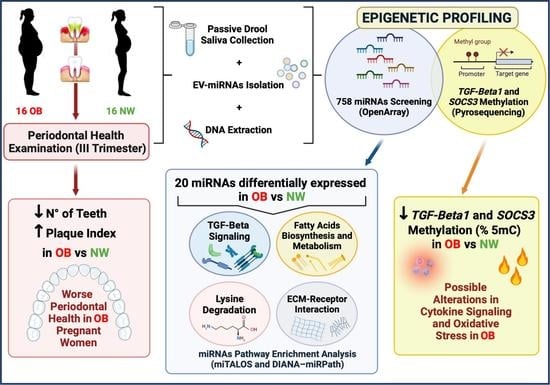
Epigenetic Profiling in the Saliva of Obese Pregnant Women
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Clinical Data and Biological Samples Collection
2.3. Isolation of Extracellular Vesicles and miRNA
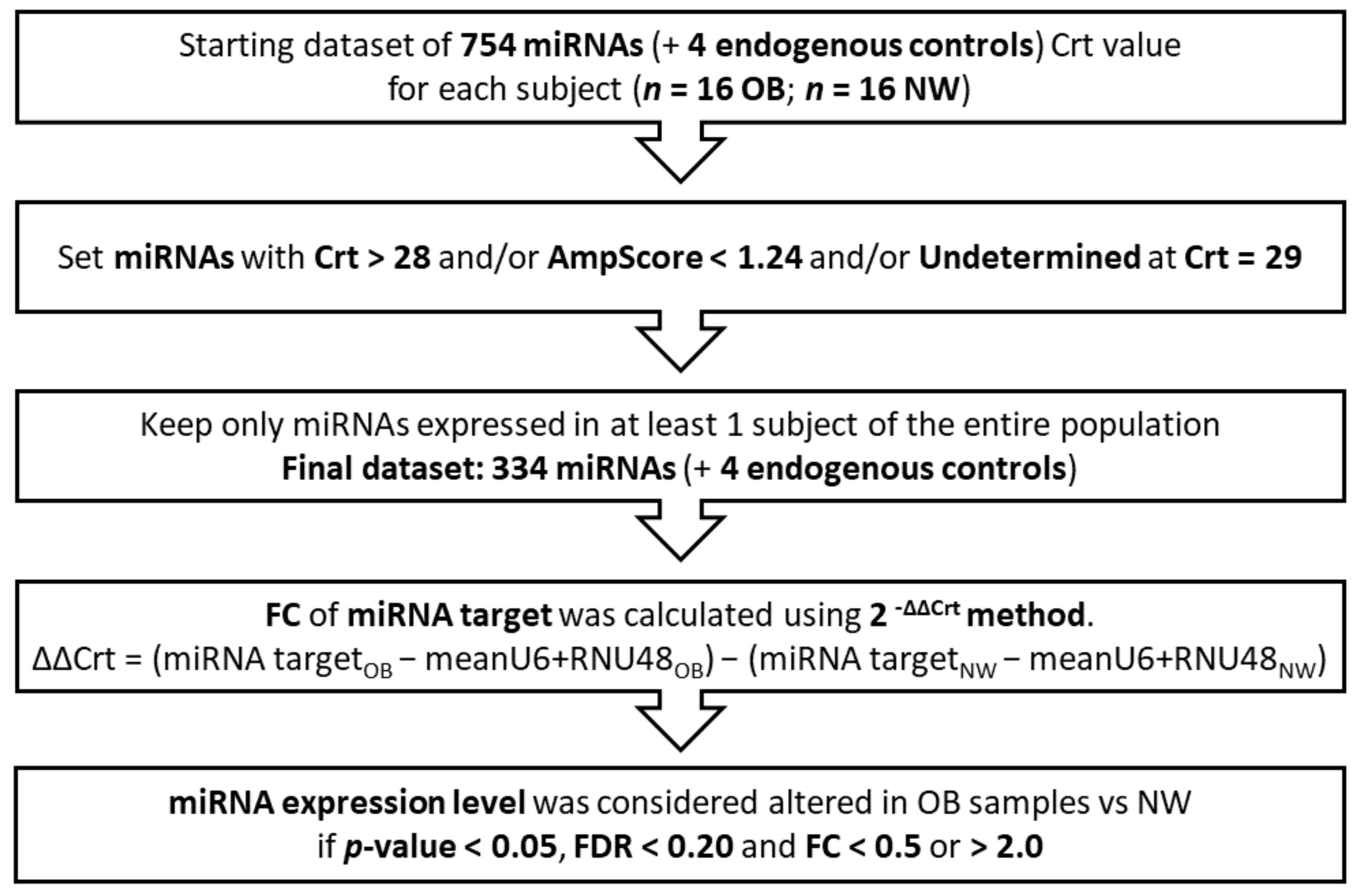
2.4. Screening of miRNA Expression
2.5. DNA Isolation
2.6. Bisulfite Conversion and DNA Methylation
2.7. Statistical Analysis and Prediction Tools
3. Results
3.1. Maternal and Oral Health Characteristics and Delivery Data
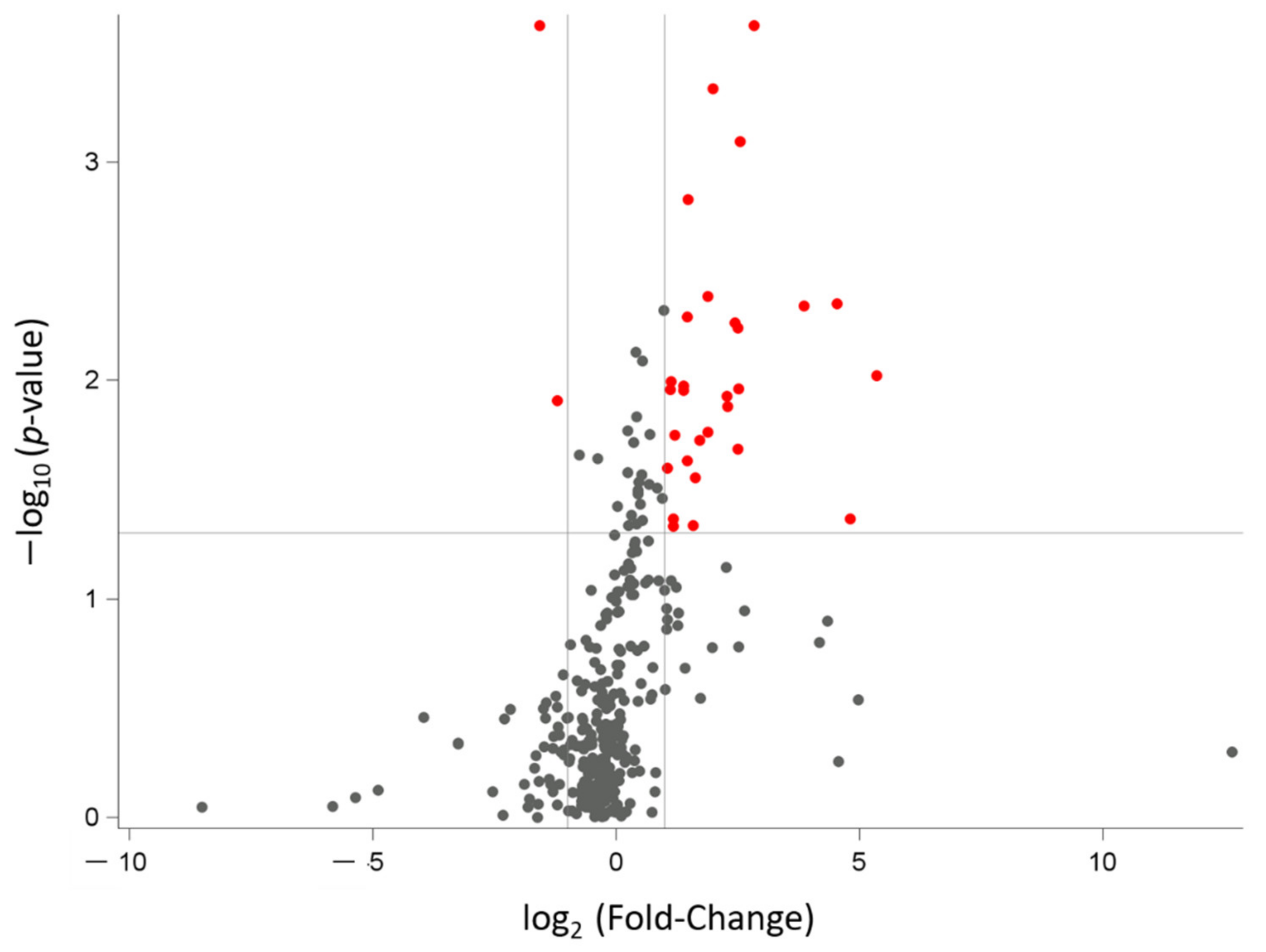
3.2. miRNAs’ Profile in Maternal Saliva
3.3. DNA Methylation in Maternal Saliva: TGF-Beta1 and SOCS3
4. Discussion
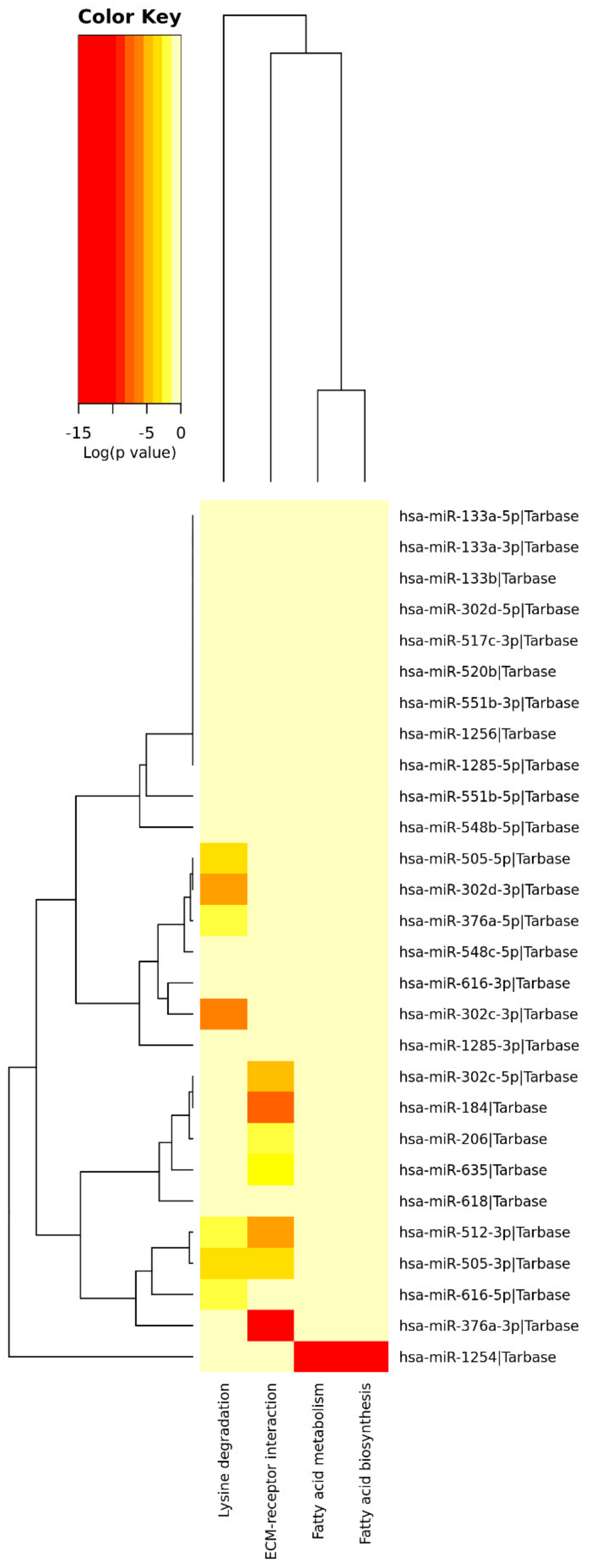
4.1. miRNAs’ Profile
4.1.1. Fatty Acids Biosynthesis and Metabolism
4.1.2. Extracellular Matrix
4.1.3. Lysine Degradation
4.2. DNA Methylation
4.2.1. TGF-Beta1
4.2.2. SOCS3
4.3. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- NCD Risk Factor Collaboration (NCD-RisC). Trends in adult body-mass index in 200 countries from 1975 to 2014: A pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet 2016, 387, 1377–1396. [Google Scholar] [CrossRef] [Green Version]
- Schaefer-Graf, U.; Napoli, A.; Nolan, C.J. Diabetic Pregnancy Study Group. Diabetes in pregnancy: A new decade of challenges ahead. Diabetologia 2018, 61, 1012–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalano, P.M.; Shankar, K. Obesity and pregnancy: Mechanisms of short term and long term adverse consequences for mother and child. BMJ 2017, 356, j1. [Google Scholar] [CrossRef]
- Poston, L.; Caleyachetty, R.; Cnattingius, S.; Corvalán, C.; Uauy, R.; Herring, S.; Gillman, M.W. Preconceptional and maternal obesity: Epidemiology and health consequences. Lancet Diabetes Endocrinol. 2016, 4, 1025–1036. [Google Scholar] [CrossRef]
- Bordin, P.; Dotto, L.; Battistella, L.; Rosso, E.; Pecci, L.; Valent, F.; Collarile, P.; Vanin, M. Gestational diabetes mellitus yesterday, today and tomorrow: A 13 year italian cohort study. Diabetes Res. Clin. Pract. 2020, 167, 108360. [Google Scholar] [CrossRef]
- Monteiro, L.J.; Norman, J.E.; Rice, G.E.; Illanes, S.E. Fetal programming and gestational diabetes mellitus. Placenta 2016, 48 (Suppl. 1), S54–S60. [Google Scholar] [CrossRef] [Green Version]
- Barker, D.J.; Thornburg, K.L.; Osmond, C.; Kajantie, E.; Eriksson, J.G. Beyond birthweight: The maternal and placental origins of chronic disease. J. Dev. Orig. Health Dis. 2010, 1, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Parisi, F.; Milazzo, R.; Savasi, V.M.; Cetin, I. Maternal Low-Grade Chronic Inflammation and Intrauterine Programming of Health and Disease. Int. J. Mol. Sci. 2021, 22, 1732. [Google Scholar] [CrossRef]
- Bianchi, C.; Taricco, E.; Cardellicchio, M.; Mandò, C.; Massari, M.; Savasi, V.; Cetin, I. The role of obesity and gestational diabetes on placental size and fetal oxygenation. Placenta 2021, 103, 59–63. [Google Scholar] [CrossRef]
- Zavatta, A.; Parisi, F.; Mandò, C.; Scaccabarozzi, C.; Savasi, V.M.; Cetin, I. Role of Inflammaging on the Reproductive Function and Pregnancy. Clin. Rev. Allergy Immunol. 2022, 1–16. [Google Scholar] [CrossRef]
- Kelly, A.C.; Powell, T.L.; Jansson, T. Placental function in maternal obesity. Clin. Sci. 2020, 134, 961–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martino, J.; Sebert, S.; Segura, M.T.; García-Valdés, L.; Florido, J.; Padilla, M.C.; Marcos, A.; Rueda, R.; McArdle, H.J.; Budge, H.; et al. Maternal Body Weight and Gestational Diabetes Differentially Influence Placental and Pregnancy Outcomes. J. Clin. Endocrinol. Metab. 2016, 101, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Mandò, C.; Anelli, G.M.; Novielli, C.; Panina-Bordignon, P.; Massari, M.; Mazzocco, M.I.; Cetin, I. Impact of Obesity and Hyperglycemia on Placental Mitochondria. Oxid. Med. Cell. Longev. 2018, 2018, 2378189. [Google Scholar] [CrossRef]
- Diceglie, C.; Anelli, G.M.; Martelli, C.; Serati, A.; Lo Dico, A.; Lisso, F.; Parisi, F.; Novielli, C.; Paleari, R.; Cetin, I.; et al. Placental Antioxidant Defenses and Autophagy-Related Genes in Maternal Obesity and Gestational Diabetes Mellitus. Nutrients 2021, 13, 1303. [Google Scholar] [CrossRef] [PubMed]
- Fattuoni, C.; Mandò, C.; Palmas, F.; Anelli, G.M.; Novielli, C.; Parejo Laudicina, E.; Savasi, V.M.; Barberini, L.; Dessì, A.; Pintus, R.; et al. Preliminary metabolomics analysis of placenta in maternal obesity. Placenta 2018, 61, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Foratori-Junior, G.A.; da Silva, B.M.; da Silva Pinto, A.C.; Honório, H.M.; Groppo, F.C.; de Carvalho Sales-Peres, S.H. Systemic and periodontal conditions of overweight/obese patients during pregnancy and after delivery: A prospective cohort. Clin. Oral Investig. 2020, 24, 157–165. [Google Scholar] [CrossRef]
- Ye, C.; Kapila, Y. Oral microbiome shifts during pregnancy and adverse pregnancy outcomes: Hormonal and Immunologic changes at play. Periodontol. 2000 2021, 87, 276–281. [Google Scholar] [CrossRef]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef]
- Byun, J.S.; Lee, H.Y.; Tian, J.; Moon, J.S.; Choi, J.; Lee, S.H.; Kim, Y.G.; Yi, H.S. Effect of Salivary Exosomal miR-25-3p on Periodontitis with Insulin Resistance. Front. Immunol. 2022, 12, 775046. [Google Scholar] [CrossRef]
- Ren, H.; Du, M. Role of Maternal Periodontitis in Preterm Birth. Front. Immunol. 2017, 8, 139. [Google Scholar] [CrossRef] [Green Version]
- Cetin, I.; Pileri, P.; Villa, A.; Calabrese, S.; Ottolenghi, L.; Abati, S. Pathogenic mechanisms linking periodontal diseases with adverse pregnancy outcomes. Reprod. Sci. 2012, 19, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, S.K.; Parry, S. Periodontal disease and pregnancy outcomes: Time to move on? J. Womens Health 2012, 21, 121–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zambon, M.; Mandò, C.; Lissoni, A.; Anelli, G.M.; Novielli, C.; Cardellicchio, M.; Leone, R.; Monari, M.N.; Massari, M.; Cetin, I.; et al. Inflammatory and Oxidative Responses in Pregnancies with Obesity and Periodontal Disease. Reprod. Sci. 2018, 25, 1474–1484. [Google Scholar] [CrossRef] [PubMed]
- Kaczor-Urbanowicz, K.E.; Martin Carreras-Presas, C.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T. Saliva diagnostics—Current views and directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Tsamou, M.; Martens, D.S.; Winckelmans, E.; Madhloum, N.; Cox, B.; Gyselaers, W.; Nawrot, T.S.; Vrijens, K. Mother’s Pre-pregnancy BMI and Placental Candidate miRNAs: Findings from the ENVIRONAGE Birth Cohort. Sci. Rep. 2017, 7, 5548, Erratum in Sci. Rep. 2018, 8, 6063. [Google Scholar] [CrossRef]
- Al-Rawi, N.H.; Al-Marzooq, F.; Al-Nuaimi, A.S.; Hachim, M.Y.; Hamoudi, R. Salivary microRNA 155; 146a/b and 203: A pilot study for potentially non-invasive diagnostic biomarkers of periodontitis and diabetes mellitus. PLoS ONE 2020, 15, e0237004. [Google Scholar] [CrossRef]
- Pedroso, J.A.B.; Ramos-Lobo, A.M.; Donato, J., Jr. SOCS3 as a future target to treat metabolic disorders. Hormones 2019, 18, 127–136. [Google Scholar] [CrossRef]
- Yener, S.; Demir, T.; Akinci, B.; Bayraktar, F.; Kebapcilar, L.; Ozcan, M.A.; Biberoglu, S.; Yesil, S. Transforming growth factor-beta 1 levels in women with prior history of gestational diabetes mellitus. Diabetes Res. Clin. Pract. 2007, 76, 193–198. [Google Scholar] [CrossRef]
- Page, R.C.; Eke, P.I. Case definitions for use in population-based surveillance of periodontitis. J. Periodontol. 2007, 78 (Suppl. 7), 1387–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pergoli, L.; Cantone, L.; Favero, C.; Angelici, L.; Iodice, S.; Pinatel, E.; Hoxha, M.; Dioni, L.; Letizia, M.; Albetti, B.; et al. Extracellular vesicle-packaged miRNA release after short-term exposure to particulate matter is associated with increased coagulation. Part. Fibre Toxicol. 2017, 14, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Bollati, V.; Baccarelli, A.; Hou, L.; Bonzini, M.; Fustinoni, S.; Cavallo, D.; Byun, H.M.; Jiang, J.; Marinelli, B.; Pesatori, A.C.; et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007, 67, 876–880. [Google Scholar] [CrossRef] [Green Version]
- Tarantini, L.; Bonzini, M.; Tripodi, A.; Angelici, L.; Nordio, F.; Cantone, L.; Apostoli, P.; Bertazzi, P.A.; Baccarelli, A.A. Blood hypomethylation of inflammatory genes mediates the effects of metal-rich airborne pollutants on blood coagulation. Occup. Environ. Med. 2013, 70, 418–425. [Google Scholar] [CrossRef] [Green Version]
- Preusse, M.; Theis, F.J.; Mueller, N.S. miTALOS v2: Analyzing Tissue Specific microRNA Function. PLoS ONE 2016, 11, e0151771. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Kostoulas, N.; Vergoulis, T.; Georgakilas, G.; Reczko, M.; Maragkakis, M.; Paraskevopoulou, M.D.; Prionidis, K.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA miRPath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012, 40, W498–W504. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Hosack, D.A.; Dennis, G., Jr.; Sherman, B.T.; Lane, H.C.; Lempicki, R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003, 4, R70. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Iorgulescu, G. Saliva between normal and pathological. Important factors in determining systemic and oral health. J. Med. Life 2009, 2, 303–307. [Google Scholar] [PubMed]
- Bertino, E.; Spada, E.; Occhi, L.; Coscia, A.; Giuliani, F.; Gagliardi, L.; Gilli, G.; Bona, G.; Fabris, C.; De Curtis, M.; et al. Neonatal anthropometric charts: The Italian neonatal study compared with other European studies. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 353–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saben, J.; Lindsey, F.; Zhong, Y.; Thakali, K.; Badger, T.M.; Andres, A.; Gomez-Acevedo, H.; Shankar, K. Maternal obesity is associated with a lipotoxic placental environment. Placenta 2014, 35, 171–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anelli, G.M.; Cardellicchio, M.; Novielli, C.; Antonazzo, P.; Mazzocco, M.I.; Cetin, I.; Mandò, C. Mitochondrial content and hepcidin are increased in obese pregnant mothers. J. Matern. Fetal Neonatal Med. 2018, 31, 2388–2395. [Google Scholar] [CrossRef] [PubMed]
- Franzago, M.; Fraticelli, F.; Stuppia, L.; Vitacolonna, E. Nutrigenetics, epigenetics and gestational diabetes: Consequences in mother and child. Epigenetics 2019, 14, 215–235. [Google Scholar] [CrossRef] [Green Version]
- Kerr, B.; Leiva, A.; Farías, M.; Contreras-Duarte, S.; Toledo, F.; Stolzenbach, F.; Silva, L.; Sobrevia, L. Foetoplacental epigenetic changes associated with maternal metabolic dysfunction. Placenta 2018, 69, 146–152. [Google Scholar] [CrossRef]
- Guarino, E.; Delli Poggi, C.; Grieco, G.E.; Cenci, V.; Ceccarelli, E.; Crisci, I.; Sebastiani, G.; Dotta, F. Circulating MicroRNAs as Biomarkers of Gestational Diabetes Mellitus: Updates and Perspectives. Int. J. Endocrinol. 2018, 2018, 6380463. [Google Scholar] [CrossRef] [Green Version]
- Jing, J.; Wang, Y.; Quan, Y.; Wang, Z.; Liu, Y.; Ding, Z. Maternal obesity alters C19MC microRNAs expression profile in fetal umbilical cord blood. Nutr. Metab. 2020, 17, 52. [Google Scholar] [CrossRef]
- Luque, A.; Farwati, A.; Crovetto, F.; Crispi, F.; Figueras, F.; Gratacós, E.; Aran, J.M. Usefulness of circulating microRNAs for the prediction of early preeclampsia at first-trimester of pregnancy. Sci. Rep. 2014, 4, 4882. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, A.E.; van Poppel, M.N.M.; Desoye, G.; Simmons, D.; Damm, P.; Jensen, D.M.; Dalgaard, L.T.; The Dali Core Investigator Group. The Temporal Profile of Circulating miRNAs during Gestation in Overweight and Obese Women with or without Gestational Diabetes Mellitus. Biomedicines 2022, 10, 482. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Badosa, G.; Bonmatí, A.; Ortega, F.J.; Mercader, J.M.; Guindo-Martínez, M.; Torrents, D.; Prats-Puig, A.; Martinez-Calcerrada, J.M.; Platero-Gutierrez, E.; De Zegher, F.; et al. Altered Circulating miRNA Expression Profile in Pregestational and Gestational Obesity. J. Clin. Endocrinol. Metab. 2015, 100, E1446–E1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirier, C.; Desgagné, V.; Guérin, R.; Bouchard, L. MicroRNAs in Pregnancy and Gestational Diabetes Mellitus: Emerging Role in Maternal Metabolic Regulation. Curr. Diabetes Rep. 2017, 17, 35. [Google Scholar] [CrossRef] [PubMed]
- Wren, M.E.; Shirtcliff, E.A.; Drury, S.S. Not all biofluids are created equal: Chewing over salivary diagnostics and the epigenome. Clin. Ther. 2015, 37, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Camejo, G.; Hurt-Camejo, E.; Olsson, U.; Bondjers, G. Lipid mediators that modulate the extracellular matrix structure and function in vascular cells. Curr. Diabetes Rep. 1999, 1, 142–149. [Google Scholar] [CrossRef]
- Olsson, U.; Bondjers, G.; Camejo, G. Fatty acids modulate the composition of extracellular matrix in cultured human arterial smooth muscle cells by altering the expression of genes for proteoglycan core proteins. Diabetes 1999, 48, 616–622. [Google Scholar] [CrossRef]
- Pessentheiner, A.R.; Ducasa, G.M.; Gordts, P.L.S.M. Proteoglycans in Obesity-Associated Metabolic Dysfunction and Meta-Inflammation. Front. Immunol. 2020, 11, 769. [Google Scholar] [CrossRef]
- Brovkina, O.; Nikitin, A.; Khodyrev, D.; Shestakova, E.; Sklyanik, I.; Panevina, A.; Stafeev, I.; Menshikov, M.; Kobelyatskaya, A.; Yurasov, A.; et al. Role of MicroRNAs in the Regulation of Subcutaneous White Adipose Tissue in Individuals with Obesity and Without Type 2 Diabetes. Front. Endocrinol. 2019, 10, 840. [Google Scholar] [CrossRef]
- Chang, Y.C.; Yang, S.F.; Lai, C.C.; Liu, J.Y.; Hsieh, Y.S. Regulation of matrix metalloproteinase production by cytokines; pharmacological agents and periodontal pathogens in human periodontal ligament fibroblast cultures. J. Periodontal Res. 2002, 37, 196–203. [Google Scholar] [CrossRef]
- Tomé, D.; Bos, C. Lysine requirement through the human life cycle. J. Nutr. 2007, 137 (Suppl. 6), 1642S–1645S. [Google Scholar] [CrossRef] [Green Version]
- Hanzu, F.A.; Vinaixa, M.; Papageorgiou, A.; Párrizas, M.; Correig, X.; Delgado, S.; Carmona, F.; Samino, S.; Vidal, J.; Gomis, R. Obesity rather than regional fat depots marks the metabolomic pattern of adipose tissue: An untargeted metabolomic approach. Obesity 2014, 22, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Desert, R.; Canlet, C.; Costet, N.; Cordier, S.; Bonvallot, N. Impact of maternal obesity on the metabolic profiles of pregnant women and their offspring at birth. Metabolomics 2015, 11, 1896–1907. [Google Scholar] [CrossRef] [Green Version]
- Scholtens, D.M.; Muehlbauer, M.J.; Daya, N.R.; Stevens, R.D.; Dyer, A.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; Bain, J.R.; Lowe, W.L., Jr.; et al. Metabolomics reveals broad-scale metabolic perturbations in hyperglycemic mothers during pregnancy. Diabetes Care 2014, 37, 158–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansson, N.; Rosario, F.J.; Gaccioli, F.; Lager, S.; Jones, H.N.; Roos, S.; Jansson, T.; Powell, T.L. Activation of placental mTOR signaling and amino acid transporters in obese women giving birth to large babies. J. Clin. Endocrinol. Metab. 2013, 98, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Jeffery, N.; Harries, L.W. miRNAs responsive to the diabetic microenvironment in the human beta cell line EndoC-βH1 may target genes in the FOXO, HIPPO and Lysine degradation pathways. Exp. Cell Res. 2019, 384, 111559, Erratum in Exp. Cell Res. 2019, 385, 111654. [Google Scholar] [CrossRef]
- Liu, H.; Chen, Y.G. The Interplay Between TGF-β Signaling and Cell Metabolism. Front. Cell Dev. Biol. 2022, 10, 846723. [Google Scholar] [CrossRef]
- Lee, J.H.; Mellado-Gil, J.M.; Bahn, Y.J.; Pathy, S.M.; Zhang, Y.E.; Rane, S.G. Protection from β-cell apoptosis by inhibition of TGF-β/Smad3 signaling. Cell Death Dis. 2020, 11, 184. [Google Scholar] [CrossRef] [Green Version]
- Toyoda, S.; Shin, J.; Fukuhara, A.; Otsuki, M.; Shimomura, I. Transforming growth factor β1 signaling links extracellular matrix remodeling to intracellular lipogenesis upon physiological feeding events. J. Biol. Chem. 2022, 298, 101748. [Google Scholar] [CrossRef]
- Yadav, H.; Devalaraja, S.; Chung, S.T.; Rane, S.G. TGF-β1/Smad3 Pathway Targets PP2A-AMPK-FoxO1 Signaling to Regulate Hepatic Gluconeogenesis. J. Biol. Chem. 2017, 292, 3420–3432. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Tian, F.; Li, H.; Zhou, Y.; Lu, J.; Ge, Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int. J. Gynecol. Obstet. 2015, 130, 49–53. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, S.J.; Bae, Y.C.; Jung, J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 2009, 27, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, T.T.; Ramos-Lobo, A.M.; Furigo, I.C.; Pedroso, J.A.; Buonfiglio, D.C.; Donato, J., Jr. SOCS3 deficiency in leptin receptor-expressing cells mitigates the development of pregnancy-induced metabolic changes. Mol. Metab. 2014, 4, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhang, X.; Wang, Z.; Hu, Y.; Sinha, R. Epigenome-wide association analysis revealed that SOCS3 methylation influences the effect of cumulative stress on obesity. Biol. Psychol. 2018, 131, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, P.; Van Obberghen, E. SOCS proteins causing trouble in insulin action. Acta Physiol. 2008, 192, 29–36. [Google Scholar] [CrossRef]
| Gene | Chromosome Position | CpG Sites | Primers: Forward (F) Reverse (R) Sequencing (S) | Sequencing Length (BP) | Annealing Temperature (°C) |
|---|---|---|---|---|---|
| TGF-Beta1 | chr17:6354421-76354821 | 2 | F: TTGGGTGATTTTTTTATAGGAGTT R: bio-TCCCCCCAAAAAAACCTATT S: GAGATGTTGAAGAGTGGTTA | 25 | 52 |
| SOCS3 | chr19:41353024-41353458 | 2 | F: GGTTTGTTTTTTGAGTTTT R:bio-CTACAAAAACTAAAAATCTCCC S: TATTTATTTTTTGGTATTAG | 23 | 54 |
| NW n = 16 | All OB n = 16 | p-Value | |
|---|---|---|---|
| Age, years B | 30.94 ± 3.89 | 33.25 ± 4.61 | ns |
| Pregestational BMI, kg/m2 B | 21.02 ± 2.25 | 36.47 ± 5.53 | <0.001 |
| GWG, kg B | 13.38 ± 4.41 | 9.13 ± 5.63 | 0.035 |
| GWG to IOM upper limit, % A | 83.61 ± 27.54 | 101.37 ± 62.57 | ns |
| Basal glycemia, mg/dL B | 81.50 ± 5.48 | 92.69 ± 15.18 | 0.034 |
| Gestational diabetes, n (%) | 0 (0) | 10 (62.5) | - |
| GA at saliva withdrawal, mL A | 33.24 ± 1.73 | 32.99 ± 2.52 | ns |
| Saliva flow rate, mL/min A | 0.48 ± 0.22 | 0.47 ± 0.17 | ns |
| Periodontal health C | |||
| Healthy, n (%) | 7 (43.8) | 5 (31.2) | ns |
| Periodontal disease, n (%) | 9 (56.2) | 11 (68.8) | |
| Number of teeth B | 27.60 ± 0.74 | 25.31 ± 3.01 | 0.027 |
| BOP, % sites B | 17.98 ± 19.76 | 39.57 ± 38.10 | ns |
| PPD, mean A | 2.38 ± 0.45 | 2.63 ± 0.88 | ns |
| Plaque index, % A | 25.61 ± 18.97 | 52.90 ± 40.32 | 0.024 |
| Calculus, % A | 27.79 ± 23.65 | 44.53 ± 35.81 | ns |
| NW n = 16 | All OB n = 16 | p-Value | |
|---|---|---|---|
| GA at delivery, wks A | 39.76 ± 0.87 | 39.33 ± 1.18 | ns |
| Neonatal weight, gr A | 3331.25 ± 294.60 | 3488.75 ± 356.62 | ns |
| Neonatal weight centile A | 47.56 ± 24.19 | 61.00 ± 28.72 | ns |
| AGA, n (%) C | 15 (93.8) | 11 (68.8) | ns |
| LGA, n (%) C | 1 (6.3) | 5 (31.3) | |
| Neonatal sex, n C | ns | ||
| Males, n (%) | 9 (56.3) | 7 (43.8) | |
| Females, n (%) | 7 (43.8) | 9 (56.3) | |
| Placental weight, gr A | 439.00 ± 79.05 | 509.29 ± 80.81 | 0.035 |
| Neonatal/placental weight B | 7.78 ± 1.66 | 6.97 ± 1.63 | ns |
| Placental area, cm2 B | 281.94 ± 64.06 | 266.16 ± 45.33 | ns |
| Placental thickness, cm A | 1.61 ± 0.37 | 1.97 ± 0.50 | 0.050 |
| miRNA Name | Fold Change (FC) | p-Value | False Discovery Rate p-Value (FDR) |
|---|---|---|---|
| hsa-miR-505 ↓ | 0.335 | 0.0002 | 0.0396 |
| hsa-miR-616 ↓ | 0.434 | 0.0123 | 0.1873 |
| hsa-miR-618 ↑ | 7.141 | 0.0002 | 0.0396 |
| hsa-miR-206 ↑ | 3.985 | 0.0005 | 0.0514 |
| hsa-miR-376a ↑ | 5.870 | 0.0008 | 0.0671 |
| hsa-miR-517c ↑ | 2.788 | 0.0015 | 0.0989 |
| hsa-miR-133a ↑ | 3.703 | 0.0041 | 0.1599 |
| hsa-miR-512 ↑ | 23.295 | 0.0044 | 0.1599 |
| hsa-miR-302d ↑ | 14.427 | 0.0046 | 0.1599 |
| hsa-miR-520b ↑ | 2.755 | 0.0051 | 0.1599 |
| hsa-miR-1254 ↑ | 5.443 | 0.0054 | 0.1599 |
| hsa-miR-133b ↑ | 5.634 | 0.0057 | 0.1599 |
| hsa-miR-1285 ↑ | 40.956 | 0.0095 | 0.1859 |
| hsa-miR-635 ↑ | 2.178 | 0.0102 | 0.1859 |
| hsa-miR-551b ↑ | 2.614 | 0.0106 | 0.1859 |
| hsa-miR-548b-5p ↑ | 5.692 | 0.0110 | 0.1859 |
| hsa-miR-1256 ↑ | 2.169 | 0.0110 | 0.1859 |
| hsa-miR-302c ↑ | 2.622 | 0.0111 | 0.1859 |
| hsa-miR-184 ↑ | 4.817 | 0.0118 | 0.1873 |
| hsa-miR-548c-5p ↑ | 4.912 | 0.0131 | 0.1909 |
| Pathways Information | p-Value | miRNAs | |
|---|---|---|---|
| Fatty acids biosynthesis | Synthesis of fatty acids. Fatty acids are generally excessive in the Western diet. | 2.1444 × 10−11 | hsa-miR-1254 |
| Fatty acids metabolism | Anabolic and catabolic processes involving fatty acids or related molecules. Fatty acids are generally excessive in the Western diet. | 0.0002 | hsa-miR-1254 |
| Lysine degradation | Catabolism of lysine (from dietary up-taken or intracellular proteins). Lysine is generally excessive in the Western diet. | 1.7153 × 10−7 | hsa-miR-505-5p; hsa-miR-302d-3p; hsa-miR-376a-5p; hsa-miR-302c-3p; hsa-miR-512-3p; hsa-miR-505-3p; hsa-miR-616-3p |
| ECM–receptor interaction | Tissue and organ morphogenesis; maintenance of cell and tissue structure and function; adhesion, migration, differentiation, proliferation, and apoptosis; force-transmitting physical link with the cytoskeleton. | <1 × 10−325 | hsa-miR-302c-5p; hsa-miR-184; hsa-miR-206; hsa-miR-635; hsa-miR-512-3p; hsa-miR-505-3p; hsa-miR-376a-3p |
| Mean OB (95% CI) | Mean NW (95% CI) | p-Value | |
|---|---|---|---|
| TGF-Beta1 (% 5mC) | 0.44 (0.24–0.63) | 0.86 (0.61–1.11) | 0.019 |
| SOCS3 (% 5mC) | 60.95 (57.20–64.70) | 68.50 (63.75–73.25) | 0.025 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandò, C.; Abati, S.; Anelli, G.M.; Favero, C.; Serati, A.; Dioni, L.; Zambon, M.; Albetti, B.; Bollati, V.; Cetin, I. Epigenetic Profiling in the Saliva of Obese Pregnant Women. Nutrients 2022, 14, 2122. https://doi.org/10.3390/nu14102122
Mandò C, Abati S, Anelli GM, Favero C, Serati A, Dioni L, Zambon M, Albetti B, Bollati V, Cetin I. Epigenetic Profiling in the Saliva of Obese Pregnant Women. Nutrients. 2022; 14(10):2122. https://doi.org/10.3390/nu14102122
Chicago/Turabian StyleMandò, Chiara, Silvio Abati, Gaia Maria Anelli, Chiara Favero, Anaïs Serati, Laura Dioni, Marta Zambon, Benedetta Albetti, Valentina Bollati, and Irene Cetin. 2022. "Epigenetic Profiling in the Saliva of Obese Pregnant Women" Nutrients 14, no. 10: 2122. https://doi.org/10.3390/nu14102122
APA StyleMandò, C., Abati, S., Anelli, G. M., Favero, C., Serati, A., Dioni, L., Zambon, M., Albetti, B., Bollati, V., & Cetin, I. (2022). Epigenetic Profiling in the Saliva of Obese Pregnant Women. Nutrients, 14(10), 2122. https://doi.org/10.3390/nu14102122