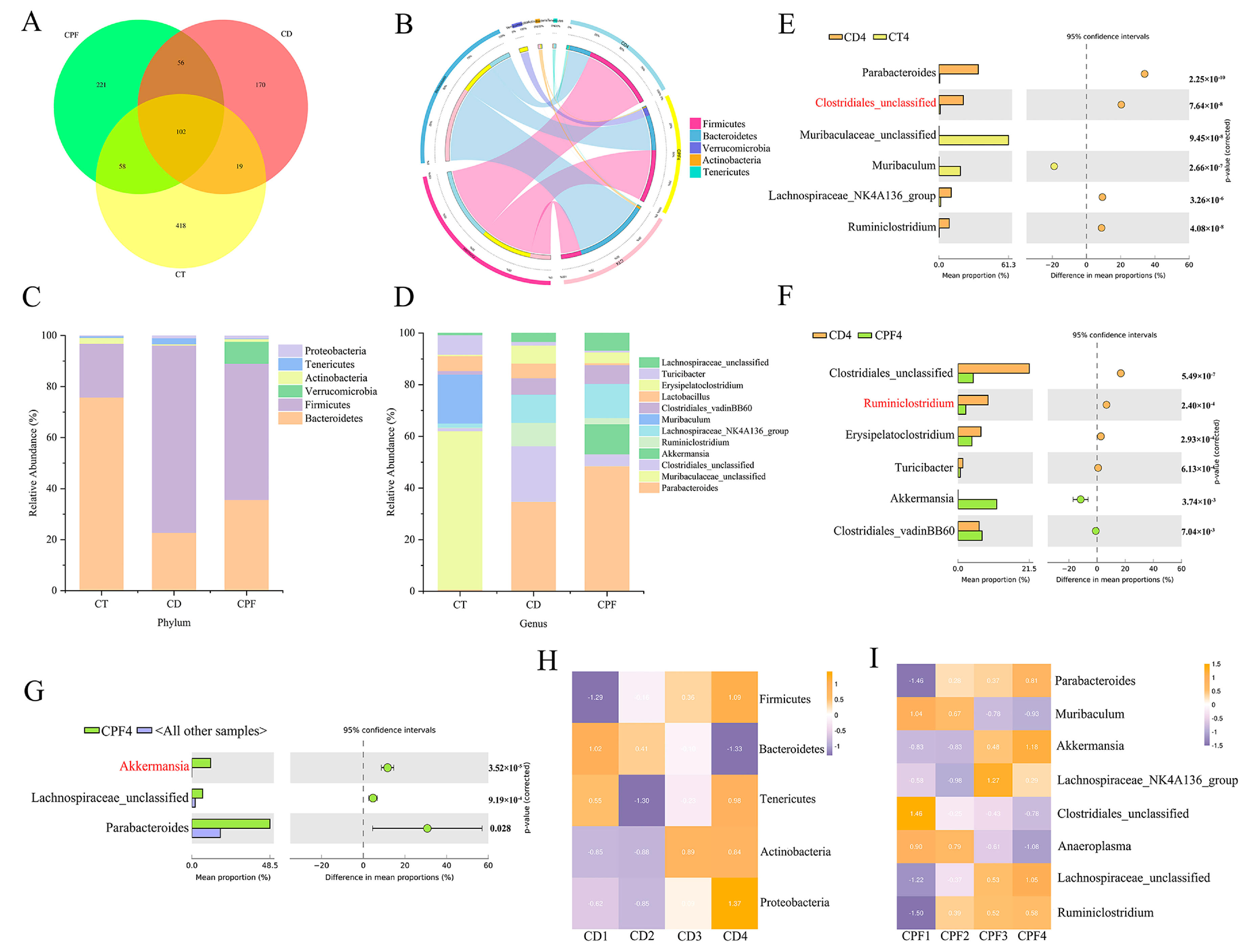
3.2. CPF Supplementation Improves the Perturbation of Gut Microbiota in Mice with Circadian Rhythm Disturbance
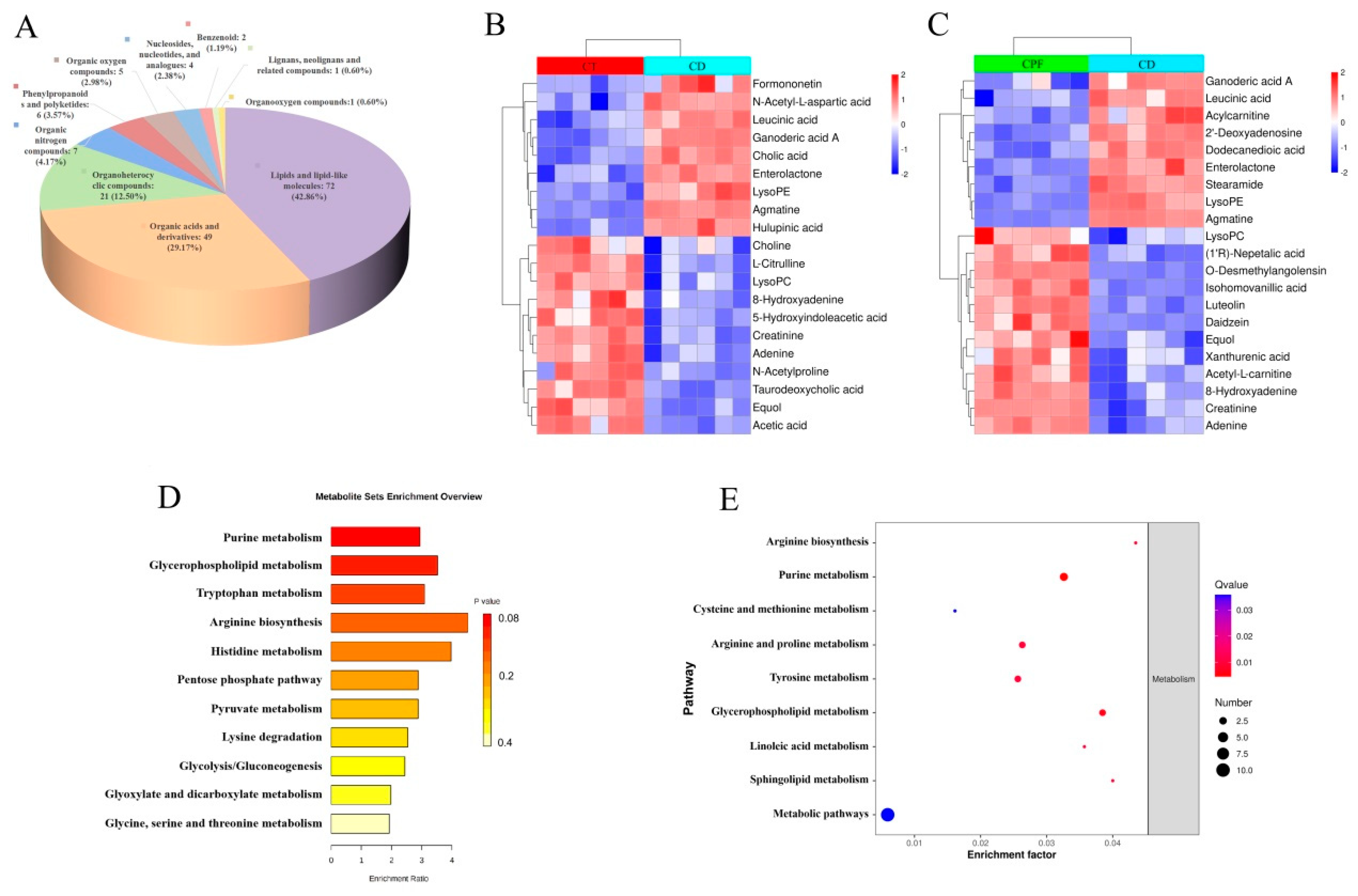
3.3. Characterization of Metabolic Differences
3.4. Effects of CPFs on Gut Metabolites in Mice with Circadian Rhythm Disturbance
3.5. Correlations between the Fecal Microbial Taxa and Intestinal Differential Metabolites
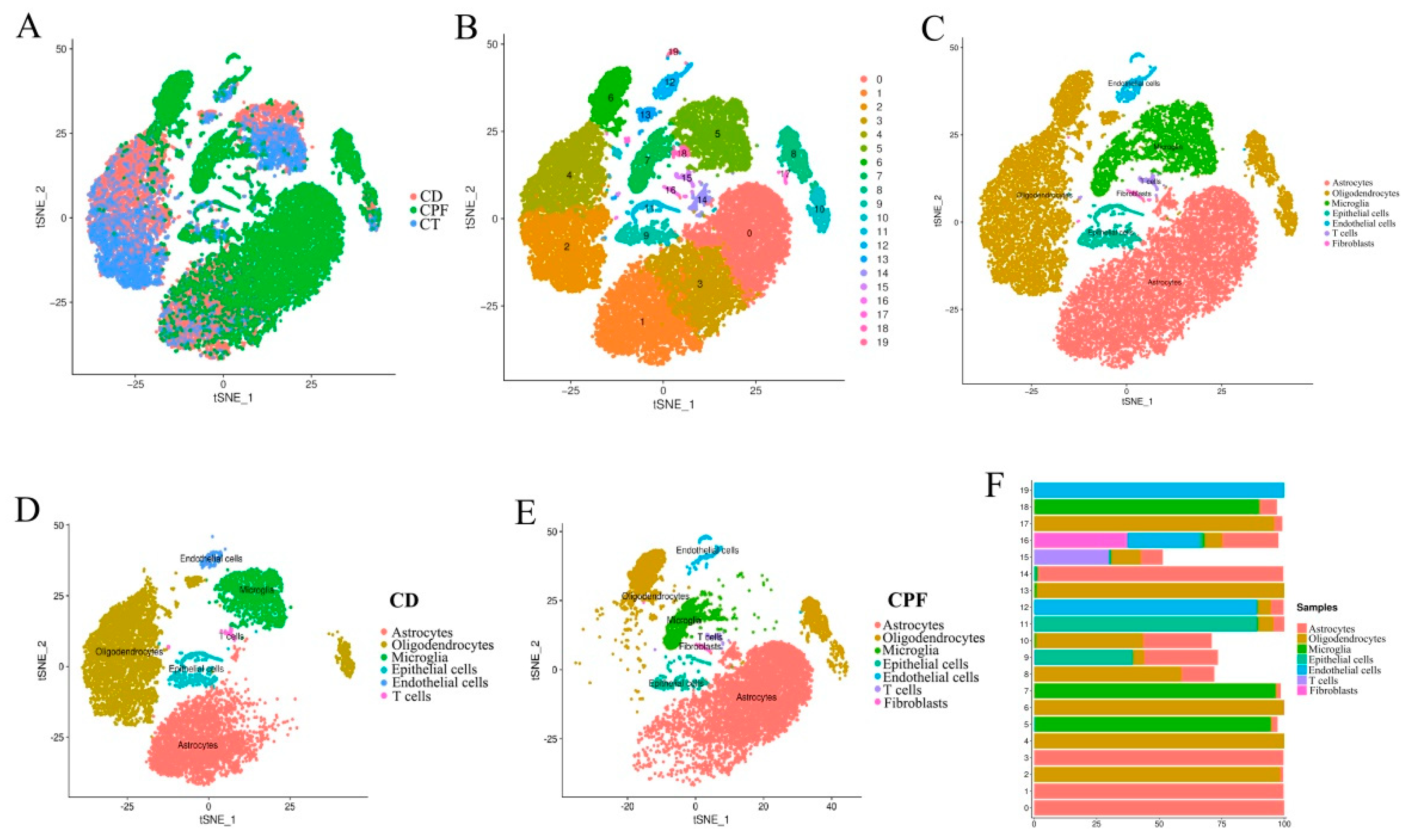
3.6. Single-Cell RNA-seq Identified Circadian-Rhythm-Associated Brain Cell Populations in the Hypothalamus of Mice
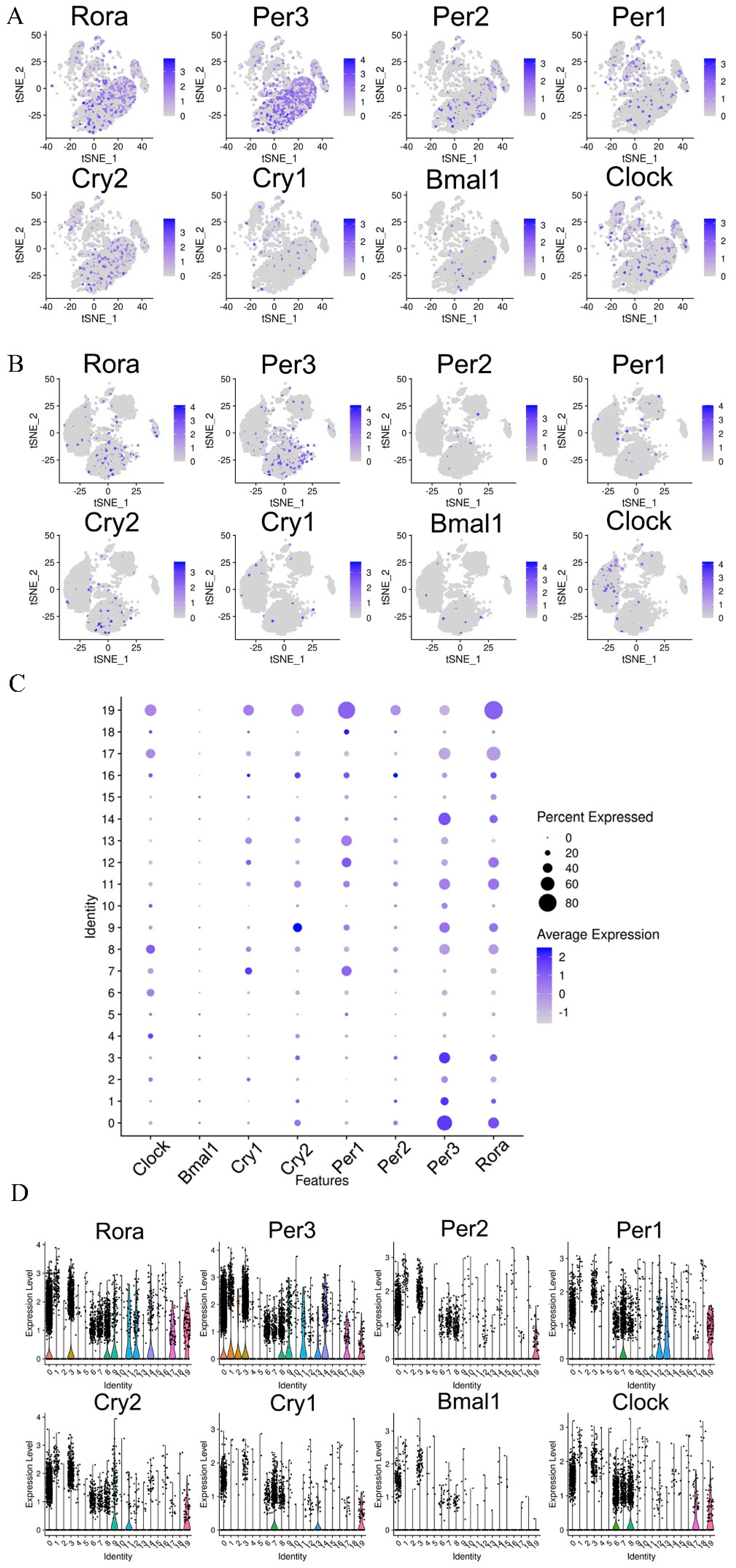
3.7. Effects of CPFs on the Expression of Circadian Clock Genes in the Hypothalamus
3.8. CPF Supplementation Regulated the Different Expression Genes (DEGs) Involved in Myelination and Neurodegenerative Diseases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Voigt, R.M.; Forsyth, C.B.; Green, S.J.; Engen, P.A.; Keshavarzian, A. Circadian rhythm and the gut microbiome. Int. Rev. Neurobiol. 2016, 131, 193–205. [Google Scholar] [PubMed]
- Man, A.W.C.; Li, H.; Xia, N. Circadian rhythm: Potential therapeutic target for atherosclerosis and thrombosis. Int. J. Mol. Sci. 2021, 22, 676. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Minguez, J.; Gomez-Abellan, P.; Garaulet, M. Circadian rhythms, food timing and obesity. Proc. Nutr. Soc. 2016, 75, 501–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkar, S.G.; Kalsbeek, A.; Cheeseman, J.F. Potential role for the gut microbiota in modulating host circadian rhythms and metabolic health. Microorganisms 2019, 7, 41. [Google Scholar] [CrossRef] [Green Version]
- Song, D.; Ho, C.T.; Zhang, X.; Wu, Z.; Cao, J. Modulatory effect of Cyclocarya paliurus flavonoids on the intestinal microbiota and liver clock genes of circadian rhythm disorder mice model. Food Res. Int. 2020, 138, 109769. [Google Scholar] [CrossRef]
- Matenchuk, B.A.; Mandhane, P.J.; Kozyrskyj, A.L. Sleep, circadian rhythm, and gut microbiota. Sleep Med. Rev. 2020, 53, 101340. [Google Scholar] [CrossRef]
- Sharma, A.; Sethi, G.; Tambuwala, M.M.; Aljabali, A.; Chellappan, D.K.; Dua, K.; Goyal, R. Circadian rhythm disruption and Alzheimer’s disease: The dynamics of a vicious cycle. Curr. Neuropharmacol. 2021, 19, 248–264. [Google Scholar] [CrossRef]
- Wulff, K.; Gatti, S.; Wettstein, J.G.; Foster, R.G. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 2010, 11, 589–599. [Google Scholar] [CrossRef]
- Guo, T.; Song, D.; Ho, C.T.; Zhang, X.; Zhang, C.; Cao, J.; Wu, Z. Omics analyses of gut microbiota in a circadian rhythm disorder mouse model fed with oolong tea polyphenols. J. Agric. Food Chem. 2019, 67, 8847–8854. [Google Scholar] [CrossRef]
- Hu, D.; Xie, Z.; Ye, Y.; Bahijri, S.; Chen, M. The beneficial effects of intermittent fasting: An update on mechanism, and the role of circadian rhythm and gut microbiota. Hepatobiliary Surg. Nutr. 2020, 9, 597–602. [Google Scholar] [CrossRef]
- Man, A.; Xia, N.; Daiber, A.; Li, H. The roles of gut microbiota and circadian rhythm in the cardiovascular protective effects of polyphenols. Br. J. Pharmacol. 2020, 177, 1278–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulet, O. Potential role of the intestinal microbiota in programming health and disease. Nutr. Rev. 2015, 73, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.; Rao, M.C.; Chang, E.B. Gut microbiota as a transducer of dietary cues to regulate host circadian rhythms and metabolism. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 679–689. [Google Scholar] [CrossRef]
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N.; et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015, 17, 681–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, T.; Ho, C.T.; Zhang, X.; Cao, J.; Wang, H.; Shao, X.; Pan, D.; Wu, Z. Oolong tea polyphenols ameliorate circadian rhythm of intestinal microbiome and liver clock genes in mouse model. J. Agric. Food Chem. 2019, 67, 11969–11976. [Google Scholar] [CrossRef]
- Li, Q.; Hu, J.; Xie, J.; Nie, S.; Xie, M.Y. Isolation, structure, and bioactivities of polysaccharides from Cyclocarya paliurus (Batal.) Iljinskaja. Ann. N. Y. Acad. Sci. 2017, 1398, 20–29. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, Y.; Jin, Q.; Zhang, D.; Gao, M.; Yao, N.; Yin, Z.; Zhang, J.; Ma, S. Cyclocarya paliurus triterpenoids improve diabetes-induced hepatic inflammation via the rho-kinase-dependent pathway. Front. Pharmacol. 2019, 10, 811. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, X.; Li, L.; Wang, H.; Chen, K.; Xu, M.; Wu, Y.; Huang, X.; Zhang, M.; Ye, X.; et al. Cyclocarya paliurus ethanol leaf extracts protect against diabetic cardiomyopathy in db/db mice via regulating PI3K/Akt/NF-kappaB signaling. Food Nutr. Res. 2020, 64. [Google Scholar] [CrossRef]
- Yao, Y.; Yan, L.; Chen, H.; Wu, N.; Wang, W.; Wang, D. Cyclocarya paliurus polysaccharides alleviate type 2 diabetic symptoms by modulating gut microbiota and short-chain fatty acids. Phytomedicine 2020, 77, 153268. [Google Scholar] [CrossRef]
- Sheng, X.; Chen, H.; Wang, J.; Zheng, Y.; Li, Y.; Jin, Z.; Li, J. Joint transcriptomic and metabolic analysis of flavonoids in Cyclocarya paliurus leaves. ACS Omega 2021, 6, 9028–9038. [Google Scholar] [CrossRef] [PubMed]
- Braune, A.; Blaut, M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 2016, 7, 216–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, L.; Chen, Y.; Zhang, X.; Zheng, X.; Cao, J.; Wu, Z.; Qin, W.; Cheng, K. A metagenomic analysis of the modulatory effect of Cyclocarya paliurus flavonoids on the intestinal microbiome in a high-fat diet-induced obesity mouse model. J. Sci. Food Agric. 2019, 99, 3967–3975. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome. Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Li, H.D.; Guo, L.; Wu, F.X.; Wang, J. Analysis of single-cell RNA-seq data by clustering approaches. Curr. Bioinform. 2019, 14, 314–322. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Kosonen, R.; Barua, S.; Kim, J.Y.; Lee, J.E. Role of agmatine in the application of neural progenitor cell in central nervous system diseases: Therapeutic potentials and effects. Anat. Cell Biol. 2021, 4, 143–151. [Google Scholar] [CrossRef]
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J.Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019, 570, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.F.; Herholz, K.; Rosa-Neto, P.; Pellerin, L.; Nordberg, A.; Zimmer, E.R. Astrocyte biomarkers in Alzheimer’s disease. Trends Mol. Med. 2019, 25, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Barateiro, A.; Brites, D.; Fernandes, A. Oligodendrocyte development and myelination in neurodevelopment: Molecular mechanisms in health and disease. Curr. Pharm. Des. 2016, 22, 656–679. [Google Scholar] [CrossRef] [PubMed]
- Kipp, M. Oligodendrocyte physiology and pathology function. Cells 2020, 9, 2078. [Google Scholar] [CrossRef]
- Subhramanyam, C.S.; Wang, C.; Hu, Q.; Dheen, S.T. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin. Cell Dev. Biol. 2019, 94, 112–120. [Google Scholar] [CrossRef]
- Walsh, J.T.; Watson, N.; Kipnis, J. T cells in the central nervous system: Messengers of destruction or purveyors of protection? Immunology 2014, 141, 340–344. [Google Scholar] [CrossRef] [Green Version]
- Haroutunian, V.; Katsel, P.; Roussos, P.; Davis, K.L.; Altshuler, L.L.; Bartzokis, G. Myelination, oligodendrocytes, and serious mental illness. Glia 2014, 62, 1856–1877. [Google Scholar] [CrossRef]
- Lake, E.M.R.; Steffler, E.A.; Rowley, C.D.; Sehmbi, M.; Minuzzi, L.; Frey, B.N.; Bock, N.A. Altered intracortical myelin staining in the dorsolateral prefrontal cortex in severe mental illness. Eur. Arch. Psychiatry Clin. Neurosci. 2017, 267, 369–376. [Google Scholar] [CrossRef]
- Bishehsari, F.; Voigt, R.M.; Keshavarzian, A. Circadian rhythms and the gut microbiota: From the metabolic syndrome to cancer. Nat. Rev. Endocrinol. 2020, 16, 731–739. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalova, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E.; et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 2016, 167, 1495–1510.e12. [Google Scholar] [CrossRef] [Green Version]
- Paulose, J.K.; Wright, J.M.; Patel, A.G.; Cassone, V.M. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS ONE 2016, 11, e0146643. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.F.; Wang, S.W.; Wang, X.X.; Weng, Y.Y.; Fan, X.Y.; Sheng, H.; Zhu, X.T.; Lou, L.J.; Zhang, F. The flavonoid-rich Quzhou Fructus Aurantii extract modulates gut microbiota and prevents obesity in high-fat diet-fed mice. Nutr. Diabetes 2019, 9, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yu, Y.; Lin, D.; Zheng, P.; Zhang, P.; Hu, M.; Wang, Q.; Pan, W.; Yang, X.; Hu, T.; et al. β-glucan attenuates cognitive impairment via the gut-brain axis in diet-induced obese mice. Microbiome 2020, 8, 143. [Google Scholar] [CrossRef]
- Zhai, Q.; Feng, S.; Arjan, N.; Chen, W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2019, 59, 3227–3236. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef] [Green Version]
- Heng, Y.; Kim, M.J.; Yang, H.J.; Kang, S.; Park, S. Lactobacillus intestinalis efficiently produces equol from daidzein and chungkookjang, short-term fermented soybeans. Arch Microbiol. 2019, 201, 1009–1017. [Google Scholar] [CrossRef]
- Mayo, B.; Vazquez, L.; Florez, A.B. Equol: A bacterial metabolite from the daidzein isoflavone and its presumed beneficial health effects. Nutrients 2019, 11, 2231. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.L.; Park, H.Y.; Vattem, D.A.; Grammas, P.; Ma, H.; Seeram, N.P. Equol, a blood-brain barrier permeable gut microbial metabolite of dietary isoflavone daidzein, exhibits neuroprotective effects against neurotoxins induced toxicity in human neuroblastoma SH-SY5Y cells and caenorhabditis elegans. Plant Foods Hum. Nutr. 2020, 75, 512–517. [Google Scholar] [CrossRef]
- Subedi, L.; Ji, E.; Shin, D.; Jin, J.; Yeo, J.H.; Kim, S.Y. Equol, a dietary daidzein gut metabolite attenuates microglial activation and potentiatesn europrotection in vitro. Nutrients 2017, 9, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Fontana, B.; Morales-Santana, S.; Diaz Navarro, C.; Rozas-Moreno, P.; Genilloud, O.; Vicente Perez, F.; Perez del Palacio, J.; Munoz-Torres, M. Metabolomic profile related to cardiovascular disease in patients with type 2 diabetes mellitus: A pilot study. Talanta 2016, 148, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Yea, K.; Kim, J.; Yoon, J.H.; Kwon, T.; Kim, J.H.; Lee, B.D.; Lee, H.J.; Lee, S.J.; Kim, J.I.; Lee, T.G.; et al. Lysophosphatidylcholine activates adipocyte glucose uptake and lowers blood glucose levels in murine models of diabetes. J. Biol. Chem. 2009, 284, 33833–33840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugasini, D.; Thomas, R.; Yalagala, P.C.R.; Tai, L.M.; Subbaiah, P.V. Dietary docosahexaenoic acid (DHA) as lysophosphatidylcholine, but not as free acid, enriches brain DHA and improves memory in adult mice. Sci. Rep. 2017, 7, 11263. [Google Scholar] [CrossRef] [PubMed]
- Kazak, F.; Yarim, G.F. Neuroprotective effects of acetyl-l-carnitine on lipopolysaccharide-induced neuroinflammation in mice: Involvement of brain-derived neurotrophic factor. Neurosci. Lett. 2017, 658, 32–36. [Google Scholar] [CrossRef]
- Garwood, C.J.; Ratcliffe, L.E.; Simpson, J.E.; Heath, P.R.; Ince, P.G.; Wharton, S.B. Review: Astrocytes in Alzheimer’s disease and other age-associated dementias: A supporting player with a central role. Neuropathol. Appl. Neurobiol. 2017, 43, 281–298. [Google Scholar] [CrossRef]
- Halassa, M.M.; Florian, C.; Fellin, T.; Munoz, J.R.; Lee, S.Y.; Abel, T.; Haydon, P.G.; Frank, M.G. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron 2009, 61, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Yeung, M.S.Y.; Djelloul, M.; Steiner, E.; Bernard, S.; Salehpour, M.; Possnert, G.; Brundin, L.; Frisen, J. Dynamics of oligodendrocyte generation in multiple sclerosis. Nature 2019, 566, 538–542. [Google Scholar] [CrossRef]
- Gokbuget, D.; Pereira, J.A.; Bachofner, S.; Marchais, A.; Ciaudo, C.; Stoffel, M.; Schulte, J.H.; Suter, U. The Lin28/let-7 axis is critical for myelination in the peripheral nervous system. Nat. Commun. 2015, 6, 8584. [Google Scholar] [CrossRef] [Green Version]
- Vogt, N. Magnificent myelin. Nat. Methods 2014, 11, 606–607. [Google Scholar] [CrossRef]


| Groups | OTUs | Chao 1 | Shannon | Simpson |
|---|---|---|---|---|
| CT | 368.67 ± 20.87 c | 369.38 ± 31.24 c | 5.50 ± 0.14 b | 0.90 ± 0.02 a |
| CD | 236.33 ± 18.55 a | 236.33 ± 18.55 a | 4.87 ± 0.07 a | 0.96 ± 0.01 b |
| CPF | 278.02 ± 15.72 b | 278.56 ± 15.64 b | 5.48 ± 0.11 b | 0.91 ± 0.02 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Ho, C.-T.; Liu, Y.; Zhan, S.; Wu, Z.; Zheng, X.; Zhang, X. The Modulatory Effect of Cyclocarya paliurus Flavonoids on Intestinal Microbiota and Hypothalamus Clock Genes in a Circadian Rhythm Disorder Mouse Model. Nutrients 2022, 14, 2308. https://doi.org/10.3390/nu14112308
Sun Y, Ho C-T, Liu Y, Zhan S, Wu Z, Zheng X, Zhang X. The Modulatory Effect of Cyclocarya paliurus Flavonoids on Intestinal Microbiota and Hypothalamus Clock Genes in a Circadian Rhythm Disorder Mouse Model. Nutrients. 2022; 14(11):2308. https://doi.org/10.3390/nu14112308
Chicago/Turabian StyleSun, Ying, Chi-Tang Ho, Yanan Liu, Shennan Zhan, Zufang Wu, Xiaojie Zheng, and Xin Zhang. 2022. "The Modulatory Effect of Cyclocarya paliurus Flavonoids on Intestinal Microbiota and Hypothalamus Clock Genes in a Circadian Rhythm Disorder Mouse Model" Nutrients 14, no. 11: 2308. https://doi.org/10.3390/nu14112308
APA StyleSun, Y., Ho, C.-T., Liu, Y., Zhan, S., Wu, Z., Zheng, X., & Zhang, X. (2022). The Modulatory Effect of Cyclocarya paliurus Flavonoids on Intestinal Microbiota and Hypothalamus Clock Genes in a Circadian Rhythm Disorder Mouse Model. Nutrients, 14(11), 2308. https://doi.org/10.3390/nu14112308







