Metagenomic Analysis Reveals a Mitigating Role for Lactobacillus paracasei and Bifidobacterium animalis in Experimental Periodontitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Groups
2.2. Bacterial Strains
2.3. Experimental Design
2.4. Clinical Indicators and Histological Assessment
2.5. Alveolar Bone Loss Assessment by Micro-CT Analysis
2.6. Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis
2.7. Enzyme-Linked Immunosorbent Assay (ELISA) of Serum
2.8. Western Immunoblotting (WB) Analysis
2.9. DNA Extraction, Library Construction, Metagenomic Sequencing, and Gene Taxonomic and Functional Annotation
2.10. Statistical Analysis
3. Results
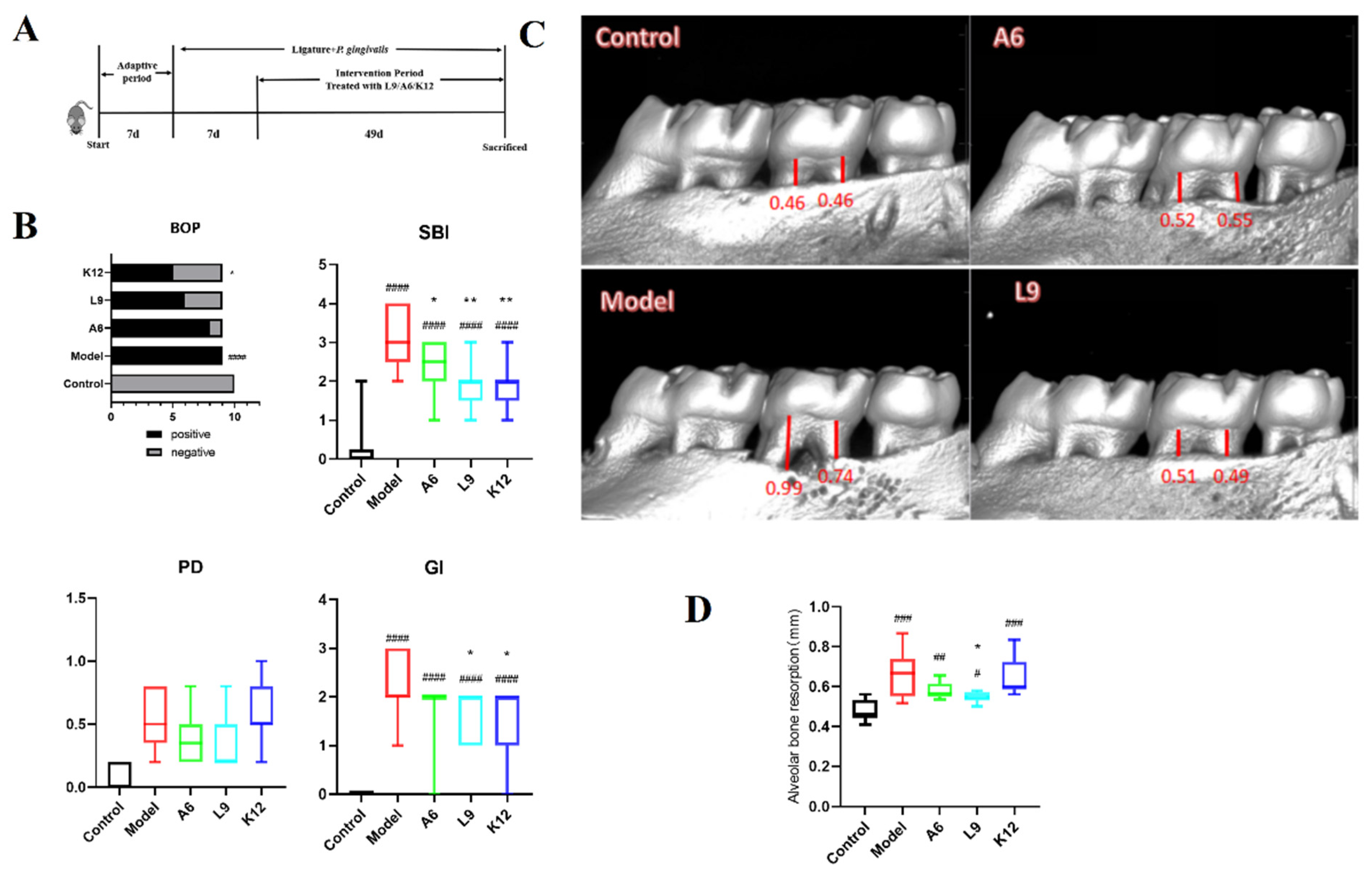
3.1. Clinical Index Score and Alveolar Bone Aspiration
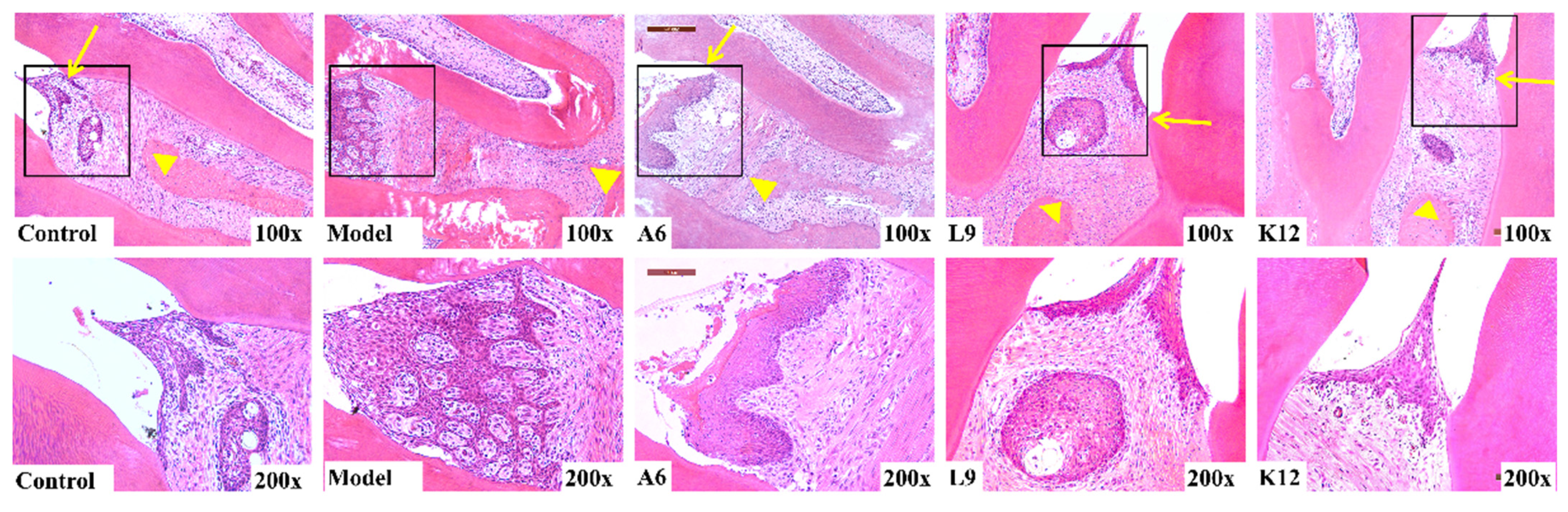
3.2. Histopathological Observations
3.3. Probiotics Downregulate Inflammatory Cytokines in the Serum
3.4. Probiotics Modulate the Expression of Pro- and Anti-Inflammatory Cytokines in Gingival Tissues
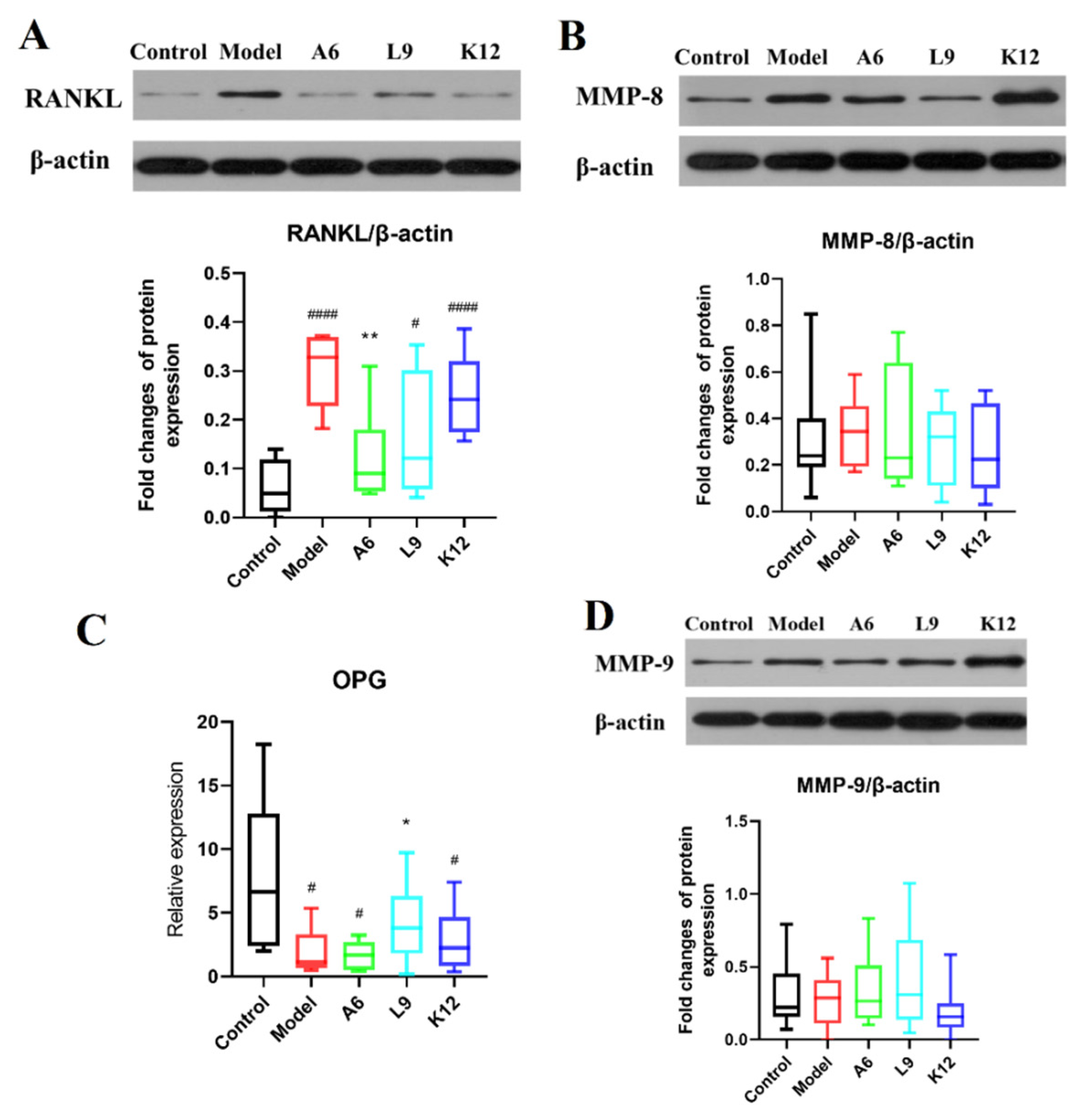
3.5. Effects of Probiotics on MMP-8 and MMP-9 Protein Expression Levels in Gingival Tissues
3.6. Effects of Probiotics on Expression Levels of RANKL/OPG in Gingival Tissues
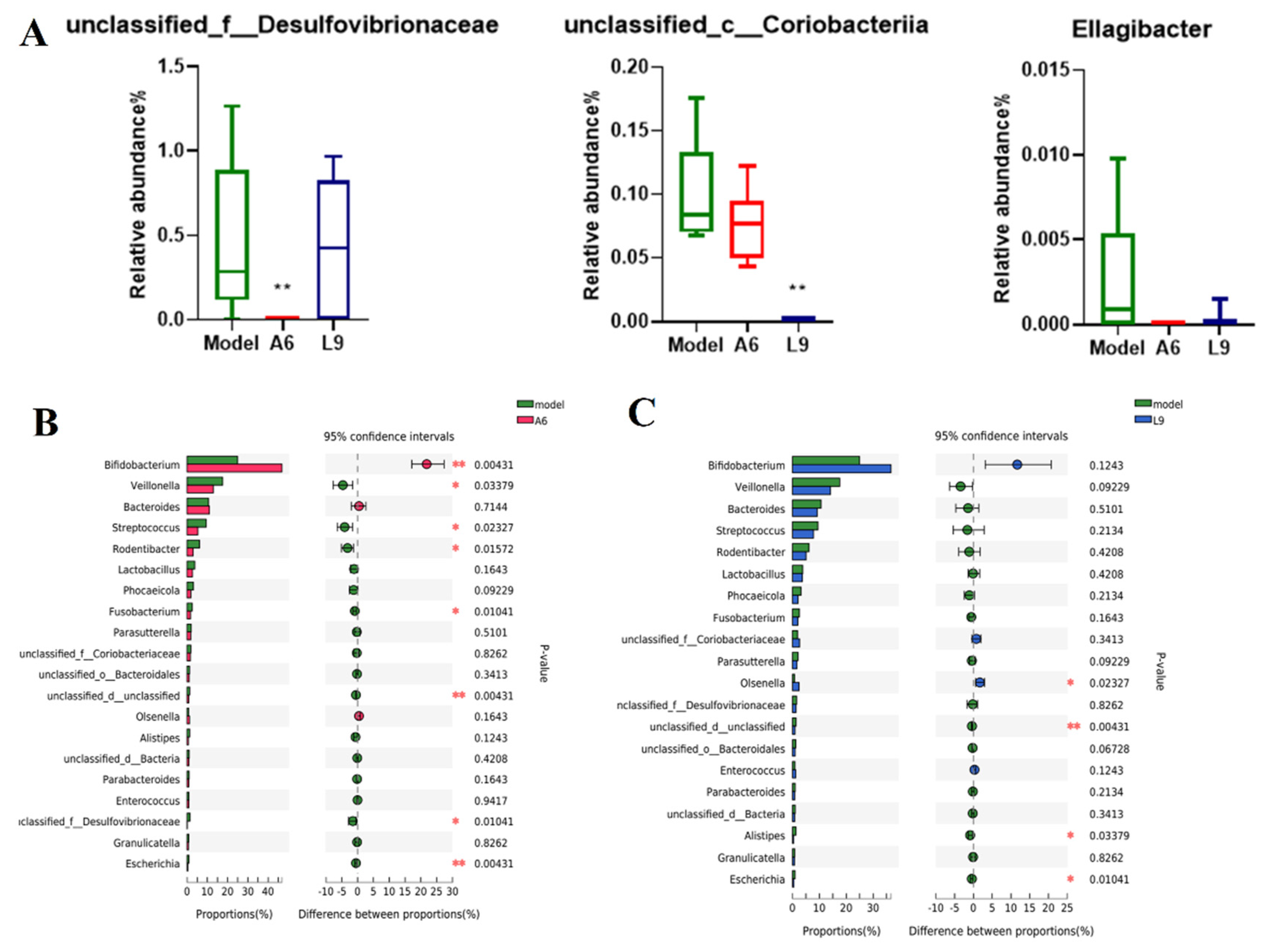
3.7. Probiotics Alter the Taxonomic Composition of the Subgingival Microbiome
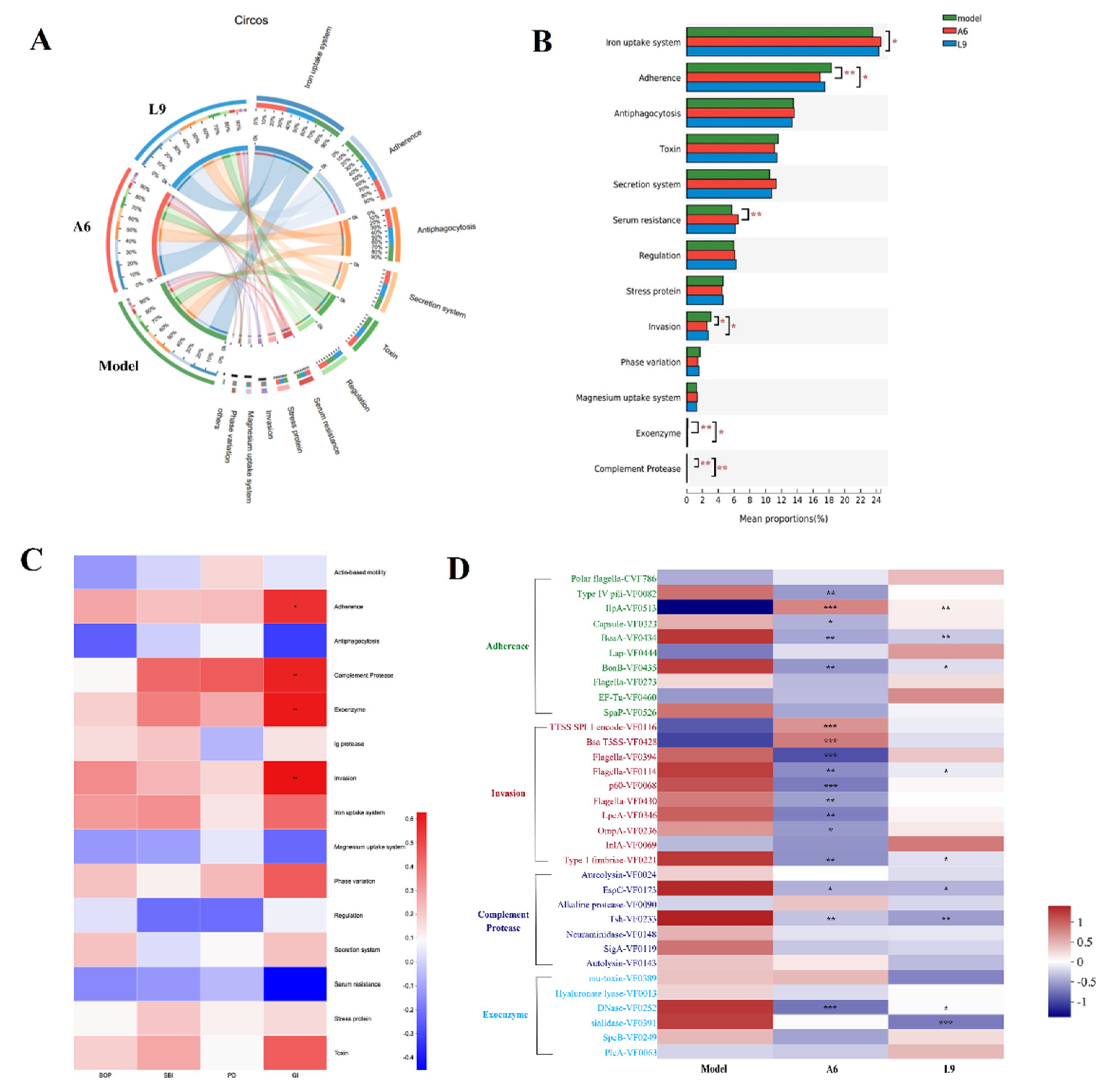
3.8. Differences in Virulence Factor Expression after Probiotic Intervention
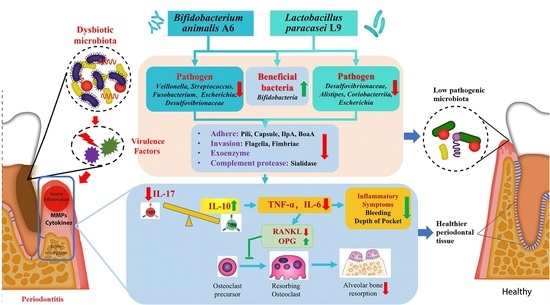
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xiao, L.; Zhang, L.; Deepal, S.; Ye, G.; Zhang, X. Epidemic trend of periodontal disease in elderly Chinese population, 1987–2015: A systematic review and meta-analysis. Sci. Rep. 2017, 7, 45000. [Google Scholar] [CrossRef] [PubMed]
- Richards, D. Review finds that severe periodontitis affects 11% of the world population. Evid.-Based Dent. 2014, 15, 70–71. [Google Scholar] [CrossRef] [PubMed]
- Frencken, J.E.; Sharma, P.; Stenhouse, L.; Green, D.; Laverty, D.; Dietrich, T. Global epidemiology of dental caries and severe periodontitis—A comprehensive review. J. Clin. Periodontol. 2017, 44, S94–S105. [Google Scholar] [CrossRef]
- Schwendicke, F.; Krois, J.; Engel, A.S.; Seidel, M.; Graetz, C. Long-term periodontitis treatment costs according to the 2018 classification of periodontal diseases. J. Dent. 2020, 99, 103417. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G. Probiotics and periodontal health. J. Med. Life 2011, 4, 387–394. [Google Scholar]
- Kar, K.; Simonian, K.; Nowzari, H. Dynamic therapeutic approach for individuals affected with aggressive periodontitis. J. Calif. Dent. Assoc. 2011, 39, 401–415. [Google Scholar]
- Ansiliero, R.; Gelinski, J.; Samistraro, Q.L.; Baratto, C.M.; Almeida, C.A.; Locatelli, C. Pathogenic microbial profile and antibiotic resistance associated with periodontitis. Indian J. Microbiol. 2021, 61, 55–65. [Google Scholar] [CrossRef]
- How, K.Y.; Song, K.P.; Chan, K.G. Porphyromonas gingivalis: An overview of periodontopathic pathogen below the gum line. Front. Microbiol. 2016, 7, 53. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- He, X.S.; Shi, W.Y. Oral microbiology: Past, present and future. Int. J. Oral. Sci. 2009, 1, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, V.H.; Bandara, H.M.; Ishikawa, K.H.; Mayer, M.P.; Samaranayake, L.P. The role of probiotic bacteria in managing periodontal disease: A systematic review. Expert Rev. Anti-Infect. Ther. 2016, 14, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.; Maeda, K.; Hidaka, K.; Nemoto, K.; Hirose, Y.; Deguchi, S. Daily intake of heat-killed Lactobacillus plantarum L-137 decreases the probing depth in patients undergoing supportive periodontal therapy. Oral Health Prev. Dent. 2016, 14, 207–214. [Google Scholar] [PubMed]
- Ikram, S.; Hassan, N.; Baig, S.; Borges, K.J.J.; Raffat, M.A.; Akram, Z. Effect of local probiotic (Lactobacillus reuteri) vs systemic antibiotic therapy as an adjunct to non-surgical periodontal treatment in chronic periodontitis. J. Investig. Clin. Dent. 2019, 10, e12393. [Google Scholar] [CrossRef]
- Invernici, M.M.; Salvador, S.L.; Silva, P.H.F.; Soares, M.S.M.; Casarin, R.; Palioto, D.B.; Souza, S.L.S.; Taba, M., Jr.; Novaes, A.B., Jr.; Furlaneto, F.A.C.; et al. Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: A randomized clinical trial. J. Clin. Periodontol. 2018, 45, 1198–1210. [Google Scholar] [CrossRef]
- Mayanagi, G.; Kimura, M.; Nakaya, S.; Hirata, H.; Sakamoto, M.; Benno, Y.; Shimauchi, H. Probiotic effects of orally administered Lactobacillus salivarius WB21-containing tablets on periodontopathic bacteria: A double-blinded, placebo-controlled, randomized clinical trial. J. Clin. Periodontol. 2009, 36, 506–513. [Google Scholar] [CrossRef]
- Han, Y.T.; Wu, Y.T.; Li, R.J.; Zhao, L.; Zhang, M. Inhibition of Lactobacillus Paracasei L9 on adhesion of oral pathogens. Food Res. Dev. 2019, 40, 15–23. [Google Scholar]
- Sun, E.N.; Zhao, L.; Ren, F.Z.; Liu, S.L.; Zhang, M.; Guo, H.Y. Complete genome sequence of Bifidobacterium animalis subsp. lactis A6, a probiotic strain with high acid resistance ability. J. Biotechnol. 2015, 200, 8–9. [Google Scholar] [CrossRef]
- Huo, Y.X.; Lu, X.H.; Wang, X.Y.; Wang, X.F.; Chen, L.L.; Guo, H.Y.; Zhang, M.; Li, Y.X. Bifidobacterium animalis subsp. lactis A6 alleviates obesity associated with promoting mitochondrial biogenesis and function of adipose tissue in Mice. Molecules 2020, 25, 1490. [Google Scholar] [CrossRef]
- Garcia, V.G.; Knoll, L.R.; Longo, M.; Novaes, V.C.N.; Assem, N.Z.; Ervolino, E.; de Toledo, B.E.C.; Theodoro, L.H. Effect of the probiotic Saccharomyces cerevisiae on ligature-induced periodontitis in rats. J. Periodontal Res. 2016, 51, 26–37. [Google Scholar] [CrossRef]
- Marchesan, J.; Girnary, M.S.; Jing, L.; Miao, M.Z.; Zhang, S.P.; Sun, L.; Morelli, T.; Schoenfisch, M.H.; Inohara, N.; Offenbacher, S.; et al. An experimental murine model to study periodontitis. Nat. Protoc. 2018, 13, 2247–2267. [Google Scholar] [CrossRef] [PubMed]
- Messora, M.R.; Pereira, L.J.; Foureaux, R.; Oliveira, L.F.; Sordi, C.G.; Alves, A.J.; Napimoga, M.H.; Nagata, M.J.; Ervolino, E.; Furlaneto, F.A. Favourable effects of Bacillus subtilis and Bacillus licheniformis on experimental periodontitis in rats. Arch. Oral Biol. 2016, 66, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Aspalli, S.; Shetty, V.S.; Devarathnamma, M.V.; Nagappa, G.; Archana, D.; Parab, P. Evaluation of antiplaque and antigingivitis effect of herbal mouthwash in treatment of plaque induced gingivitis: A randomized, clinical trial. J. Indian Soc. Periodontol. 2014, 18, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Mazza, J.E.; Newman, M.G.; Sims, T.N. Clinical and antimicrobial effect of stannous fluoride on periodontitis. J. Clin. Periodontol. 1981, 8, 203–212. [Google Scholar] [CrossRef]
- Huang, S.G.; Huang, Q.L.; Huang, B.; Lu, F.L. The effect of Scutellaria baicalensis Georgi on immune response in mouse model of experimental periodontitis. J. Dent. Sci. 2013, 8, 405–411. [Google Scholar] [CrossRef][Green Version]
- Zheng, X.; Tizzano, M.; Redding, K.; He, J.; Peng, X.; Jiang, P.; Xu, X.; Zhou, X.; Margolskee, R.F. Gingival solitary chemosensory cells are immune sentinels for periodontitis. Nat. Commun. 2019, 10, 4496. [Google Scholar] [CrossRef]
- Araujo, A.A.; Pereira, A.; Medeiros, C.; Brito, G.A.C.; Leitao, R.F.C.; Araujo, L.S.; Guedes, P.M.M.; Hiyari, S.; Pirih, F.Q.; Araujo Junior, R.F. Effects of metformin on inflammation, oxidative stress, and bone loss in a rat model of periodontitis. PLoS ONE 2017, 12, e0183506. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, D.H.; Liu, C.M.; Luo, R.B.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.; Zhou, H.W. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef]
- Liu, G.J.; Luan, Q.X.; Chen, F.; Chen, Z.B.; Zhang, Q.; Yu, X.Q. Shift in the subgingival microbiome following scaling and root planning in generalized aggressive periodontitis. J. Clin. Periodontol. 2018, 45, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C. The non-canonical NF-kappa B pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Sanchavanakit, N.; Saengtong, W.; Manokawinchoke, J.; Pavasant, P. TNF-alpha stimulates MMP-3 production via PGE(2) signalling through the NF-kB and p38 MAPK pathway in a murine cementoblast cell line. Arch. Oral Biol. 2015, 60, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, J.; Chao, J. Porphyromonas gingivalis promotes monocyte migration by activating MMP-9. J. Periodontal Res. 2012, 47, 236–242. [Google Scholar] [CrossRef]
- Chen, W.; Gao, B.; Hao, L.; Zhu, G.; Jules, J.; MacDougall, M.J.; Wang, J.; Han, X.; Zhou, X.; Li, Y.P. The silencing of cathepsin K used in gene therapy for periodontal disease reveals the role of cathepsin K in chronic infection and inflammation. J. Periodontal Res. 2016, 51, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Casarin, R.C.; Casati, M.Z.; Pimentel, S.P.; Cirano, F.R.; Algayer, M.; Pires, P.R.; Ghiraldini, B.; Duarte, P.M.; Ribeiro, F.V. Resveratrol improves bone repair by modulation of bone morphogenetic proteins and osteopontin gene expression in rats. Int. J. Oral Maxillofac. Surg. 2014, 43, 900–906. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Gruner, D.; Paris, S.; Schwendicke, F. Probiotics for managing caries and periodontitis: Systematic review and meta-analysis. J. Dent. 2016, 48, 16–25. [Google Scholar] [CrossRef]
- Chen, Y.T.; Hsieh, P.S.; Ho, H.H.; Hsieh, S.H.; Kuo, Y.W.; Yang, S.F.; Lin, C.W. Antibacterial activity of viable and heat-killed probiotic strains against oral pathogens. Lett. Appl. Microbiol. 2020, 70, 310–317. [Google Scholar] [CrossRef]
- Jasberg, H.; Soderling, E.; Endo, A.; Beighton, D.; Haukioja, A. Bifidobacteria inhibit the growth of Porphyromonas gingivalis but not of Streptococcus mutans in an in vitro biofilm model. Eur. J. Oral Sci. 2016, 124, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.F.F.; Salvador, S.L.; Silva, P.H.F.; Furlaneto, F.A.C.; Figueiredo, L.; Casarin, R.; Ervolino, E.; Palioto, D.B.; Souza, S.L.S.; Taba, M.; et al. Benefits of Bifidobacterium animalis subsp lactis Probiotic in Experimental Periodontitis. J. Periodontol. 2017, 88, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, S.; Kolenbrander, P.E. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J. Bacteriol. 2010, 192, 2965–2972. [Google Scholar] [CrossRef] [PubMed]
- Silva-Boghossian, C.M.; Neves, A.B.; Resende, F.A.R.; Colombo, A.P.V. Suppuration-associated bacteria in patients with chronic and aggressive periodontitis. J. Periodontol. 2013, 84, E9–E16. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qi, J.; Zhao, H.; He, S.; Zhang, Y.; Wei, S.; Zhao, F. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci. Rep. 2013, 3, 1843. [Google Scholar] [CrossRef]
- Yamanaka, T.; Sumita-Sasazaki, Y.; Sugimori, C.; Matsumoto-Mashimo, C.; Yamane, K.; Mizukawa, K.; Yoshida, M.; Hayashi, H.; Nambu, T.; Leung, K.P.; et al. Biofilm-like structures and pathogenicity of Escherichia hermannii YS-11, a clinical isolate from a persistent apical periodontitis lesion. Fems Immunol. Med. Mic. 2010, 59, 456–465. [Google Scholar] [CrossRef]
- Ng, E.; Tay, J.R.H.; Balan, P.; Ong, M.M.A.; Bostanci, N.; Belibasakis, G.N.; Seneviratne, C.J. Metagenomic sequencing provides new insights into the subgingival bacteriome and aetiopathology of periodontitis. J. Periodontal Res. 2021, 56, 205–218. [Google Scholar] [CrossRef]
- Kushkevych, I.; Coufalova, M.; Vitezova, M.; Rittmann, S.K.M.R. Sulfate-reducing bacteria of the oral cavity and their relation with periodontitis-Recent advances. J. Clin. Med. 2020, 9, 2347. [Google Scholar] [CrossRef]
- Pirih, F.Q.; Monajemzadeh, S.; Singh, N.; Sinacola, R.S.; Shin, J.M.; Chen, T.; Fenno, J.C.; Kamarajan, P.; Rickard, A.H.; Travan, S.; et al. Association between metabolic syndrome and periodontitis: The role of lipids, inflammatory cytokines, altered host response, and the microbiome. Periodontol. 2000 2021, 87, 50–75. [Google Scholar] [CrossRef]
- Inaba, H.; Nomura, R.; Kato, Y.; Takeuchi, H.; Amano, A.; Asai, F.; Nakano, K.; Lamont, R.J.; Matsumoto-Nakano, M. Adhesion and invasion of gingival epithelial cells by Porphyromonas gulae. PLoS ONE 2019, 14, e0213309. [Google Scholar] [CrossRef] [PubMed]
- Lux, R.; Shi, W.Y. Chemotaxis-guided movements in bacteria. Crit. Rev. Oral Biol. Med. 2004, 15, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Altabtbaei, K.; Maney, P.; Ganesan, S.M.; Dabdoub, S.M.; Nagaraja, H.N.; Kumar, P.S. Anna Karenina and the subgingival microbiome associated with periodontitis. Microbiome 2021, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Merz, A.J.; So, M.; Sheetz, M.P. Pilus retraction powers bacterial twitching motility. Nature 2000, 407, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhou, W.; Wang, H.; Liang, S. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv. Protein Chem. Struct. Biol. 2020, 120, 45–84. [Google Scholar]
- Roy, S.; Douglas, C.W.I.; Stafford, G.P. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythia. J. Bacteriol. 2010, 192, 2285–2293. [Google Scholar] [CrossRef]
- Vimr, E.R. Microbial sialidases: Does bigger always mean better? Trends Microbiol. 1994, 2, 271–277. [Google Scholar] [CrossRef]
- Cantley, M.D.; Bartold, P.M.; Marino, V.; Reid, R.C.; Fairlie, D.P.; Wyszynski, R.N.; Zilm, P.S.; Haynes, D.R. The use of live-animal micro-computed tomography to determine the effect of a novel phospholipase A2 inhibitor on alveolar bone loss in an in vivo mouse model of periodontitis. J. Periodontal Res. 2009, 44, 317–322. [Google Scholar] [CrossRef]
- Cantley, M.D.; Haynes, D.R.; Marino, V.; Bartold, P.M. Pre-existing periodontitis exacerbates experimental arthritis in a mouse model. J. Clin. Periodontol. 2011, 38, 532–541. [Google Scholar] [CrossRef]
- Benatti, B.B.; Campos-Junior, J.C.; Silva-Filho, V.J.; Alves, P.M.; Rodrigues, I.R.; Uber-Bucek, E.; Vieira, S.M.; Napimoga, M.H. Effects of a Mikania laevigata extract on bone resorption and RANKL expression during experimental periodontitis in rats. J. Appl. Oral Sci. 2012, 20, 340–346. [Google Scholar] [CrossRef][Green Version]
- Bostanci, N.; Abe, T.; Belibasakis, G.N.; Hajishengallis, G. TREM-1 is upregulated in experimental periodontitis, and its blockade inhibits IL-17A and RANKL expression and suppresses Bone loss. J. Clin. Med. 2019, 8, 1579. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, T.; Hajishengallis, G. Topical treatment with probiotic Lactobacillus brevis CD2 inhibits experimental periodontal inflammation and bone loss. J. Periodontal Res. 2014, 49, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Correa, J.D.; Pereira, D.S.; Madeira, M.F.M.; Queiroz, C.M.; Souza, D.G.; Teixeira, M.M.; Costa, J.E.; Teixeira, A.L.; da Silva, T.A. Brain-derived neurotrophic factor in chronic periodontitis. Mediat. Inflamm. 2014, 2014, 373765. [Google Scholar] [CrossRef] [PubMed]
- Koide, M.; Kobayashi, Y.; Ninomiya, T.; Nakamura, M.; Yasuda, H.; Arai, Y.; Okahashi, N.; Yoshinari, N.; Takahashi, N.; Udagawa, N. Osteoprotegerin-deficient male mice as a model for severe alveolar bone loss: Comparison with RANKL-overexpressing transgenic male mice. Endocrinology 2013, 154, 773–782. [Google Scholar] [CrossRef]
- Liu, W.X.; Li, Z.J.; Niu, X.L.; Yao, Z.; Deng, W.M. The role of T Helper 17 cells and other IL-17-producing cells in bone resorption and remodeling. Int. Rev. Immunol. 2015, 34, 332–347. [Google Scholar] [CrossRef]


| Primer | Primer sequences(5′to3′) |
|---|---|
| TNF-α F′ | TGAACTTCGGGGTGATCGGT |
| TNF-α R′ | GGCTACGGGCTTGTCACTCG |
| IL10 F′ | GCAGGACTTTAAGGGTTACTTGG |
| IL10 R′ | TGCTCCACTGCCTTGCTTTT |
| IL6 F′ | GATTGTATGAACAGCGATGATGC |
| IL6 R′ | AGAAACGGAACTCCAGAAGACC |
| OPG F′ | AGACGAGATTGAGAGAACGAGAA |
| OPG R′ | ACGGTTTTGGGAAAGTGGTAT |
| GAPDH F′ | TTCCTACCCCCAATGTATCCG |
| GAPDH R′ | CCACCCTGTTGCTGTAGCCATA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; Fang, B.; Wuri, G.; Zhao, L.; Liu, F.; Zhang, M. Metagenomic Analysis Reveals a Mitigating Role for Lactobacillus paracasei and Bifidobacterium animalis in Experimental Periodontitis. Nutrients 2022, 14, 2125. https://doi.org/10.3390/nu14102125
Wu F, Fang B, Wuri G, Zhao L, Liu F, Zhang M. Metagenomic Analysis Reveals a Mitigating Role for Lactobacillus paracasei and Bifidobacterium animalis in Experimental Periodontitis. Nutrients. 2022; 14(10):2125. https://doi.org/10.3390/nu14102125
Chicago/Turabian StyleWu, Fang, Bing Fang, Guna Wuri, Liang Zhao, Fudong Liu, and Ming Zhang. 2022. "Metagenomic Analysis Reveals a Mitigating Role for Lactobacillus paracasei and Bifidobacterium animalis in Experimental Periodontitis" Nutrients 14, no. 10: 2125. https://doi.org/10.3390/nu14102125
APA StyleWu, F., Fang, B., Wuri, G., Zhao, L., Liu, F., & Zhang, M. (2022). Metagenomic Analysis Reveals a Mitigating Role for Lactobacillus paracasei and Bifidobacterium animalis in Experimental Periodontitis. Nutrients, 14(10), 2125. https://doi.org/10.3390/nu14102125