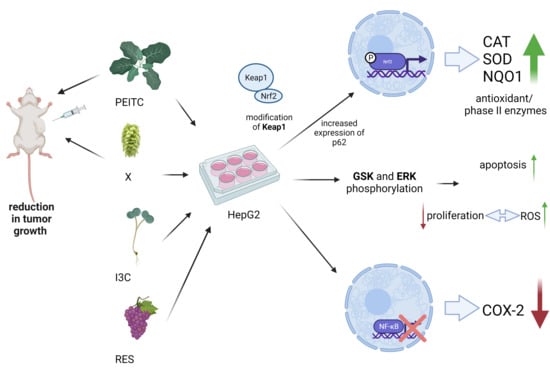
Comparison of the Impact of Xanthohumol and Phenethyl Isothiocyanate and Their Combination on Nrf2 and NF-κB Pathways in HepG2 Cells In Vitro and Tumor Burden In Vivo
Abstract
:1. Introduction
2. Materials and Methods
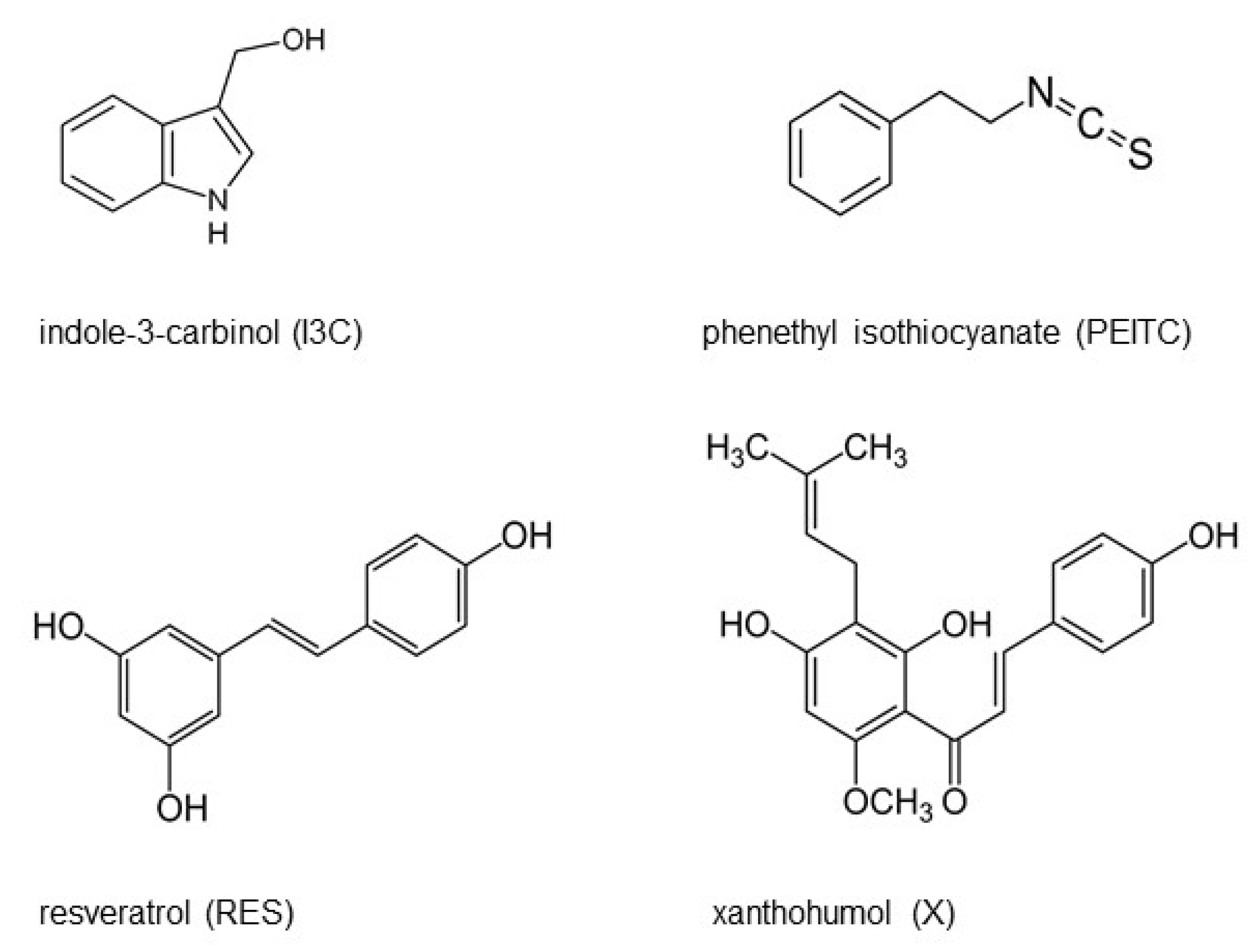
2.1. Chemicals
2.2. Cell Culture and Viability Assay
2.3. MUSE® Cell Flow Cytometry Analysis
2.3.1. Cell Cycle Distribution
2.3.2. Proliferation
2.3.3. Apoptosis
2.3.4. MAPK Pathway Activation
2.3.5. Level of ROS
2.4. Preparation of RNA, Cytosolic, and Nuclear Extracts
2.5. Quantitative Real-Time PCR
2.6. Transcriptions Factors Binding Assays
2.7. Western Blotting
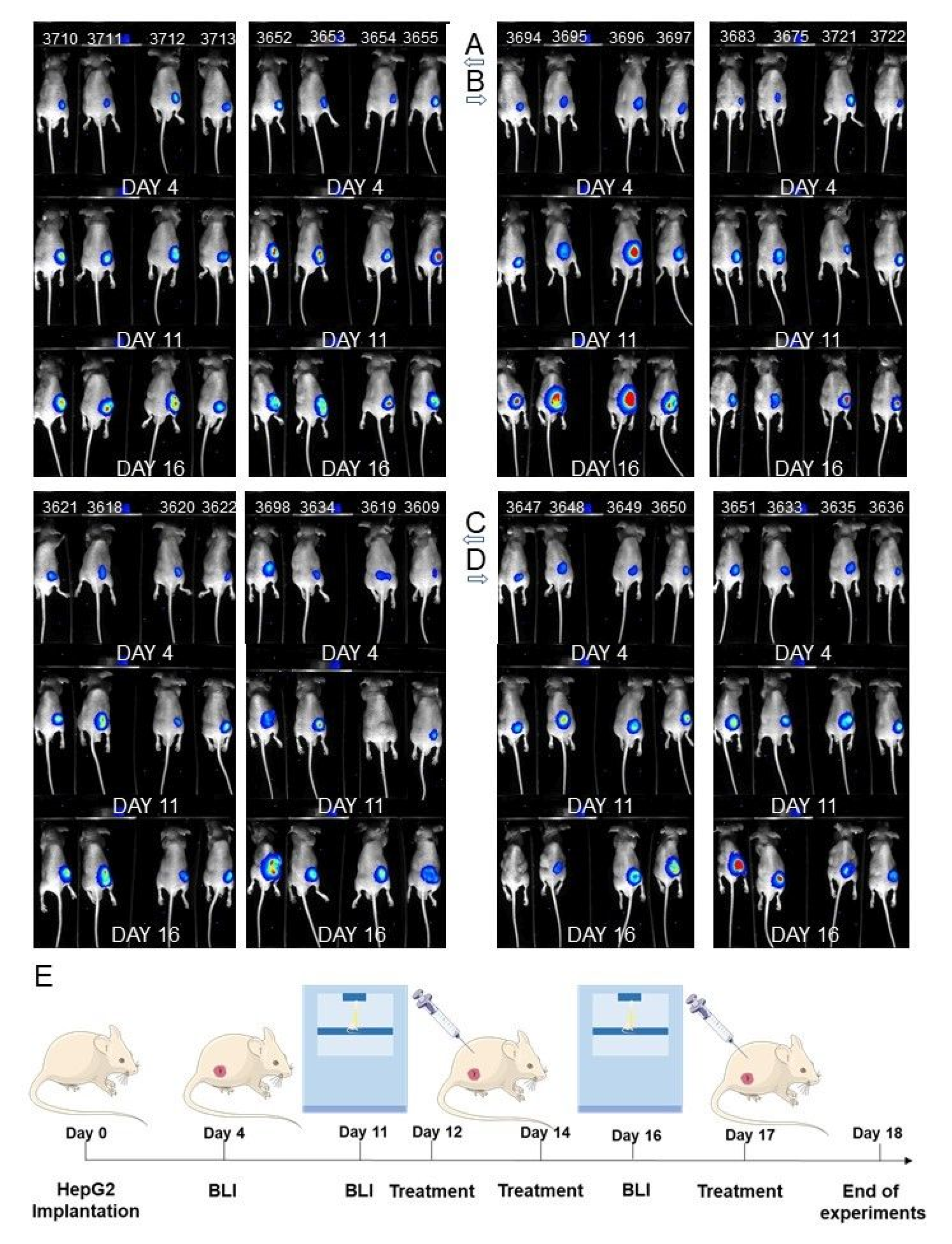
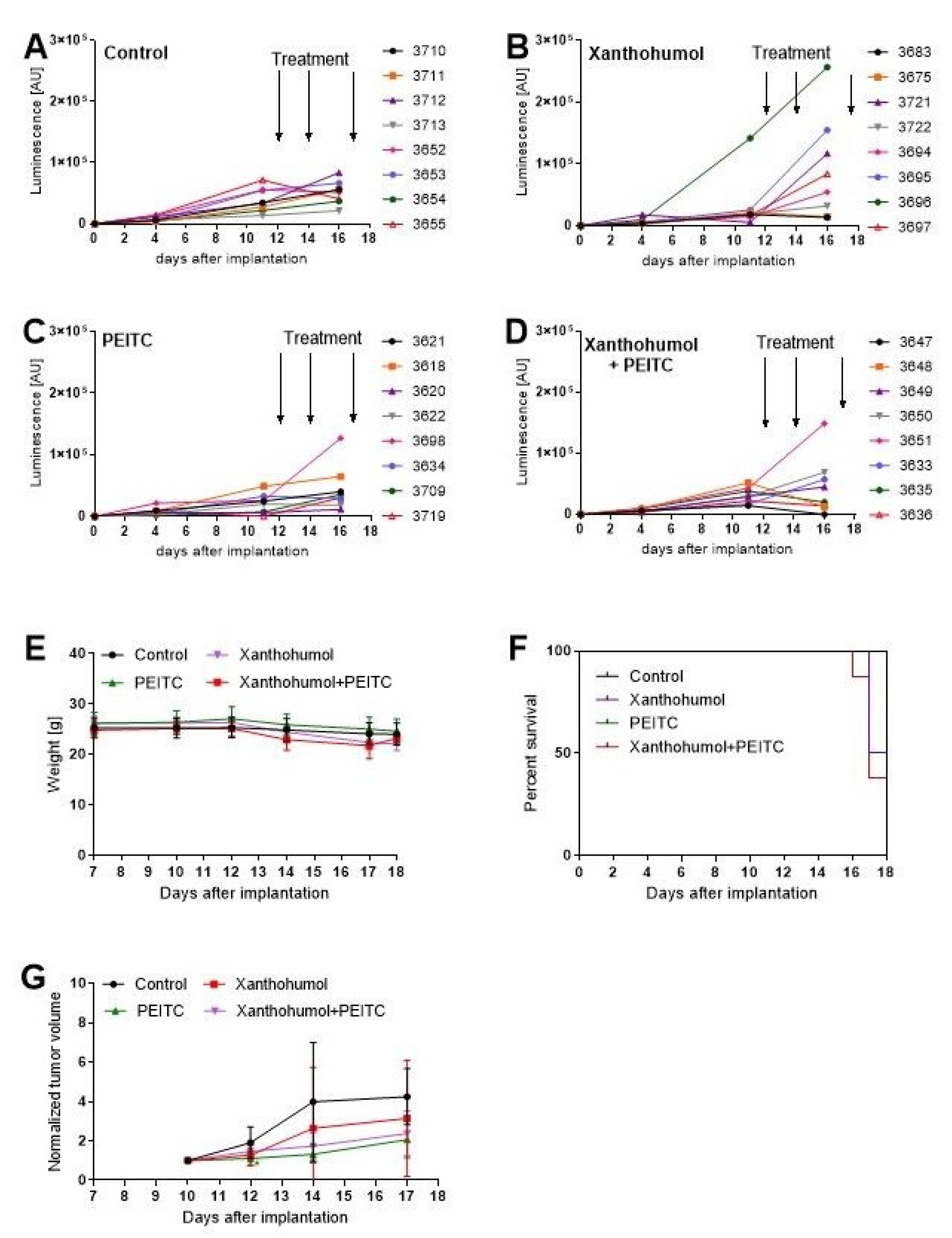
2.8. In Vivo Tumor Growth
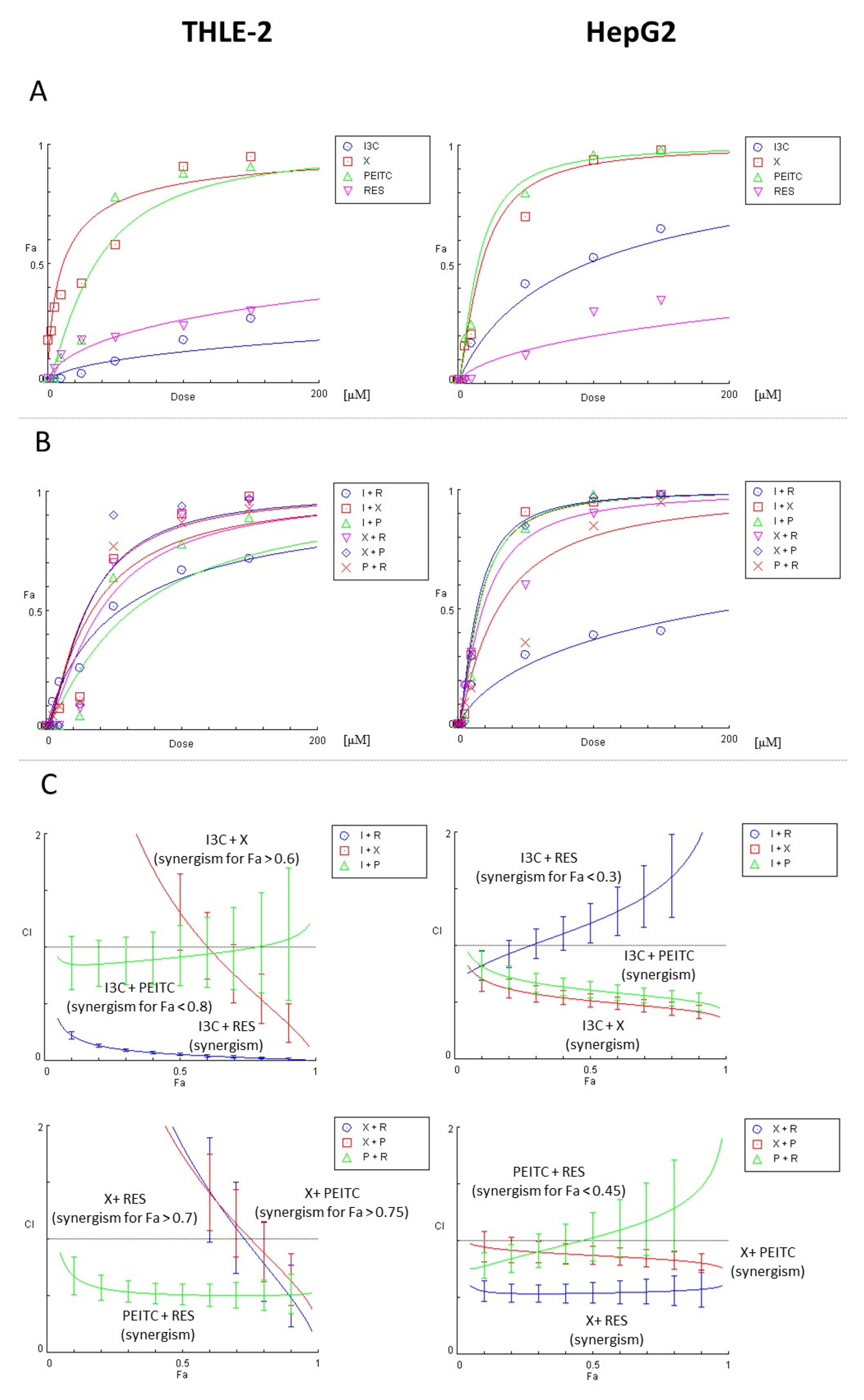
2.9. The Assessment of the Type of Interaction between the Compounds
2.10. Statistical Analysis
3. Results
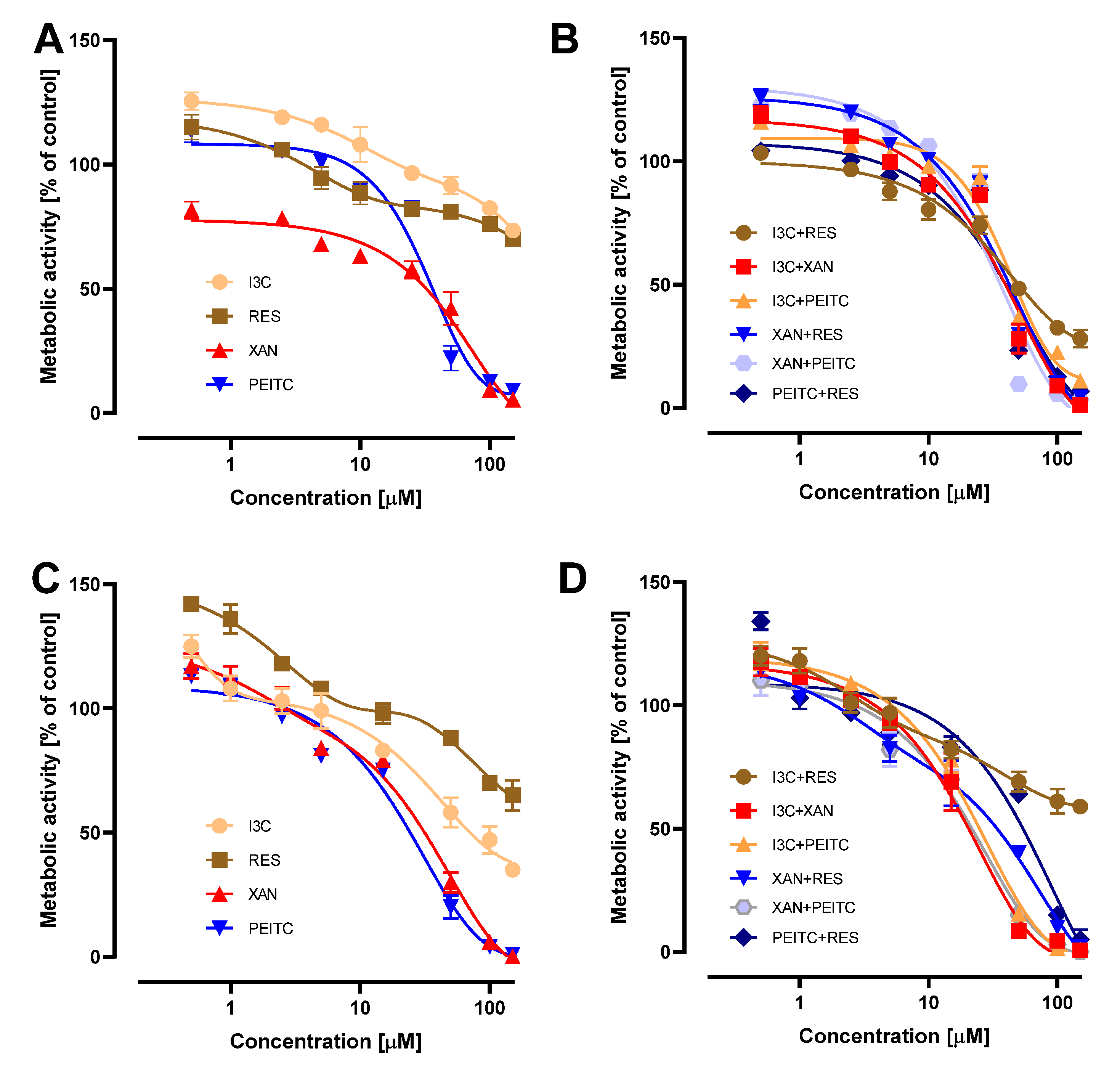
3.1. Impact of X, PEITC, RES, I3C and Their Mixtures on the Cell Viability and Determination of the Type of Interactions between Phytochemicals
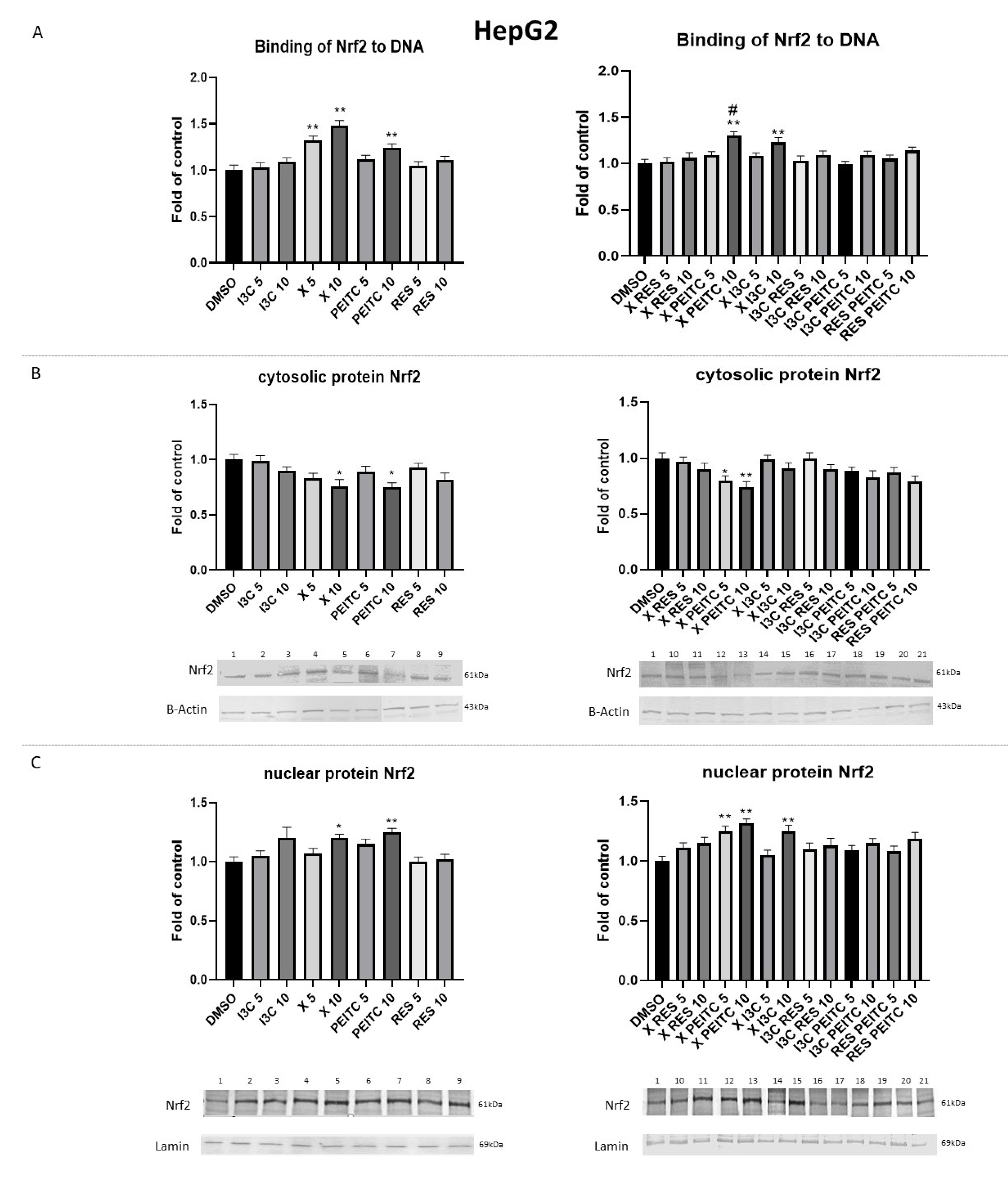
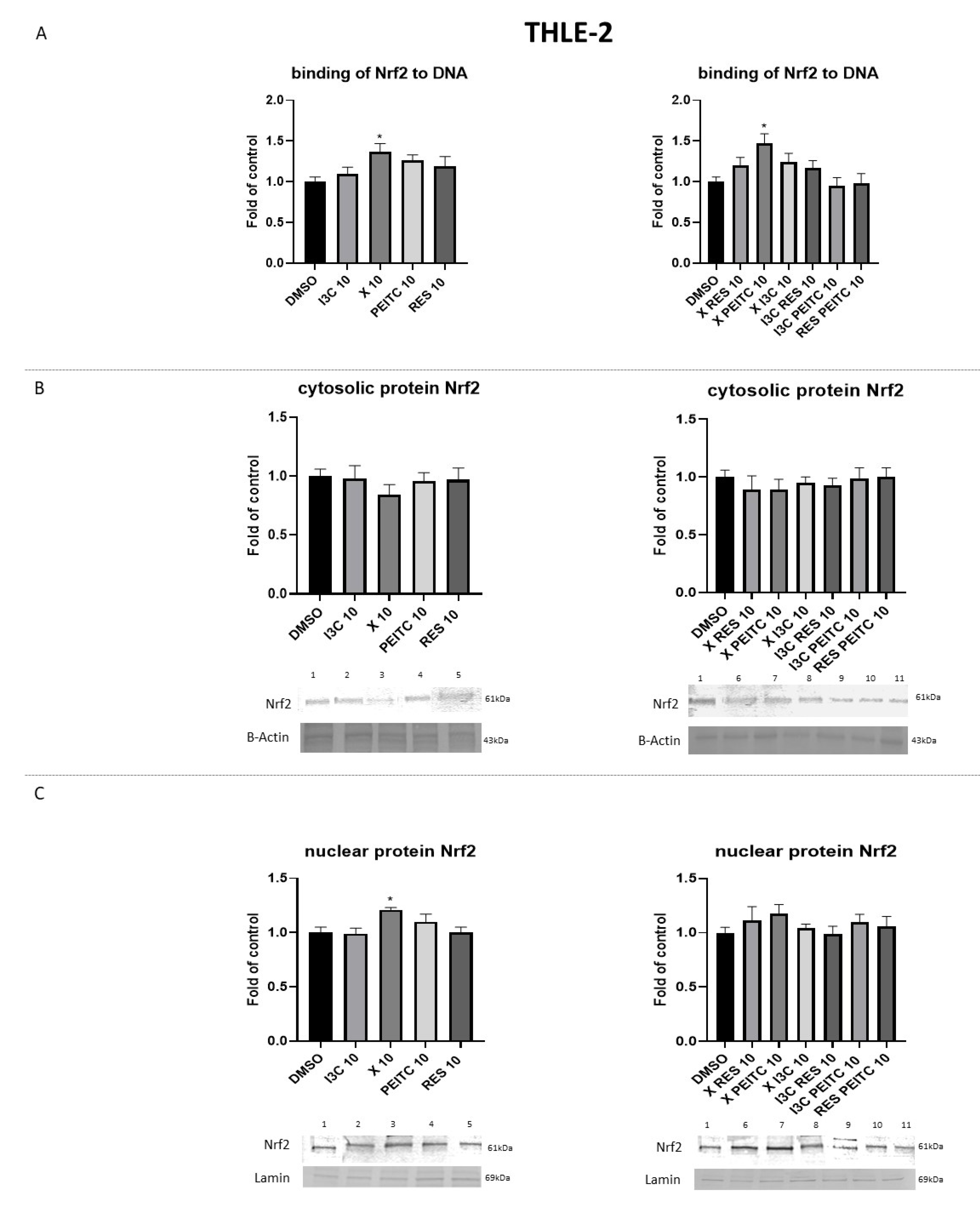
3.2. X, PEITC, RES, I3C, and Their Mixtures Affect the Activation of Nrf2
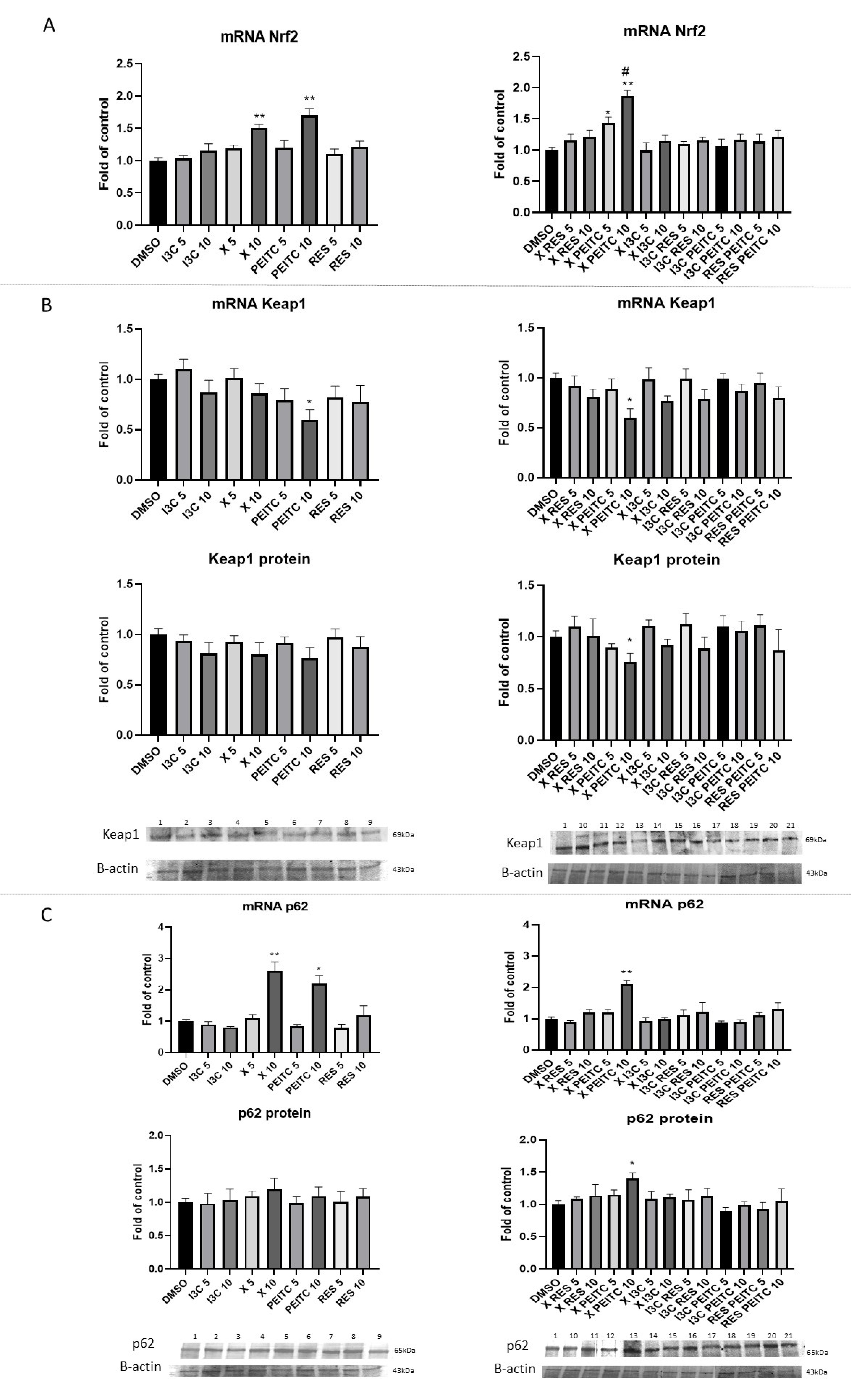
3.3. X, PEITC, RES, I3C, and Their Mixtures Affect the Expression of Nrf2 Target Genes
3.4. X, PEITC, RES, I3C, and Their Mixtures Affect the Expression of Nrf2, Keap1, and p62
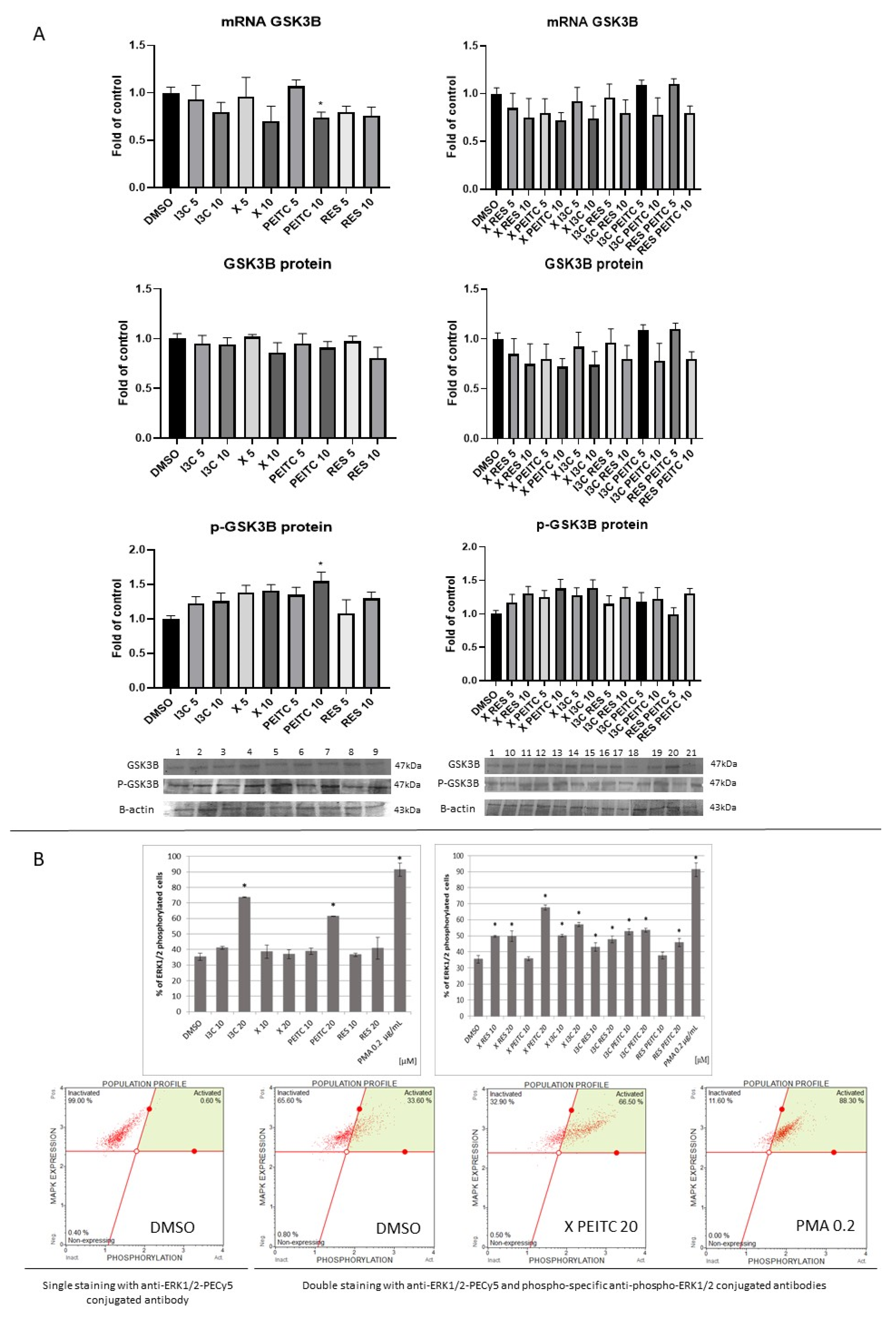
3.5. X, PEITC, RES, I3C and Their Mixtures Affect GSK-3β Expression and Phosphorylation and ERK1/2 Phosphorylation
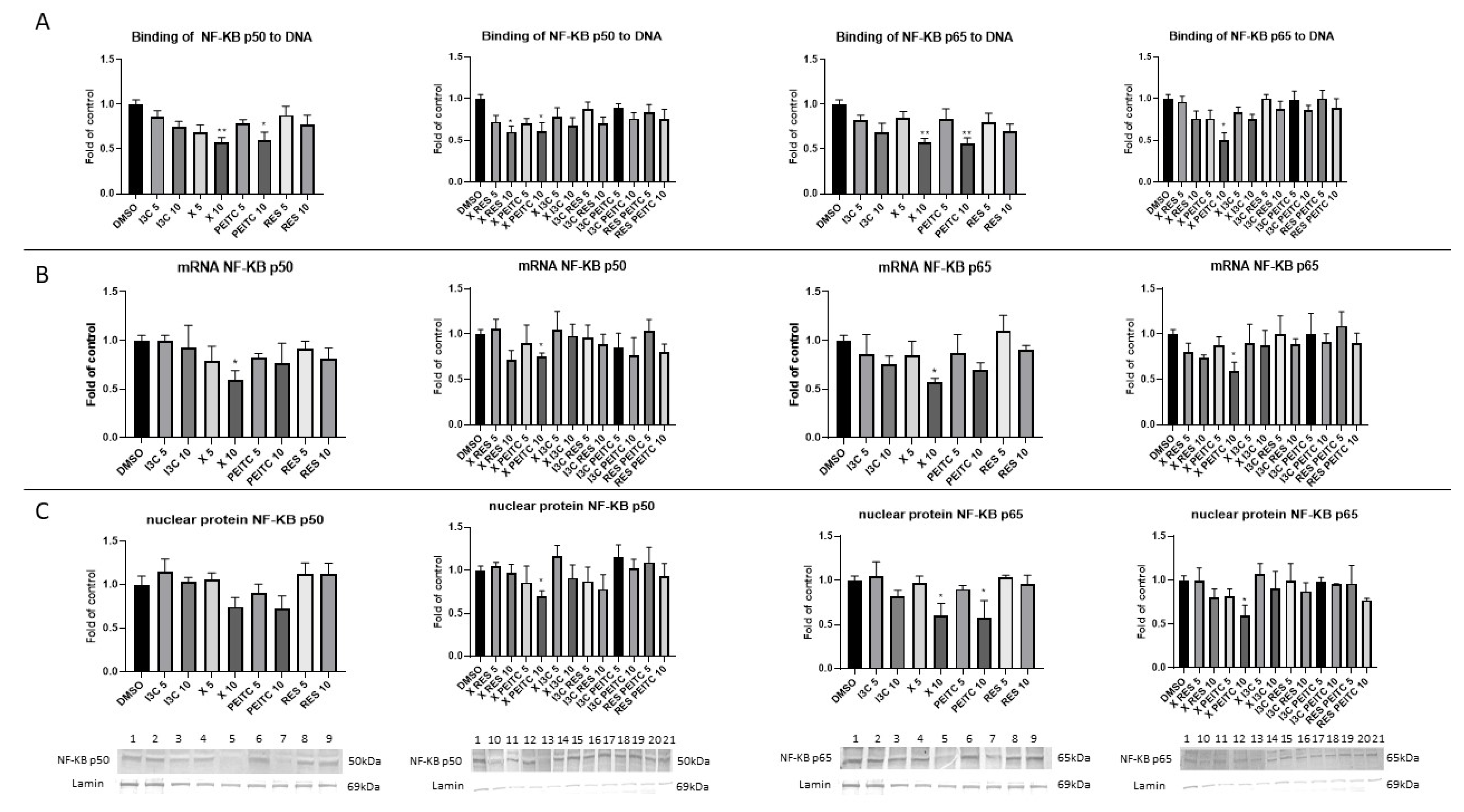
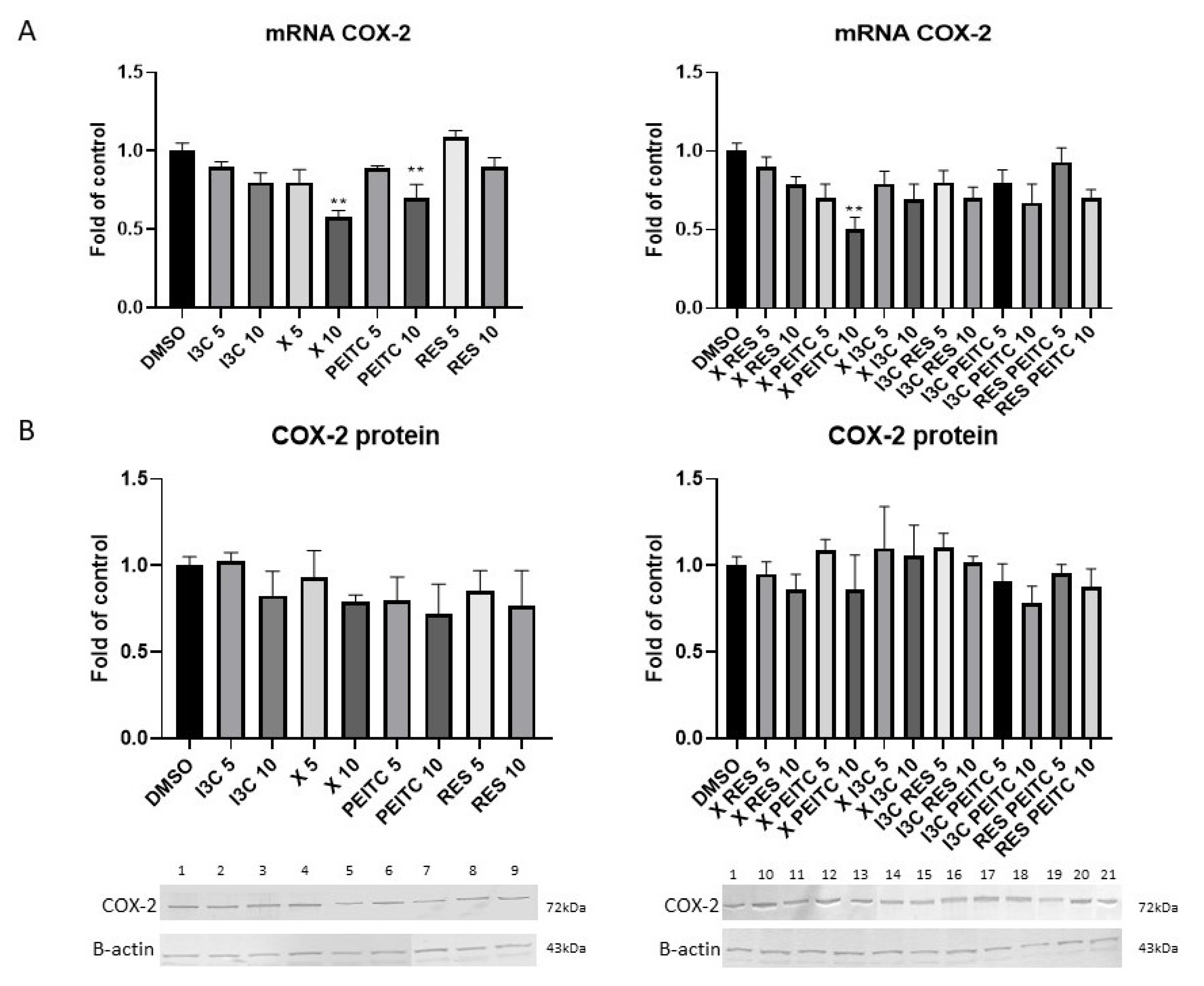
3.6. X, PEITC, RES, I3C, and Their Mixtures Affect the NF-κB Activation and Expression of Its Active Subunits and COX-2 Gene
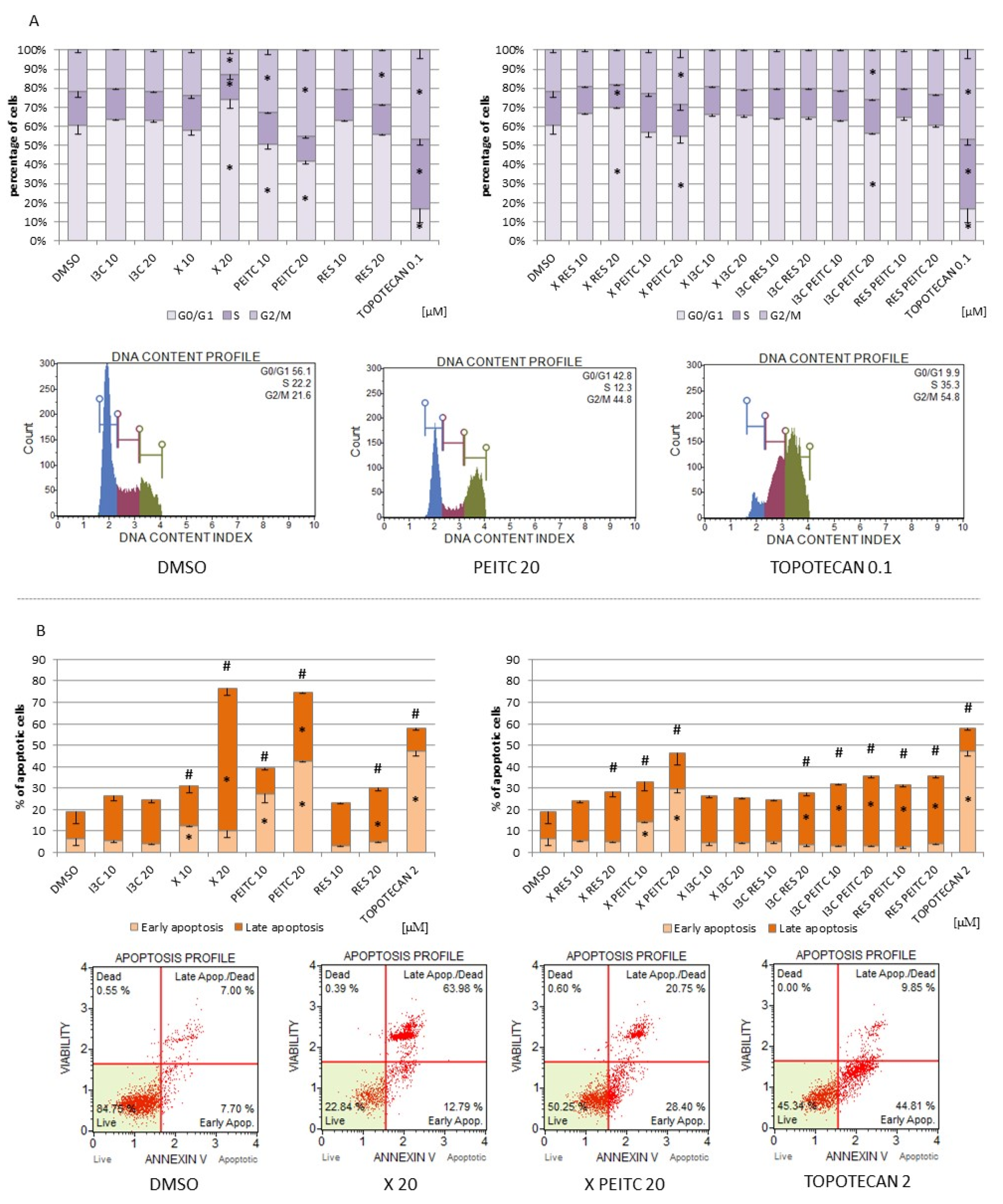
3.7. Impact of X, PEITC, RES, I3C, and Their Mixtures on Cell Cycle Distribution and Apoptosis Induction
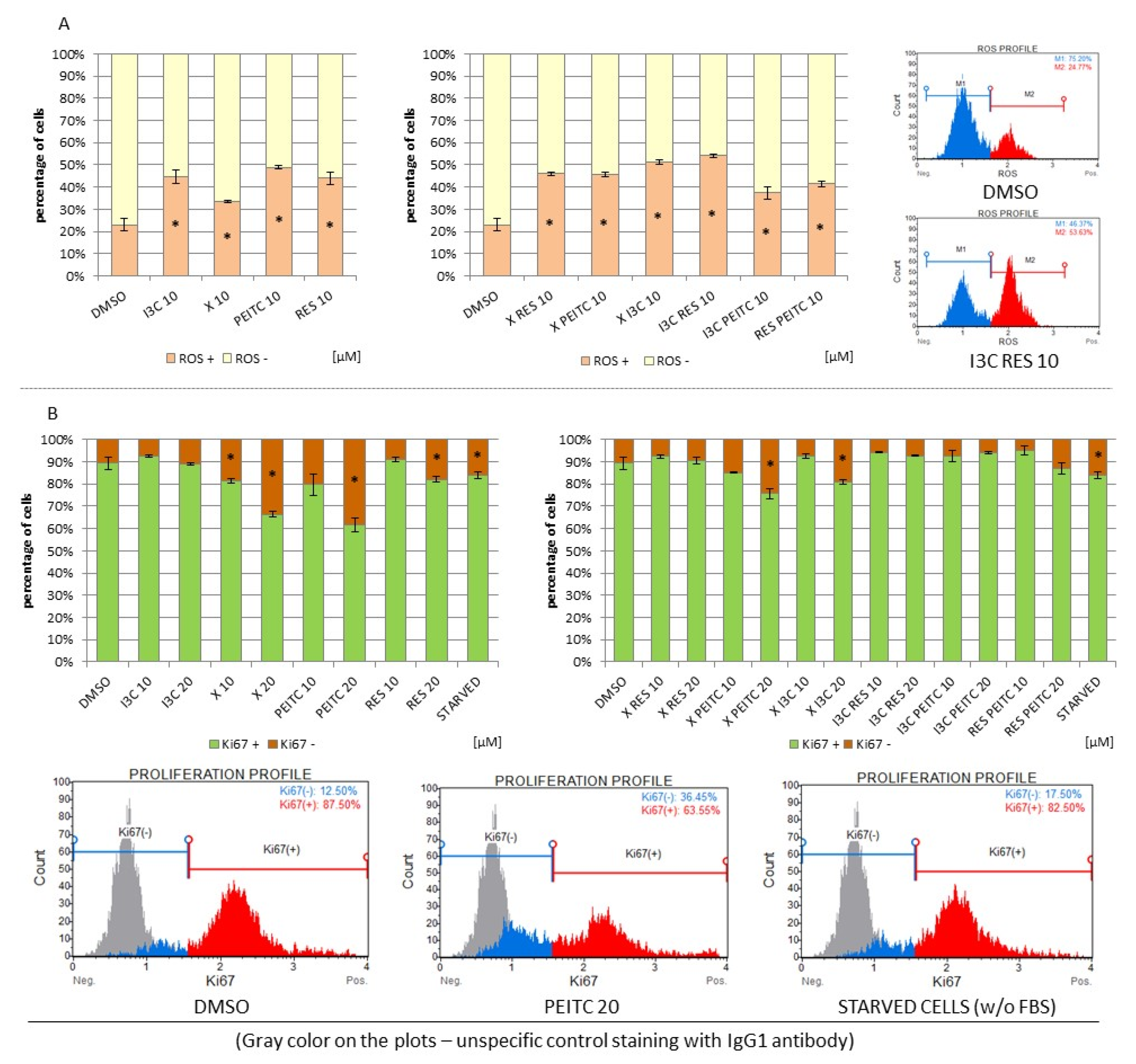
3.8. Impact of X, PEITC, RES, I3C, and Their Mixtures on the Level of Reactive Oxygen Species and Cell Proliferation
3.9. Antitumor Efficacy of PEITC, X and Their Combination in a Human Hepatic Tumor Xenograft Model
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, G.; Karin, M. NF-κB and STAT3—key players in liver inflammation and cancer. Cell Res. 2010, 21, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Napetschnig, J.; Wu, H. Molecular Basis of NF-κB Signaling. Annu. Rev. Biophys. 2013, 42, 443–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.M.; Yang, J.H.; Ki, S.H. Role of the Nrf2-are pathway in liver diseases. Oxidative Med. Cell. Longev. 2013, 2013, 763257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodson, M.; Zhang, D.D. Non-Canonical Activation of NRF2: New Insights and Its Relevance to Disease. Curr. Pathobiol. Rep. 2017, 5, 171–176. [Google Scholar] [CrossRef]
- Gao, A.-M.; Ke, Z.-P.; Shi, F.; Sun, G.-C.; Chen, H. Chrysin enhances sensitivity of BEL-7402/ADM cells to doxorubicin by suppressing PI3K/Akt/Nrf2 and ERK/Nrf2 pathway. Chem. Interact. 2013, 206, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.-M.; Ke, Z.-P.; Wang, J.-N.; Yang, J.-Y.; Chen, S.-Y.; Chen, H. Apigenin sensitizes doxorubicin-resistant hepatocellular carcinoma BEL-7402/ADM cells to doxorubicin via inhibiting PI3K/Akt/Nrf2 pathway. Carcinogenesis 2013, 34, 1806–1814. [Google Scholar] [CrossRef]
- Lee, S.E.; Yang, H.; Jeong, S.I.; Jin, Y.-H.; Park, C.-S.; Park, Y.S. Induction of heme oxygenase-1 inhibits cell death in crotonaldehyde-stimulated HepG2 cells via the PKC-δ-p38-Nrf2 pathway. PLoS ONE 2012, 7, e41676. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef] [Green Version]
- Lau, A.; Zheng, Y.; Tao, S.; Wang, H.; Whitman, S.A.; White, E.; Zhang, D.D. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol. Cell. Biol. 2013, 33, 2436–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.-S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojo, I.A.; Rada, P.; Egea, J.; da Rosa, A.; Lopez, M.G.; Cuadrado, A. Functional interference between glycogen synthase kinase-3 beta and the transcription factor Nrf2 in protection against kainate-induced hippocampal cell death. Mol. Cell. Neurosci. 2008, 39, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Inami, Y.; Waguri, S.; Sakamoto, A.; Kouno, T.; Nakada, K.; Hino, O.; Watanabe, S.; Ando, J.; Iwadate, M.; Yamamoto, M.; et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011, 193, 275–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, Y.; Waguri, S.; Sou, Y.-S.; Kageyama, S.; Hasegawa, J.; Ishimura, R.; Saito, T.; Yang, Y.; Kouno, T.; Fukutomi, T.; et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 2013, 51, 618–631. [Google Scholar] [CrossRef] [Green Version]
- Naidu, S.D.; Suzuki, T.; Yamamoto, M.; Fahey, J.W.; Dinkova-Kostova, A.T. Phenethyl isothiocyanate, a dual activator of transcription factors NRF2 and HSF1. Mol. Nutr. Food Res. 2018, 62, e1700908. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, F.; Paredes-Gonzalez, X.; Kong, A.-N.T. Dietary glucosinolates sulforaphane, phenethyl isothiocyanate, indole-3-carbinol/3,3′-diindolylmethane: Antioxidative stress/inflammation, Nrf2, epigenetics/epigenomics and in vivo cancer chemopreventive efficacy. Curr. Pharmacol. Rep. 2015, 1, 179–196. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Matsuo, Y.; Imafuji, H.; Okubo, T.; Maeda, Y.; Sato, T.; Shamoto, T.; Tsuboi, K.; Morimoto, M.; Takahashi, H.; et al. Xanthohumol inhibits angiogenesis by suppressing nuclear factor-κB activation in pancreatic cancer. Cancer Sci. 2017, 109, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Krajka-Kuźniak, V.; Paluszczak, J.; Baer-Dubowska, W. Xanthohumol induces phase II enzymes via Nrf2 in human hepatocytes in vitro. Toxicol. In Vitro 2013, 27, 149–156. [Google Scholar] [CrossRef]
- Gupta, V.K.; Singh, R.; Sharma, B. Phytochemicals mediated signalling pathways and their implications in cancer chemotherapy: Challenges and opportunities in phytochemicals based drug development: A review. Biochem. Compd. 2017, 5, 2. [Google Scholar] [CrossRef]
- Singh, A.A.; Patil, M.P.; Kang, M.-J.; Niyonizigiye, I.; Kim, G.-D. Biomedical application of indole-3-carbinol: A mini-review. Phytochem. Lett. 2020, 41, 49–54. [Google Scholar] [CrossRef]
- Harish, V.; Haque, E.; Śmiech, M.; Taniguchi, H.; Jamieson, S.; Tewari, D.; Bishayee, A. Xanthohumol for human malignancies: Chemistry, pharmacokinetics and molecular targets. Int. J. Mol. Sci. 2021, 22, 4478. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Zielińska-Przyjemska, M.; Wierzchowski, M.; Kleszcz, R.; Studzińska-Sroka, E.; Kaczmarek, M.; Paluszczak, J.; Cielecka-Piontek, J.; Krajka-Kuźniak, V. Methoxy-stilbenes downregulate the transcription of Wnt/β-catenin-dependent genes and lead to cell cycle arrest and apoptosis in human T98G glioblastoma cells. Adv. Med. Sci. 2020, 66, 6–20. [Google Scholar] [CrossRef]
- Sundaram, M.K.; Preetha, R.; Haque, S.; Akhter, N.; Khan, S.; Ahmed, S.; Hussain, A. Dietary isothiocyanates inhibit cancer progression by modulation of epigenome. Semin. Cancer Biol. 2021, in press. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Cykowiak, M.; Szaefer, H.; Kleszcz, R.; Baer-Dubowska, W. Combination of xanthohumol and phenethyl isothiocyanate inhibits NF-κB and activates Nrf2 in pancreatic cancer cells. Toxicol. In Vitro 2020, 65, 104799. [Google Scholar] [CrossRef] [PubMed]
- Cykowiak, M.; Krajka-Kuźniak, V.; Baer-Dubowska, W. Combinations of phytochemicals more efficiently than single components activate Nrf2 and induce the expression of antioxidant enzymes in pancreatic cancer cells. Nutr. Cancer 2021, 1–16. [Google Scholar] [CrossRef]
- Zhang, L.; Virgous, C.; Si, H. Synergistic anti-inflammatory effects and mechanisms of combined phytochemicals. J. Nutr. Biochem. 2019, 69, 19–30. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Baer-Dubowska, W. Modulation of Nrf2 and NF-κB signaling pathways by naturally occurring compounds in relation to cancer prevention and therapy are combinations better than single compounds? Int. J. Mol. Sci. 2021, 22, 8223. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krajka-Kuźniak, V.; Paluszczak, J.; Szaefer, H.; Baer-Dubowska, W. The activation of the Nrf2/ARE pathway in HepG2 hepatoma cells by phytochemicals and subsequent modulation of phase II and antioxidant enzyme expression. J. Physiol. Biochem. 2015, 71, 227–238. [Google Scholar] [CrossRef]
- Morris, M.E.; Dave, R.A. Pharmacokinetics and pharmacodynamics of phenethyl isothiocyanate: Implications in breast cancer prevention. AAPS J. 2014, 16, 705–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loureiro, P.V.; Jiménez, I.H.; Sendón, R.; De Quirós, A.R.-B.; Barbosa-Pereira, L. Determination of xanthohumol in hops, food supplements and beers by HPLC. Foods 2019, 8, 435. [Google Scholar] [CrossRef] [Green Version]
- Lister, A.; Nedjadi, T.; Kitteringham, N.R.; Campbell, F.; Costello, E.; Lloyd, B.; Copple, I.M.; Williams, S.; Owen, A.; Neoptolemos, J.P.; et al. Nrf2 is overexpressed in pancreatic cancer: Implications for cell proliferation and therapy. Mol. Cancer 2011, 10, 37. [Google Scholar] [CrossRef] [Green Version]
- Bogen, K.T. Low-dose dose–response for in vitro Nrf2-ARE activation in human HepG2 cells. Dose-Response 2017, 15, 1559325817699696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinkova-Kostova, A.; Talalay, P. NAD(P)H: Quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010, 501, 116–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.Z.; Ke, Y.; Misra, H.P.; Trush, M.A.; Li, Y.R.; Zhu, H.; Jia, Z. Mechanistic studies of cancer cell mitochondria- and NQO1-mediated redox activation of beta-lapachone, a potentially novel anticancer agent. Toxicol. Appl. Pharmacol. 2014, 281, 285–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na, H.-K.; Surh, Y.-J. Modulation of Nrf2-mediated antioxidant and detoxifying enzyme induction by the green tea polyphenol EGCG. Food Chem. Toxicol. 2008, 46, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Baeuerle, P. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 1991, 10, 3805–3817. [Google Scholar] [CrossRef]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef] [Green Version]
- Raghunath, A.; Sundarraj, K.; Arfuso, F.; Sethi, G.; Perumal, E. Dysregulation of Nrf2 in hepatocellular carcinoma: Role in cancer progression and chemoresistance. Cancers 2018, 10, 481. [Google Scholar] [CrossRef] [Green Version]
- Khandelwal, N.; Simpson, J.; Taylor, G.; Rafique, S.; Whitehouse, A.; Hiscox, J.; Stark, A.L. Nucleolar NF-κB/RelA mediates apoptosis by causing cytoplasmic relocalization of nucleophosmin. Cell Death Differ. 2011, 18, 1889–1903. [Google Scholar] [CrossRef] [Green Version]


| Temperature | Time | |
|---|---|---|
| Enzyme activation | 95 °C | 5 min |
| 50 cycles | 95 °C | 15 s |
| 56 °C | 20 s | |
| 72 °C | 40 s | |
| Final extension | 72 °C | 5 min |
| Forward Primer | Reverse Primer | |
|---|---|---|
| PBGD | 5′TCAGATAGCATACAAGAGACC | 5′TGGAATGTTACGAGCAGTG |
| TBP | 5′GGCACCACTCCACTGTATC | 5′GGGATTATATTCGGCGTTTCG |
| Nrf2 | 5′ATTGCTACTAATCAGGCTCAG | 5′GTTTGGCTTCTGGACTTGG |
| SOD | 5′CGACAGAAGGAAAGTAATG | 5′TGGATAGAGGATTAAAGTGAGG |
| CAT | 5′TGGACAAGTACAATGCTGAG | 5′TTACACGGATGAACGCTAAG |
| NQO1 | 5′CAATTCAGAGTGGCATTC | 5′GAAGTTTAGGTCAAAGAGG |
| KEAP1 | 5′ATGGGCGAGAAGTGTGTC | 5′TCTGCTCAGCGAAGTTGG |
| p62 | 5′TCTGGGCATTGAAGTTGA | 5′CTCTGTGCTGGAACTCTC |
| GSK-3B | 5′ACCCAAATGTAAACTACCAAATG | 5′TCCACGGTCTCCAGTATTAGC |
| NF-κB p50 | 5′ATCATCCACCTTCATTCTCAA | 5′AATCCTCCACCACATCTTCC |
| NF-κB p65 | 5′CGCCTGTCCTTTCTCATC | 5′ACCTCAATGTCCTCTTTCTG |
| COX-2 | 5′CCTGTGCCTGATGATTGC | 5′CAGCCCGTTGGTGAAAGC |
| HepG2 | THLE-2 | |||
|---|---|---|---|---|
| IC50 | ±SEM | IC50 | ±SEM | |
| I3C | N/A | N/A | ||
| X | 34.2 | 2.3 | 37.0 | 3.0 |
| PEITC | 31.0 | 2.0 | 38.0 | 2.0 |
| RES | N/A | N/A | ||
| X + I3C | 30.2 | 0.6 | 40.7 | 1.0 |
| X + PEITC | 26.5 | 1.6 | 37.5 | 1.2 |
| X + RES | 38.5 | 5.5 | 42.0 | 0.8 |
| I3C + PEITC | 32.2 | 1.0 | 44.5 | 0.8 |
| I3C + RES | N/A | 48.5 | 0.5 | |
| RES + PEITC | 63.8 | 2.0 | 39.5 | 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cykowiak, M.; Krajka-Kuźniak, V.; Kleszcz, R.; Kucińska, M.; Szaefer, H.; Piotrowska-Kempisty, H.; Plewiński, A.; Murias, M.; Baer-Dubowska, W. Comparison of the Impact of Xanthohumol and Phenethyl Isothiocyanate and Their Combination on Nrf2 and NF-κB Pathways in HepG2 Cells In Vitro and Tumor Burden In Vivo. Nutrients 2021, 13, 3000. https://doi.org/10.3390/nu13093000
Cykowiak M, Krajka-Kuźniak V, Kleszcz R, Kucińska M, Szaefer H, Piotrowska-Kempisty H, Plewiński A, Murias M, Baer-Dubowska W. Comparison of the Impact of Xanthohumol and Phenethyl Isothiocyanate and Their Combination on Nrf2 and NF-κB Pathways in HepG2 Cells In Vitro and Tumor Burden In Vivo. Nutrients. 2021; 13(9):3000. https://doi.org/10.3390/nu13093000
Chicago/Turabian StyleCykowiak, Marta, Violetta Krajka-Kuźniak, Robert Kleszcz, Małgorzata Kucińska, Hanna Szaefer, Hanna Piotrowska-Kempisty, Adam Plewiński, Marek Murias, and Wanda Baer-Dubowska. 2021. "Comparison of the Impact of Xanthohumol and Phenethyl Isothiocyanate and Their Combination on Nrf2 and NF-κB Pathways in HepG2 Cells In Vitro and Tumor Burden In Vivo" Nutrients 13, no. 9: 3000. https://doi.org/10.3390/nu13093000
APA StyleCykowiak, M., Krajka-Kuźniak, V., Kleszcz, R., Kucińska, M., Szaefer, H., Piotrowska-Kempisty, H., Plewiński, A., Murias, M., & Baer-Dubowska, W. (2021). Comparison of the Impact of Xanthohumol and Phenethyl Isothiocyanate and Their Combination on Nrf2 and NF-κB Pathways in HepG2 Cells In Vitro and Tumor Burden In Vivo. Nutrients, 13(9), 3000. https://doi.org/10.3390/nu13093000