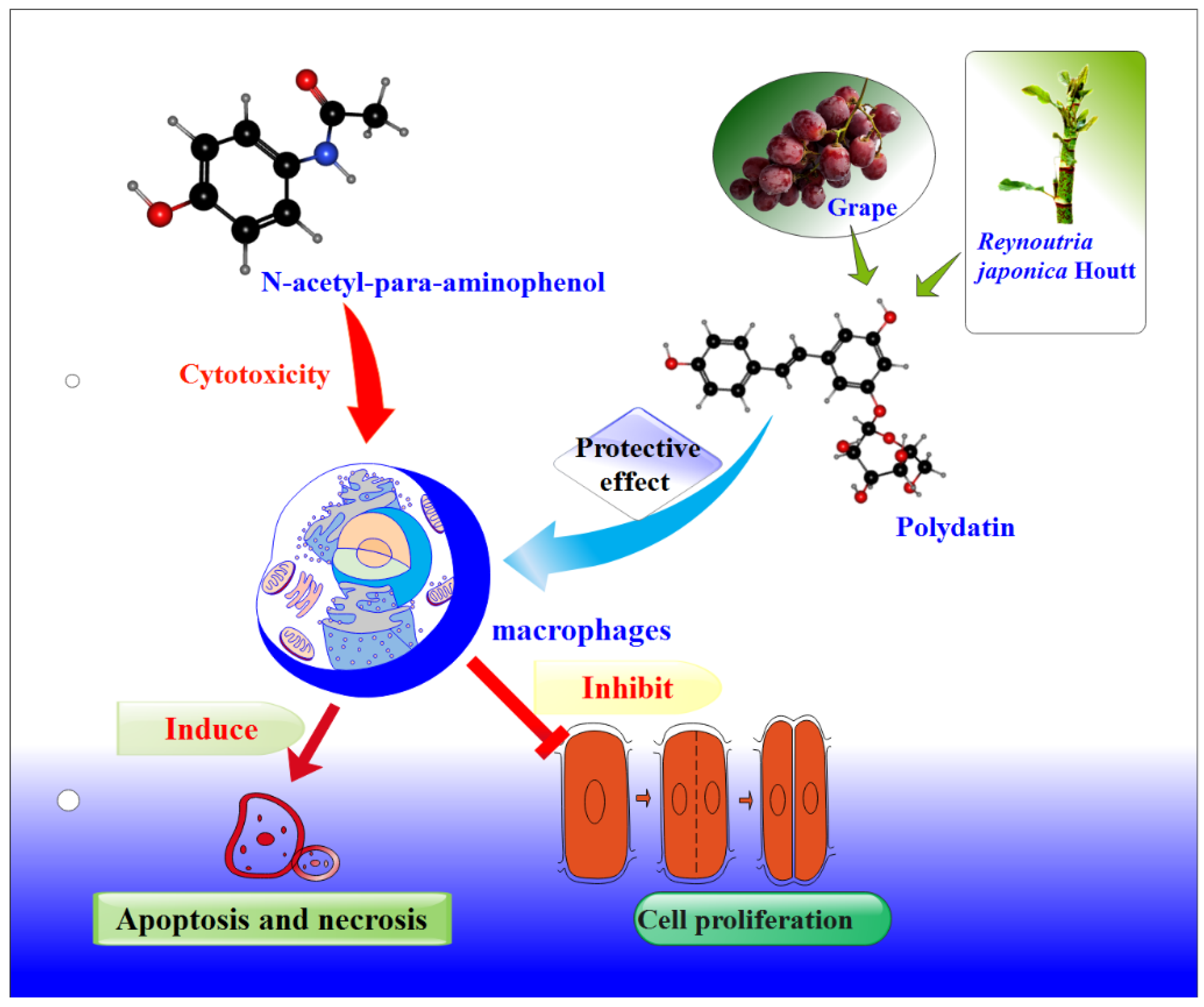
Protective Effects of Polydatin from Grapes and Reynoutria japonica Houtt. on Damaged Macrophages Treated with Acetaminophen
Abstract
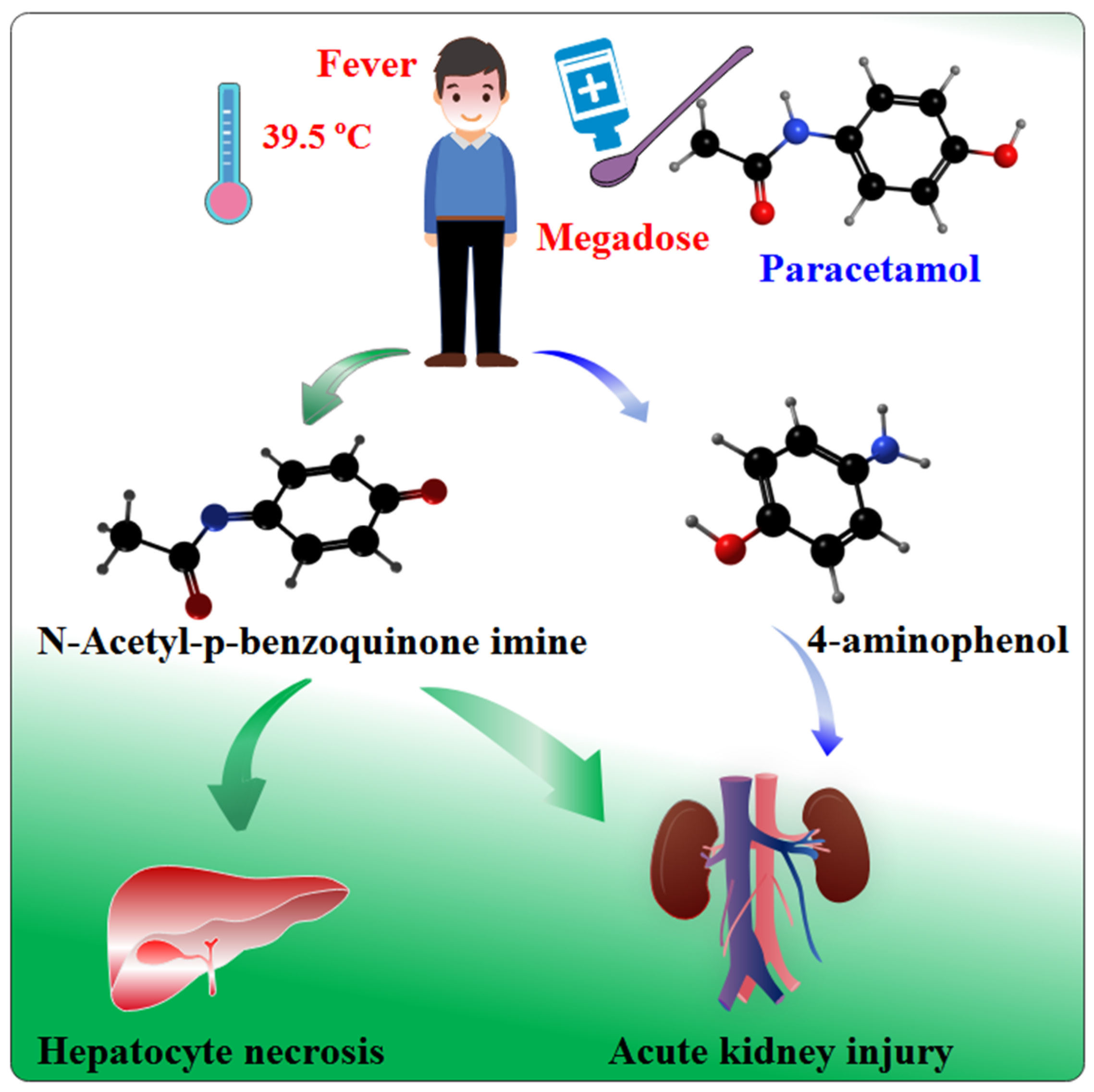
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Cell Culture
2.2.2. Cell Injury Model Construction and Drug Treatment
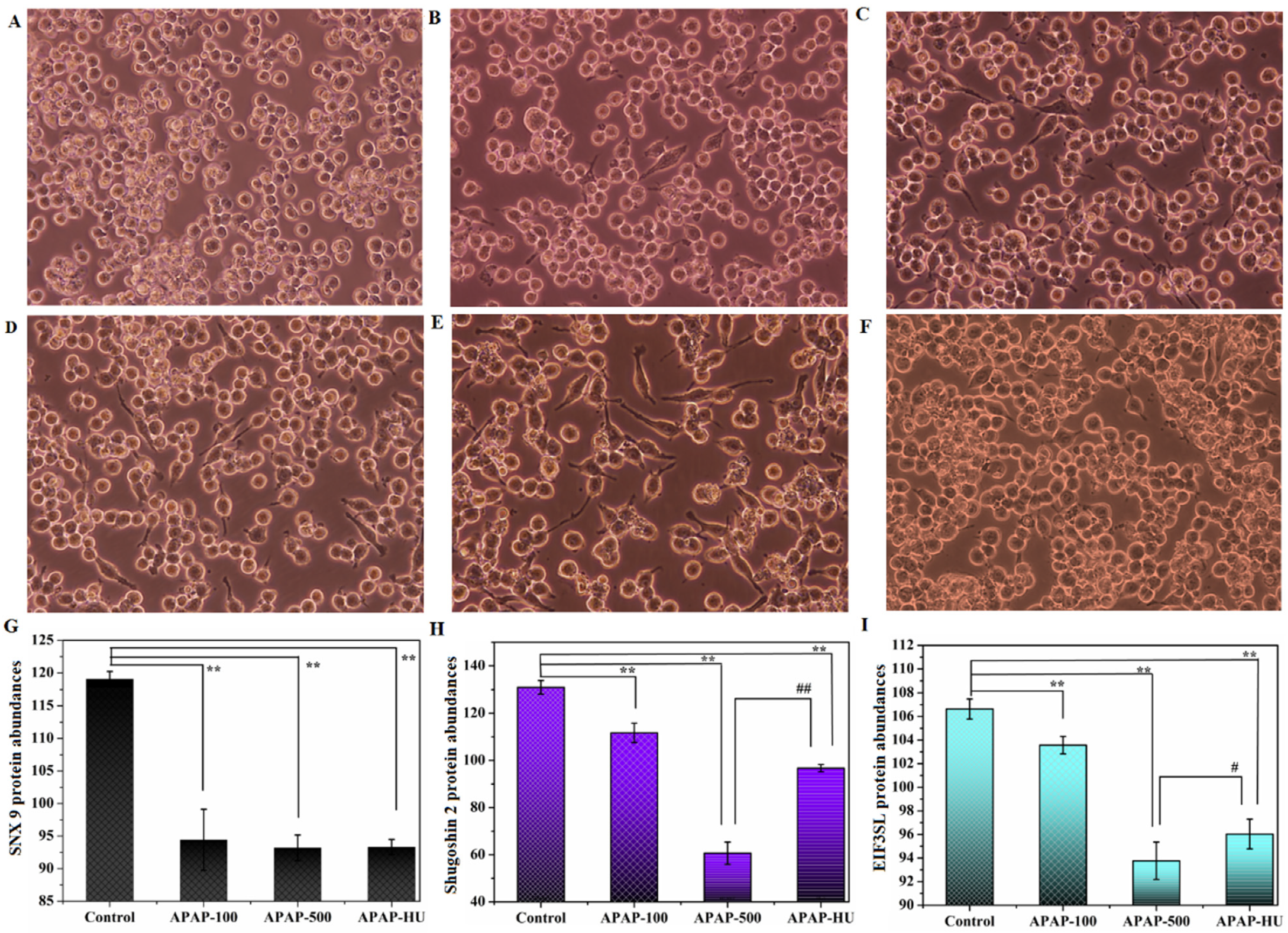
2.2.3. Cell Morphology Observation
2.2.4. MTT Assay for Cell Viability
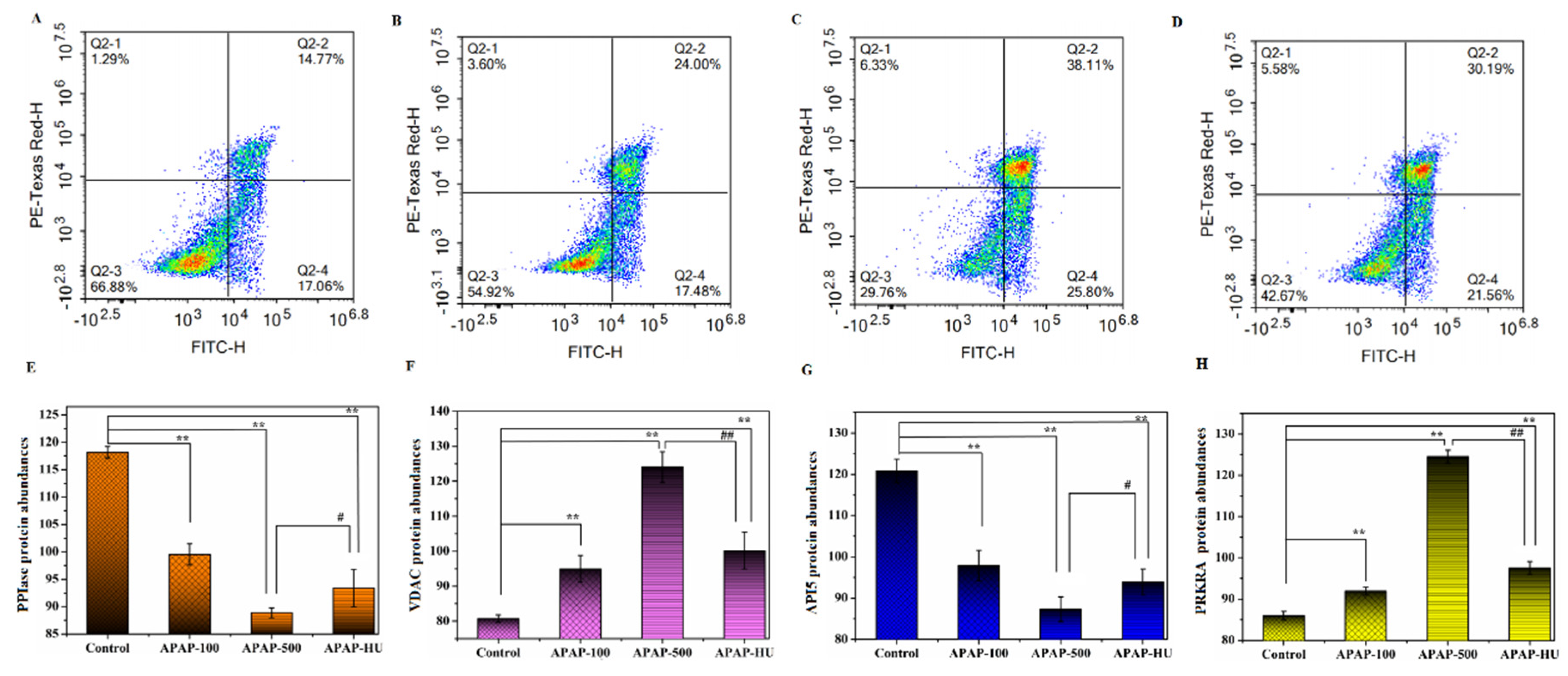
2.2.5. Apoptosis Assay
2.2.6. Preparation and Digestion of Proteins
2.2.7. LC-MS/MS Analysis
2.2.8. Data Analysis
3. Results and Discussion
3.1. Effect of Drugs on Cell Proliferation
3.2. Effect of Drugs on Apoptosis
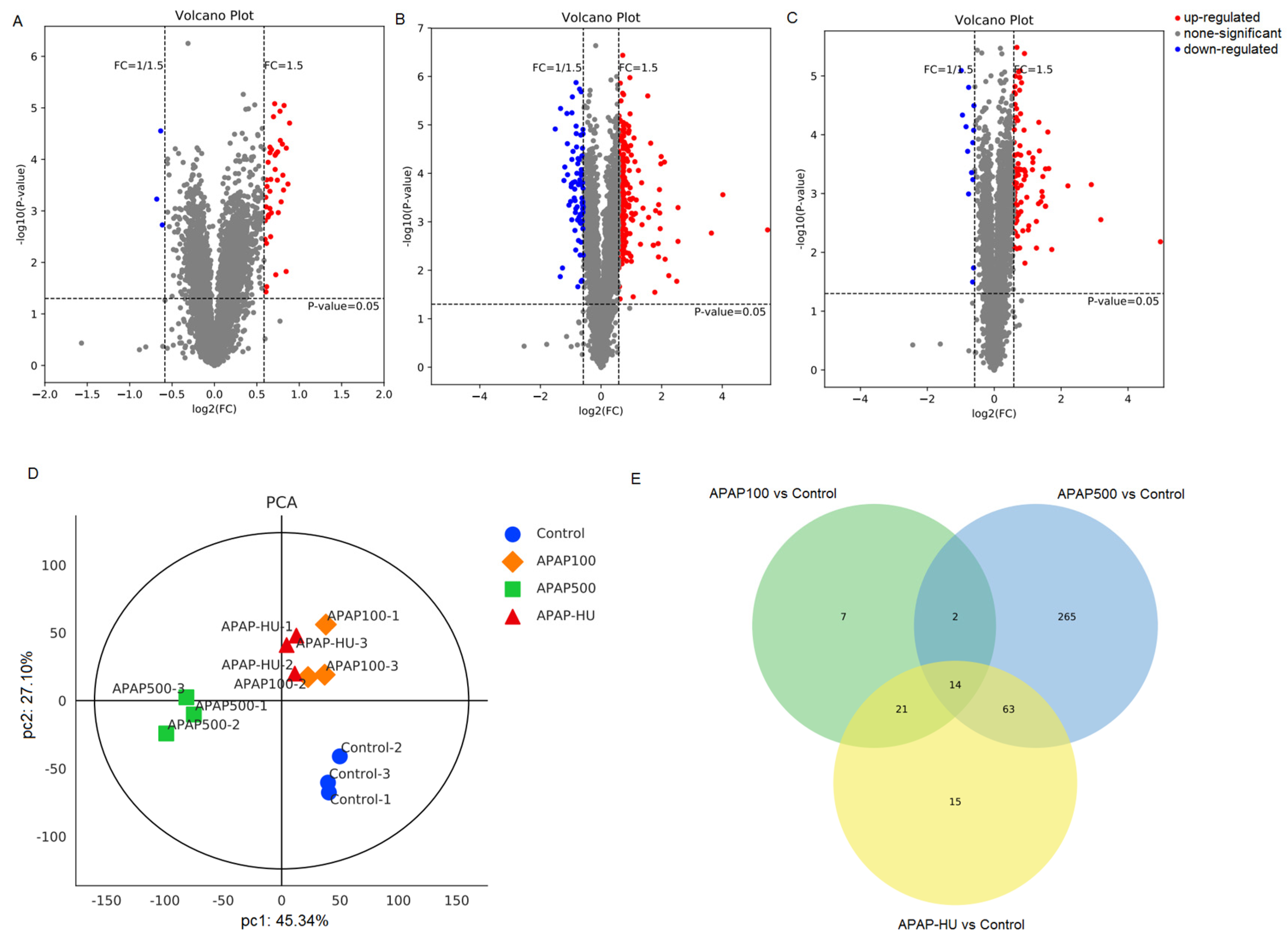
3.3. Differential Protein Screening
3.4. Principal Component Analysis (PCA)
3.5. Venn Diagram Analysis
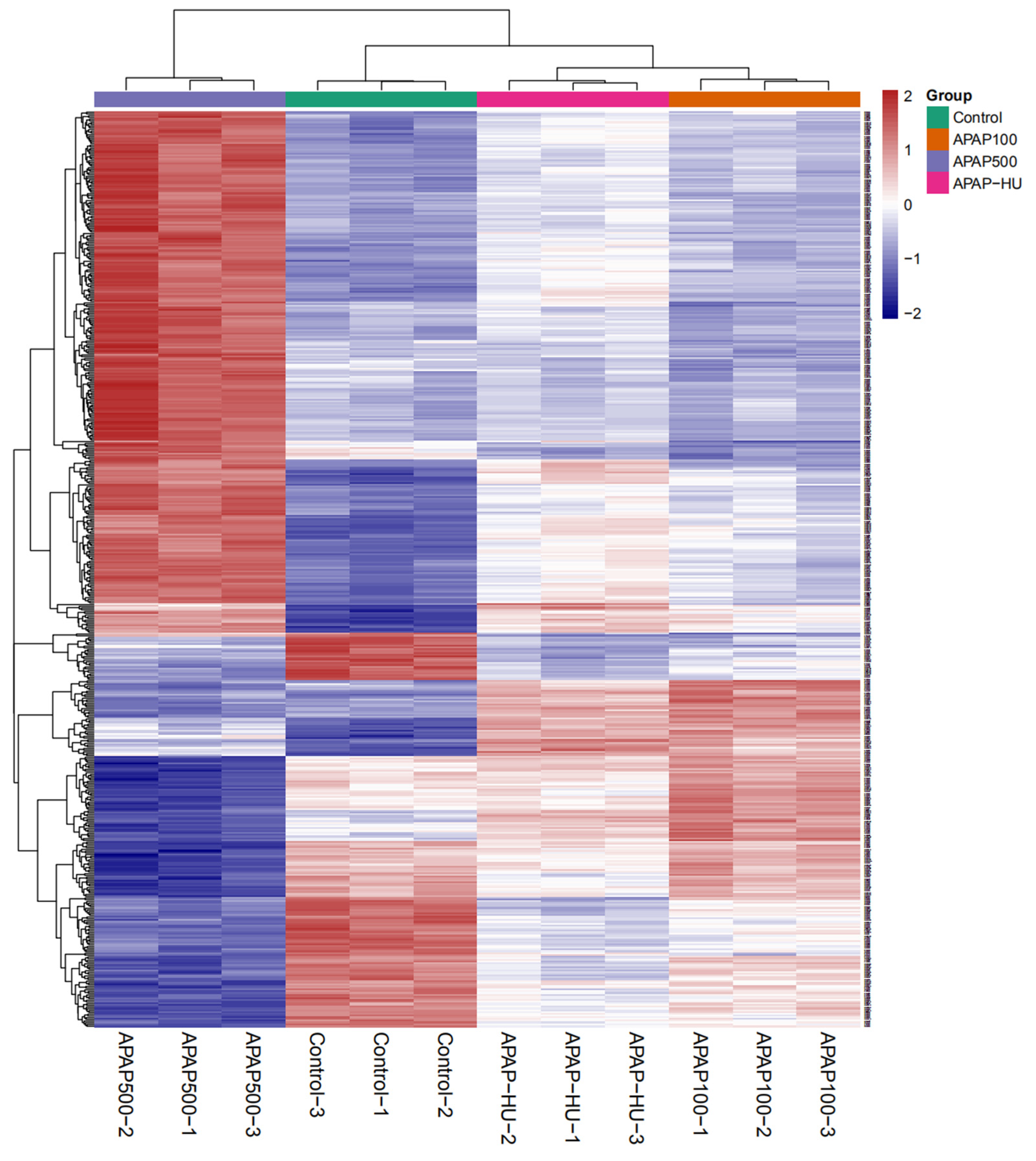
3.6. Clustering Analysis of Differential Protein Expression Levels
3.7. Protein Interaction Network Diagram
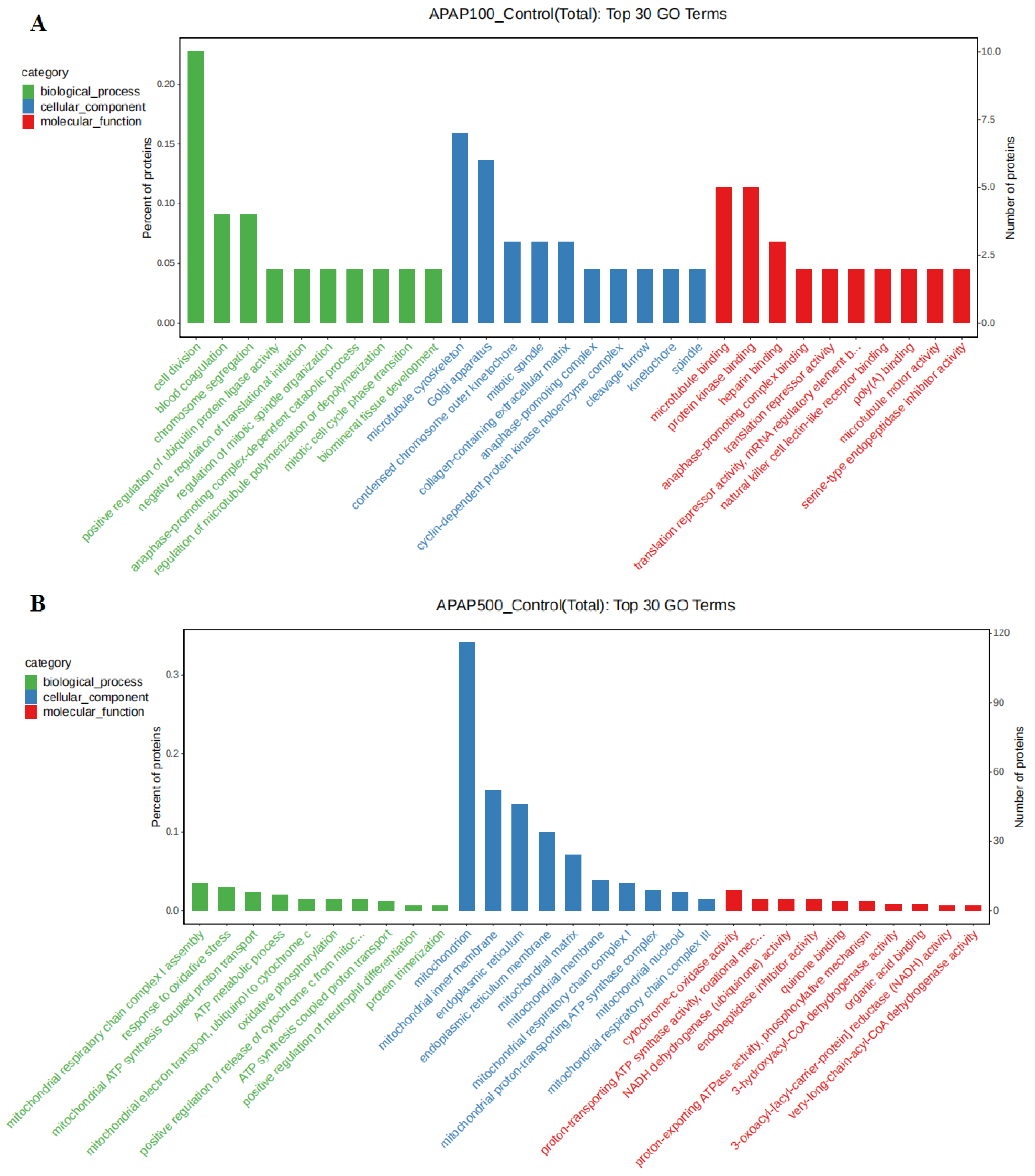
3.8. Gene Ontology (GO) Analysis
3.9. Polydatin Can Repair the Damage Caused by Acetaminophen
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ishitsuka, Y.; Kondo, Y.; Kadowaki, D. Toxicological property of acetaminophen: The dark side of a safe antipyretic/analgesic drug? Biol. Pharm. Bull. 2020, 43, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, I.M.; Walson, P.D. Acetaminophen and ibuprofen in the treatment of pediatric fever: A narrative review. Curr. Med. Res. Opin. 2021, 37, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Azarmehr, N.; Afshar, P.; Moradi, M.; Sadeghi, H.; Sadeghi, H.; Alipoor, B.; Khalvati, B.; Barmoudeh, Z.; Abbaszadeh-Goudarzi, K.; Doustimotlagh, A.H. Hepatoprotective and antioxidant activity of watercress extract on acetaminophen-induced hepatotoxicity in rats. Heliyon 2019, 5, e02072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiew, A.L.; James, L.P.; Isbister, G.K.; Pickering, J.W.; McArdle, K.; Chan, B.S.; Buckley, N.A. Early acetaminophen-protein adducts predict hepatotoxicity following overdose (ATOM-5). J. Hepatol. 2020, 72, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, B.; Apte, U. Liver regeneration after acetaminophen hepatotoxicity: Mechanisms and therapeutic opportunities. Am. J. Pathol. 2019, 189, 719–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, A.; Jaeschke, H. Acetaminophen hepatotoxicity. Semin. Liver Dis. 2019, 39, 221–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, M.; Huo, Y.; Yin, S.; Hu, H. Mechanisms of acetaminophen-induced liver injury and its implications for therapeutic interventions. Redox. Biol. 2018, 17, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Athersuch, T.J.; Antoine, D.J.; Boobis, A.R.; Coen, M.; Daly, A.K.; Possamai, L.; Nicholson, J.K.; Wilson, I.D. Paracetamol metabolism, hepatotoxicity, biomarkers and therapeutic interventions: A perspective. Toxicol. Res. 2018, 7, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Na, J.D.; Park, J.H. Silymarin prevents acetaminophen-induced hepatotoxicity via up-regulation of the glutathione conjugation capacity in mice. J. Funct. Foods 2018, 49, 235–240. [Google Scholar] [CrossRef]
- Hiragi, S.; Yamada, H.; Tsukamoto, T.; Yoshida, K.; Kondo, N.; Matsubara, T.; Yanagita, M.; Tamura, H.; Kuroda, T. Acetaminophen administration and the risk of acute kidney injury: A self-controlled case series study. Clin. Epidemiol. 2018, 10, 265. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-G.; Lin, C.-L.; Dai, M.-S.; Chang, P.-Y.; Chen, J.-H.; Huang, T.-C.; Wu, Y.-Y.; Kao, C.-H. Risk of acute kidney injury and long-term outcome in patients with acetaminophen intoxication: A nationwide population-based retrospective cohort study. Medicine 2015, 94, e2040. [Google Scholar] [CrossRef] [PubMed]
- Stollings, J.L.; Wheeler, A.P.; Rice, T.W. Incidence and characterization of acute kidney injury after acetaminophen overdose. J. Crit. Care 2016, 35, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Jaeschke, H.; Murray, F.J.; Monnot, A.D.; Jacobson-Kram, D.; Cohen, S.M.; Hardisty, J.F.; Atillasoy, E.; Hermanowski-Vosatka, A.; Kuffner, E.; Wikoff, D. Assessment of the biochemical pathways for acetaminophen toxicity: Implications for its carcinogenic hazard potential. Regul. Toxicol. Pharm. 2021, 120, 104859. [Google Scholar] [CrossRef] [PubMed]
- Varricchi, G.; Marone, G.; Kovanen, P.T. Cardiac Mast Cells: Underappreciated Immune Cells in Cardiovascular Homeostasis and Disease. Trends Immunol. 2020, 41, 734–746. [Google Scholar] [CrossRef]
- Dimitrov, D.H.; Braida, N.; Walss-Bass, C. The link between immune system dysregulation and schizophrenia: A look at the genetic evidence. Psychiatr. Times 2011, 28, 23–27. [Google Scholar]
- Murray, R.Z. Skin Wound Healing: Normal Macrophage Function and Macrophage Dysfunction in Diabetic Wounds. Molecules 2021, 26, 4917. [Google Scholar] [CrossRef]
- Collisson, E.; Griggs, L.; Drechsler, Y. Macrophages from disease resistant B2 haplotype chickens activate T lymphocytes more effectively than macrophages from disease susceptible B19 birds. Dev. Comp. Immunol. 2017, 67, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Tengvall, S.; Che, K.F.; Lindén, A. Interleukin-26: An Emerging Player in Host Defense and Inflammation. J. Innate Immun. 2016, 8, 15–22. [Google Scholar] [CrossRef]
- Grenha, A.; Rodrigues, S. Activation of Macrophages: Establishing a Role for Polysaccharides in Drug Delivery Strategies Envisaging Antibacterial Therapy. Curr. Pharm. Des. 2015, 21, 4869–4887. [Google Scholar] [CrossRef]
- Du, Q.-H.; Peng, C.; Zhang, H. Polydatin: A review of pharmacology and pharmacokinetics. Pharm. Biol. 2013, 51, 1347–1354. [Google Scholar] [CrossRef]
- García-Martínez, D.J.; Arroyo-Hernández, M.; Posada-Ayala, M.; Santos, C. The High Content of Quercetin and Catechin in Airen Grape Juice Supports Its Application in Functional Food Production. Foods 2021, 10, 1532. [Google Scholar] [CrossRef] [PubMed]
- Şöhretoğlu, D.; Baran, M.Y.; Arroo, R.; Kuruüzüm-Uz, A. Recent advances in chemistry, therapeutic properties and sources of polydatin. Phytochem. Rev. 2018, 17, 973–1005. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Yan, T.; Wang, F.; Li, J.; Jia, L.; Jia, J.; Hu, G. Screening of an endophyte transforming polydatin to resveratrol from Reynoutria japonica houtt and the optimization of its transformation parameters. Molecules 2020, 25, 4830. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, E.; Fusco, R.; Biundo, F.; D’Amico, R.; Benedetto, F.; Di Paola, R.; Cuzzocrea, S. Palmitoylethanolamide and Polydatin combination reduces inflammation and oxidative stress in vascular injury. Pharmacol. Res. 2017, 123, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xu, J.; Zeng, Y.; Chen, L.; Le Xu, X. Polydatin attenuates atherosclerosis in apolipoprotein E-deficient mice: Role of reverse cholesterol transport. Phytomedicine 2019, 62, 152935. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Alvi, S.S.; Iqbal, D.; Khan, M.S. Insights into pharmacological mechanisms of polydatin in targeting risk factors-mediated atherosclerosis. Life Sci. 2020, 254, 117756. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, D.-q.; Liao, Z.; Wang, B.; Gong, S.; Wang, C.; Zhang, M.-z.; Wang, G.-h.; Cai, H.; Liao, F.-F. Anti-oxidant polydatin (piceid) protects against substantia nigral motor degeneration in multiple rodent models of Parkinson’s disease. Mol. Neurodegener. 2015, 10, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Piao, H.; Jiang, J.; Jin, G.; Zheng, M.; Yang, J.; Jin, X.; Sun, T.; Choi, Y.H.; Li, L. Polydatin inhibits mast cell-mediated allergic inflammation by targeting PI3K/Akt, MAPK, NF-κB and Nrf2/HO-1 pathways. Sci. Rep. 2017, 7, 11895. [Google Scholar] [CrossRef]
- Wang, Q.; Kaan, H.Y.K.; Hooda, R.N.; Goh, S.L.; Sondermann, H. Structure and plasticity of Endophilin and Sorting Nexin 9. Structure 2008, 16, 1574–1587. [Google Scholar] [CrossRef] [Green Version]
- Kao, Y.; Tsai, W.-C.; Chen, S.-H.; Hsu, S.-Y.; Huang, L.-C.; Chang, C.-J.; Huang, S.-M.; Hueng, D.-Y. Shugoshin 2 is a biomarker for pathological grading and survival prediction in patients with gliomas. Sci. Rep. 2021, 11, 18541. [Google Scholar] [CrossRef]
- Xiong, H.; Hu, M.; Huang, H.; Gong, J.; Wu, J.; Zhang, H. Eukaryotic translation initiation factor 3B downregulation inhibits cell proliferation and promotes cell apoptosis through negatively regulating tumor necrosis factor receptor superfamily member 21 in gastric cancer. Transl. Cancer Res. 2019, 8, 2242. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, Y.; Shimizu, S. Role of the mitochondrial membrane permeability transition in cell death. Apoptosis 2007, 12, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Innes, B.T.; Sowole, M.A.; Gyenis, L.; Dubinsky, M.; Konermann, L.; Litchfield, D.W.; Brandl, C.J.; Shilton, B.H. Peroxide-mediated oxidation and inhibition of the peptidyl-prolyl isomerase Pin1. BBA-Mol. Basis Dis. 2015, 1852, 905–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manganelli, V.; Salvatori, I.; Costanzo, M.; Capozzi, A.; Caissutti, D.; Caterino, M.; Valle, C.; Ferri, A.; Sorice, M.; Ruoppolo, M. Overexpression of Neuroglobin Promotes Energy Metabolism and Autophagy Induction in Human Neuroblastoma SH-SY5Y Cells. Cells 2021, 10, 3394. [Google Scholar] [CrossRef] [PubMed]
- Keinan, N.; Tyomkin, D.; & Shoshan-Barmatz, V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol. Cell Biol. 2010, 30, 5698–5709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoshan-Barmatz, V.; De, S.; Meir, A. The mitochondrial voltage-dependent anion channel 1, Ca2+ transport, apoptosis, and their regulation. Front. Oncol. 2017, 7, 60. [Google Scholar] [CrossRef] [Green Version]
- Han, B.-G.; Kim, K.H.; Lee, S.J.; Jeong, K.-C.; Cho, J.-W.; Noh, K.H.; Kim, T.W.; Kim, S.-J.; Yoon, H.-J.; Suh, S.W. Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules. J. Biol. Chem. 2012, 287, 10727–10737. [Google Scholar] [CrossRef] [Green Version]
- Clemens, M.J.; ELIA, A. The double-stranded RNA-dependent protein kinase PKR: Structure and function. J. Interf. Cytok. Res. 1997, 17, 503–524. [Google Scholar] [CrossRef]
- Vandersluis, B.; Costanzo, M.; Billmann, M.; Ward, H.N.; Myers, C.L.; Andrews, B.J.; Boone, C. Integrating genetic and protein–protein interaction networks maps a functional wiring diagram of a cell. Curr. Opin. Microbiol. 2018, 45, 170–179. [Google Scholar] [CrossRef]
- Iovanella, A.; Cinelli, M.; Ferraro, G. Evaluating relevance and redundancy to quantify how binary node metadata interplay with the network structure. Sci. Rep. 2019, 9, 11404. [Google Scholar] [CrossRef]
- Mccarter, J.P.; Mitreva, M.; Martin, J.; Dante, M. Analysis and functional classification of transcripts from the nematode Meloidogyne incognita. Genome Biol. 2003, 4, R26. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Wang, W.; Zhang, K.; Liu, Q.; Ma, T.; Tan, L.; Ma, L. Protective Effects of Polydatin from Grapes and Reynoutria japonica Houtt. on Damaged Macrophages Treated with Acetaminophen. Nutrients 2022, 14, 2077. https://doi.org/10.3390/nu14102077
Liu C, Wang W, Zhang K, Liu Q, Ma T, Tan L, Ma L. Protective Effects of Polydatin from Grapes and Reynoutria japonica Houtt. on Damaged Macrophages Treated with Acetaminophen. Nutrients. 2022; 14(10):2077. https://doi.org/10.3390/nu14102077
Chicago/Turabian StyleLiu, Can, Wenyi Wang, Kaixin Zhang, Qiudi Liu, Tongyao Ma, Li Tan, and Lanqing Ma. 2022. "Protective Effects of Polydatin from Grapes and Reynoutria japonica Houtt. on Damaged Macrophages Treated with Acetaminophen" Nutrients 14, no. 10: 2077. https://doi.org/10.3390/nu14102077
APA StyleLiu, C., Wang, W., Zhang, K., Liu, Q., Ma, T., Tan, L., & Ma, L. (2022). Protective Effects of Polydatin from Grapes and Reynoutria japonica Houtt. on Damaged Macrophages Treated with Acetaminophen. Nutrients, 14(10), 2077. https://doi.org/10.3390/nu14102077




