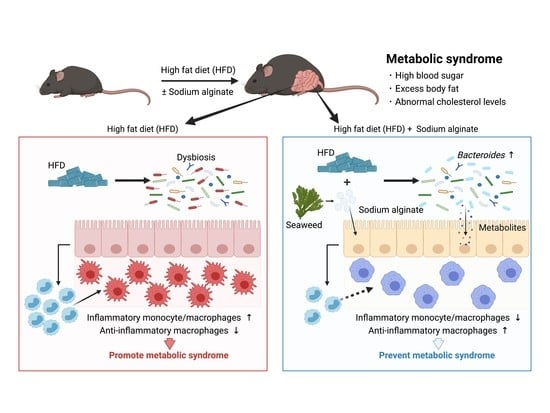
Seaweed Dietary Fiber Sodium Alginate Suppresses the Migration of Colonic Inflammatory Monocytes and Diet-Induced Metabolic Syndrome via the Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Oral Glucose Tolerance Test (OGTT)
2.3. Total Blood Cholesterol
2.4. Histological Analysis
2.5. Quantification of Hepatic Lipid Droplets in Hematoxylin–Eosin (H&E)-Stained Sections
2.6. Sequencing and Processing of Bacterial 16S rRNA Genes in Fecal DNA
2.7. Isolation of Colonic Lamina Propria Cells
2.8. Flow Cytometry
2.9. CE-TOFMS Metabolome Analysis
2.10. Statistical Analyses
3. Results
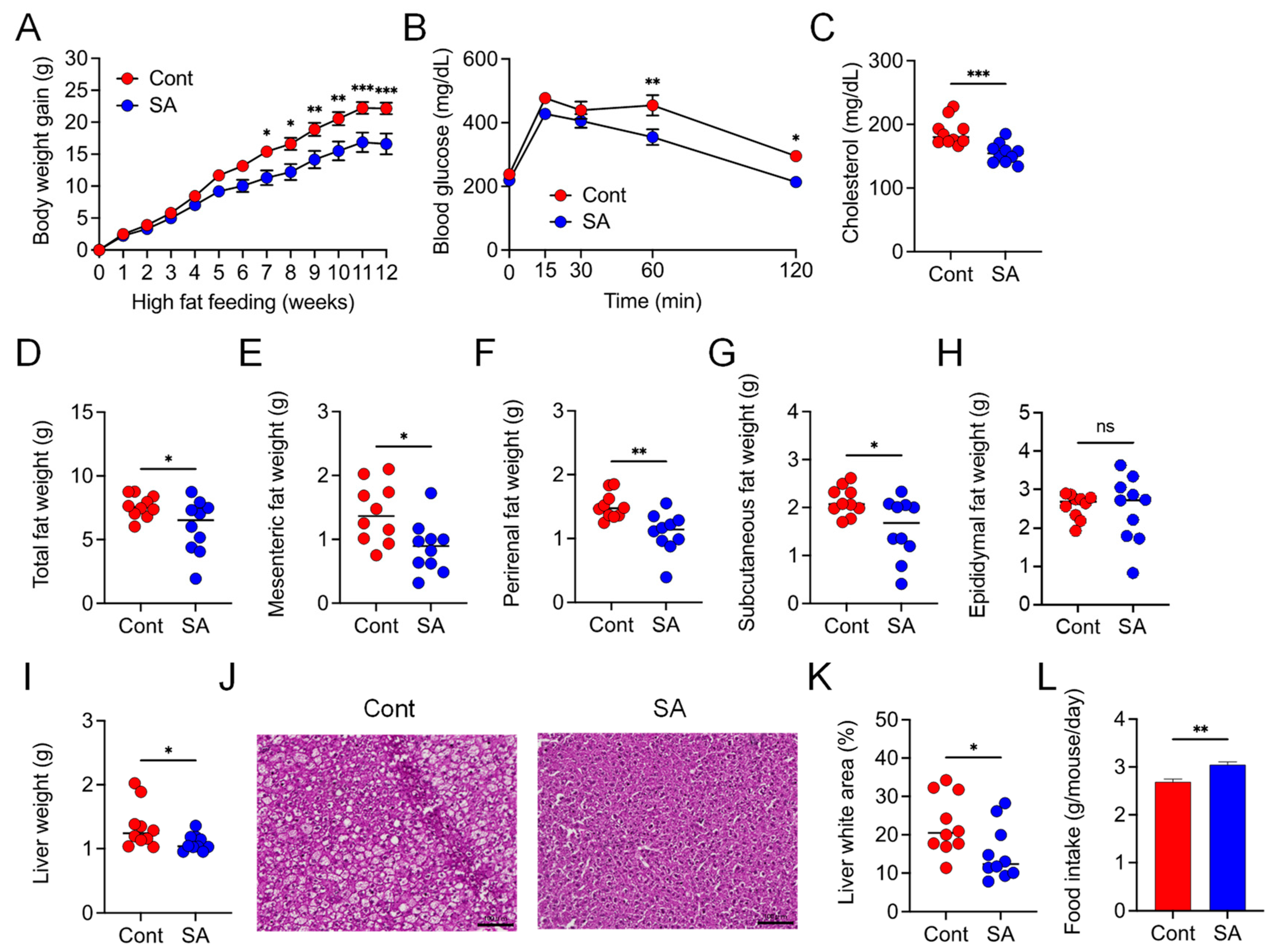
3.1. Dietary Supplementation with SA Suppresses HFD-Induced MetS
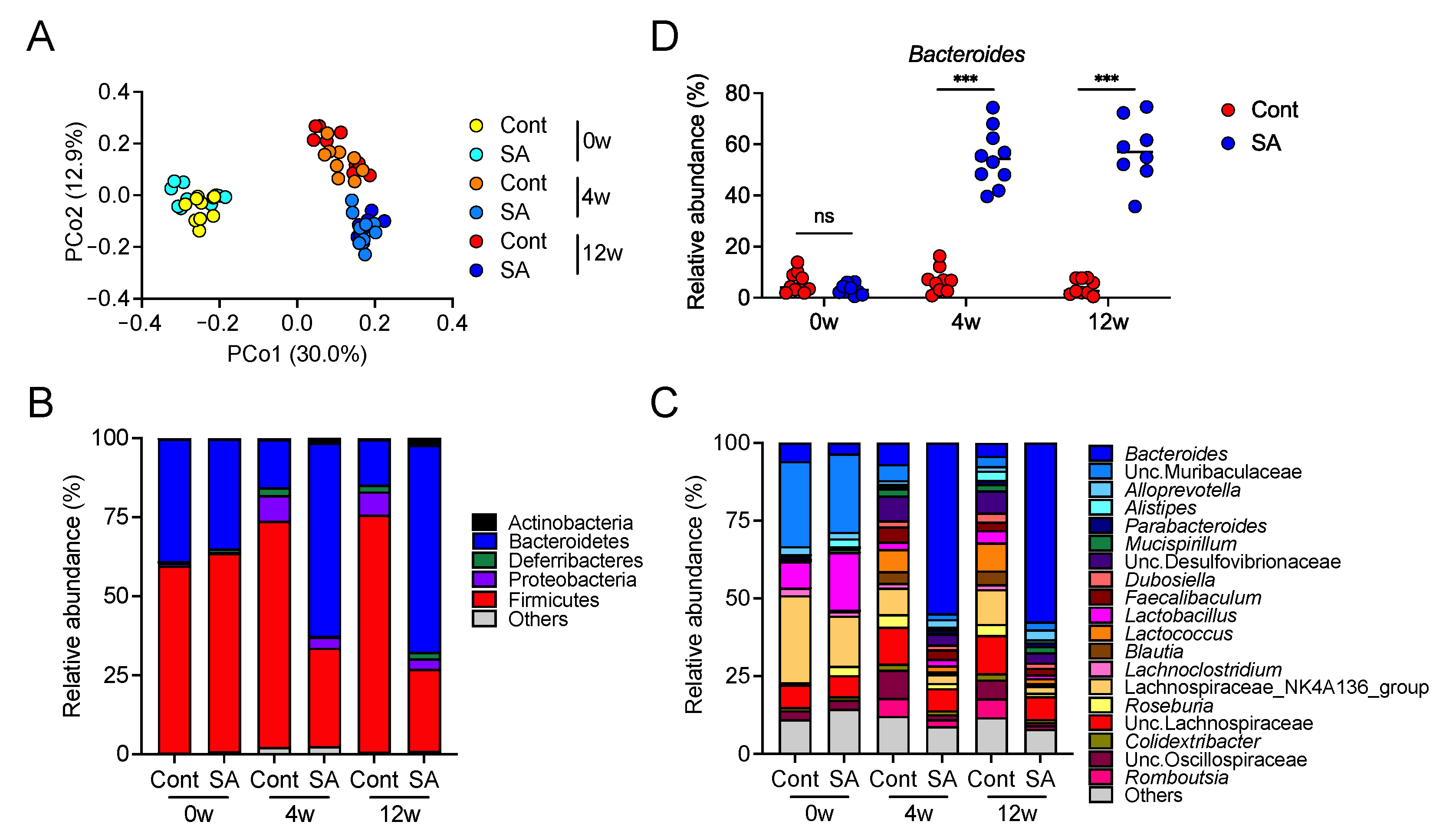
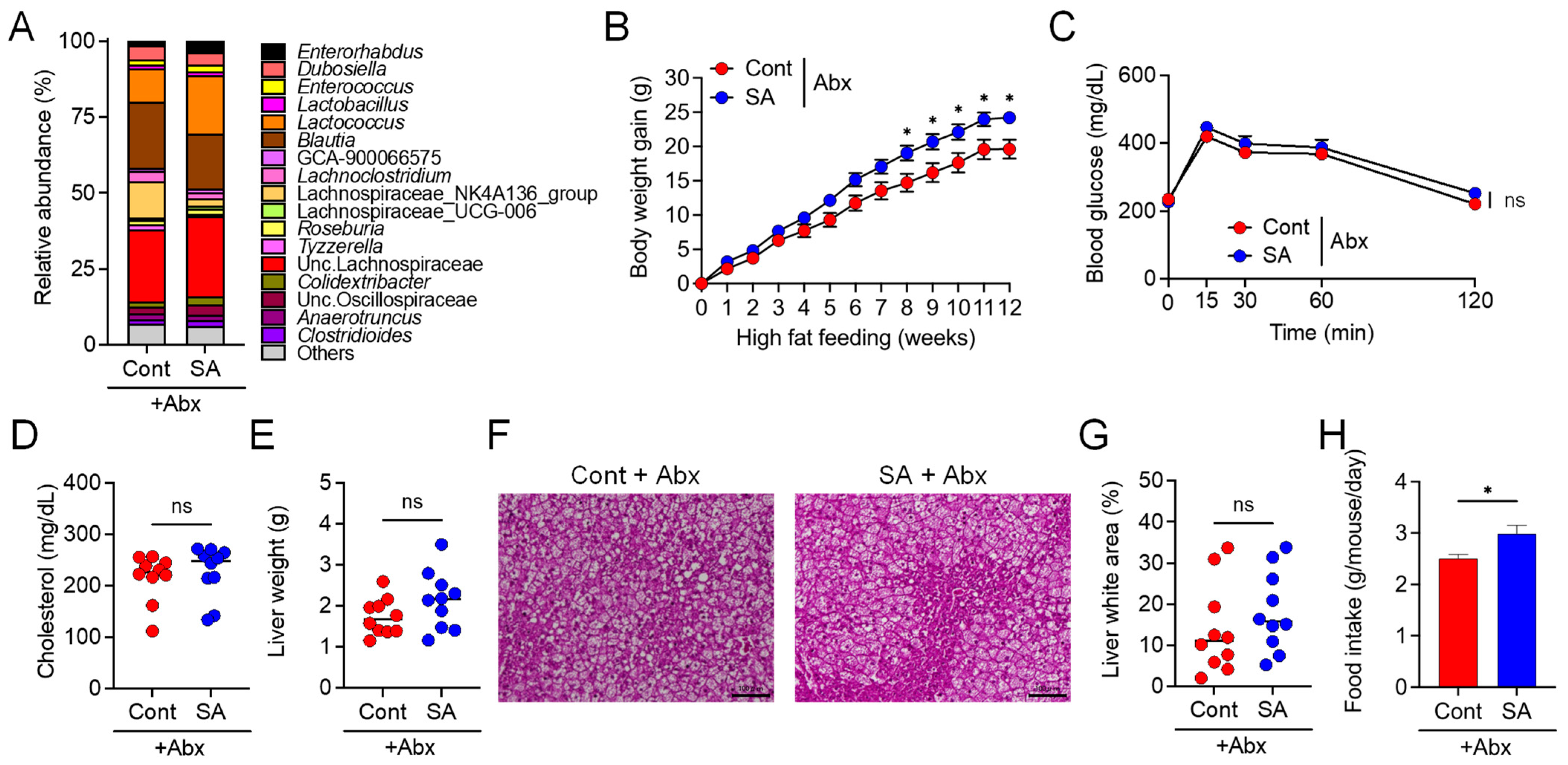
3.2. Suppression of Diet-Induced Obesity by SA Depends on the Gut Microbiota
3.3. SA Reduces Colonic Inflammatory Monocytes in a Gut Microbiota-Dependent Manner
3.4. Increase in Bacteroides Is Negatively Correlated with the Abundance of Colonic Inflammatory Monocytes
3.5. SA Increases Metabolites That Are Positively Correlated with the Abundance of Bacteroides
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Correa, E.; Gonzalez-Perez, I.; Clavel-Perez, P.I.; Contreras-Vargas, Y.; Carvajal, K. Biochemical and nutritional overview of diet-induced metabolic syndrome models in rats: What is the best choice? Nutr. Diabetes 2020, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Cani, P.D. Metabolism in 2013: The gut microbiota manages host metabolism. Nat. Rev. Endocrinol. 2014, 10, 74–76. [Google Scholar] [CrossRef]
- Safari, Z.; Monnoye, M.; Abuja, P.M.; Mariadassou, M.; Kashofer, K.; Gerard, P.; Zatloukal, K. Steatosis and gut microbiota dysbiosis induced by high-fat diet are reversed by 1-week chow diet administration. Nutr. Res. 2019, 71, 72–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, E.A.; Velazquez, K.T.; Herbert, K.M. Influence of high-fat diet on gut microbiota: A driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 515–520. [Google Scholar] [CrossRef]
- Hibberd, A.A.; Yde, C.C.; Ziegler, M.L.; Honore, A.H.; Saarinen, M.T.; Lahtinen, S.; Stahl, B.; Jensen, H.M.; Stenman, L.K. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Benef. Microbes 2019, 10, 121–135. [Google Scholar] [CrossRef]
- Park, D.Y.; Ahn, Y.T.; Park, S.H.; Huh, C.S.; Yoo, S.R.; Yu, R.; Sung, M.K.; McGregor, R.A.; Choi, M.S. Supplementation of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS ONE 2013, 8, e59470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenman, L.K.; Waget, A.; Garret, C.; Klopp, P.; Burcelin, R.; Lahtinen, S. Potential probiotic Bifidobacterium animalis ssp. lactis 420 prevents weight gain and glucose intolerance in diet-induced obese mice. Benef. Microbes 2014, 5, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Everard, A.; Matamoros, S.; Geurts, L.; Delzenne, N.M.; Cani, P.D. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. mBio 2014, 5, e01011–e01014. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.R.; Probert, H.M.; Loo, J.V.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004, 17, 259–275. [Google Scholar] [CrossRef] [Green Version]
- Hay, I.D.; Ur Rehman, Z.; Moradali, M.F.; Wang, Y.; Rehm, B.H. Microbial alginate production, modification and its applications. Microb. Biotechnol. 2013, 6, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brownlee, I.A.; Allen, A.; Pearson, J.P.; Dettmar, P.W.; Havler, M.E.; Atherton, M.R.; Onsoyen, E. Alginate as a source of dietary fiber. Crit. Rev. Food Sci. Nutr. 2005, 45, 497–510. [Google Scholar] [CrossRef]
- Wang, X.; Liu, F.; Gao, Y.; Xue, C.H.; Li, R.W.; Tang, Q.J. Transcriptome analysis revealed anti-obesity effects of the Sodium Alginate in high-fat diet -induced obese mice. Int. J. Biol. Macromol. 2018, 115, 861–870. [Google Scholar] [CrossRef]
- Light, J.M.; Stookey, G.K.; Muhler, J.C. Effect of dietary sodium alginate on radiostrontium absorption and retention in rats. J. Dent. Res. 1970, 49, 442–453. [Google Scholar] [CrossRef]
- Fujii, H.; Baba, T.; Ishida, Y.; Kondo, T.; Yamagishi, M.; Kawano, M.; Mukaida, N. Ablation of the Ccr2 gene exacerbates polyarthritis in interleukin-1 receptor antagonist-deficient mice. Arthritis Rheum. 2011, 63, 96–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimizuka, T.; Seki, N.; Yamaguchi, G.; Akiyama, M.; Higashi, S.; Hase, K.; Kim, Y.G. Amino Acid-Based Diet Prevents Lethal Infectious Diarrhea by Maintaining Body Water Balance in a Murine Citrobacter rodentium Infection Model. Nutrients 2021, 13, 1896. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vazquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [Green Version]
- Goodyear, A.W.; Kumar, A.; Dow, S.; Ryan, E.P. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J. Immunol. Methods 2014, 405, 97–108. [Google Scholar] [CrossRef]
- Hirayama, A.; Nakashima, E.; Sugimoto, M.; Akiyama, S.; Sato, W.; Maruyama, S.; Matsuo, S.; Tomita, M.; Yuzawa, Y.; Soga, T. Metabolic profiling reveals new serum biomarkers for differentiating diabetic nephropathy. Anal. Bioanal. Chem. 2012, 404, 3101–3109. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [Green Version]
- Kawano, Y.; Nakae, J.; Watanabe, N.; Kikuchi, T.; Tateya, S.; Tamori, Y.; Kaneko, M.; Abe, T.; Onodera, M.; Itoh, H. Colonic Pro-inflammatory Macrophages Cause Insulin Resistance in an Intestinal Ccl2/Ccr2-Dependent Manner. Cell Metab. 2016, 24, 295–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bain, C.C.; Scott, C.L.; Uronen-Hansson, H.; Gudjonsson, S.; Jansson, O.; Grip, O.; Guilliams, M.; Malissen, B.; Agace, W.W.; Mowat, A.M. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal. Immunol. 2013, 6, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Plantinga, M.; Guilliams, M.; Vanheerswynghels, M.; Deswarte, K.; Branco-Madeira, F.; Toussaint, W.; Vanhoutte, L.; Neyt, K.; Killeen, N.; Malissen, B.; et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity 2013, 38, 322–335. [Google Scholar] [CrossRef] [Green Version]
- Zigmond, E.; Varol, C.; Farache, J.; Elmaliah, E.; Satpathy, A.T.; Friedlander, G.; Mack, M.; Shpigel, N.; Boneca, I.G.; Murphy, K.M.; et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 2012, 37, 1076–1090. [Google Scholar] [CrossRef] [Green Version]
- Weisberg, S.P.; Hunter, D.; Huber, R.; Lemieux, J.; Slaymaker, S.; Vaddi, K.; Charo, I.; Leibel, R.L.; Ferrante, A.W., Jr. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J. Clin. Investig. 2006, 116, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.Y.; Lee, Y.S.; Kim, Y.; Lee, S.H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.S.; Lim, H.S.; Kim, M.S.; et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauffin Cano, P.; Santacruz, A.; Moya, A.; Sanz, Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS ONE 2012, 7, e41079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Almela, I.; Romani-Perez, M.; Bullich-Vilarrubias, C.; Benitez-Paez, A.; Gomez Del Pulgar, E.M.; Frances, R.; Liebisch, G.; Sanz, Y. Bacteroides uniformis combined with fiber amplifies metabolic and immune benefits in obese mice. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Bao, L.; Wang, K.; Sun, S.; Liao, M.; Liu, C.; Zhou, N.; Ma, K.; Zhang, Y.; Chen, Y.; et al. Activation of a Specific Gut Bacteroides-Folate-Liver Axis Benefits for the Alleviation of Nonalcoholic Hepatic Steatosis. Cell Rep. 2020, 32, 108005. [Google Scholar] [CrossRef] [PubMed]
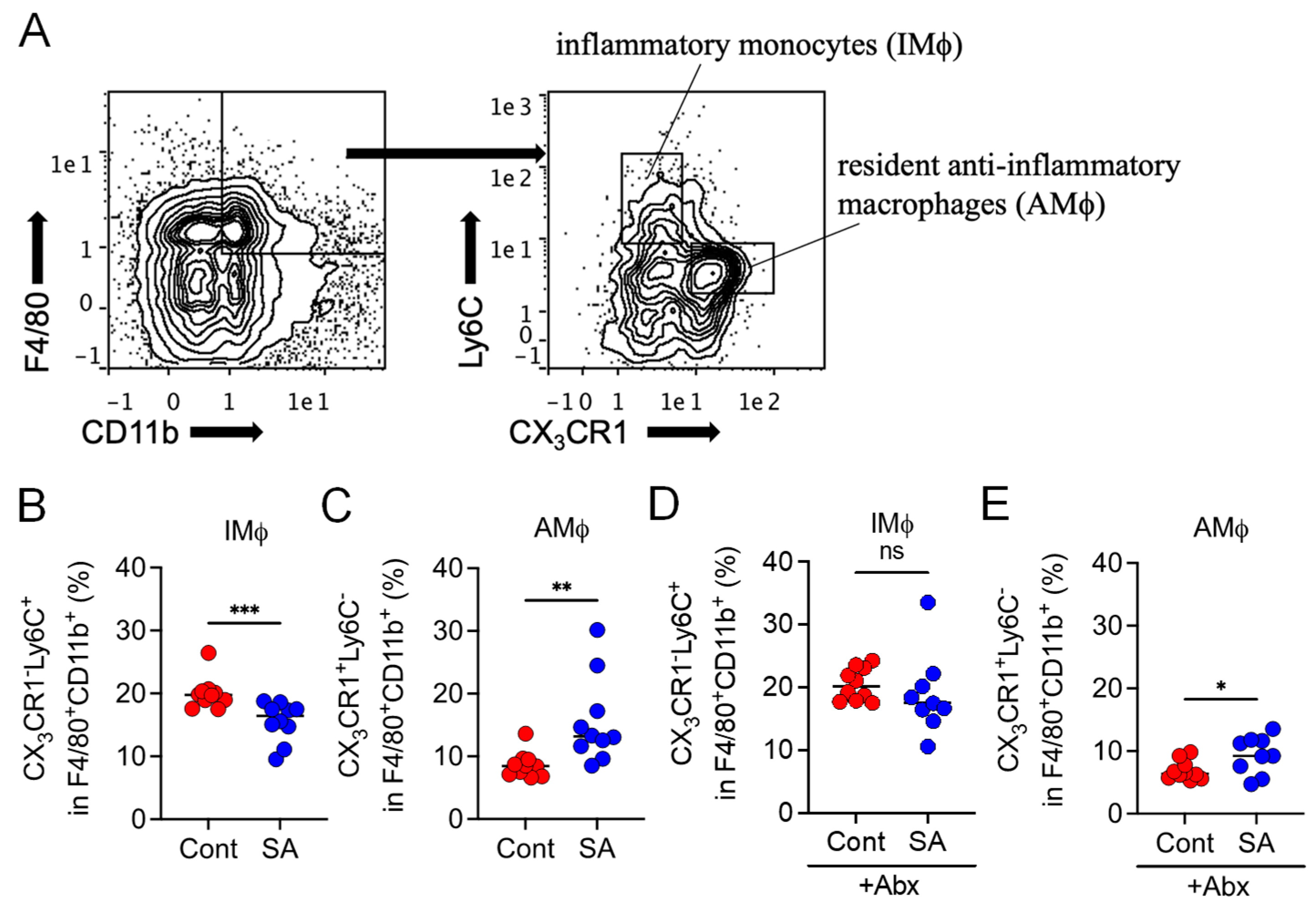
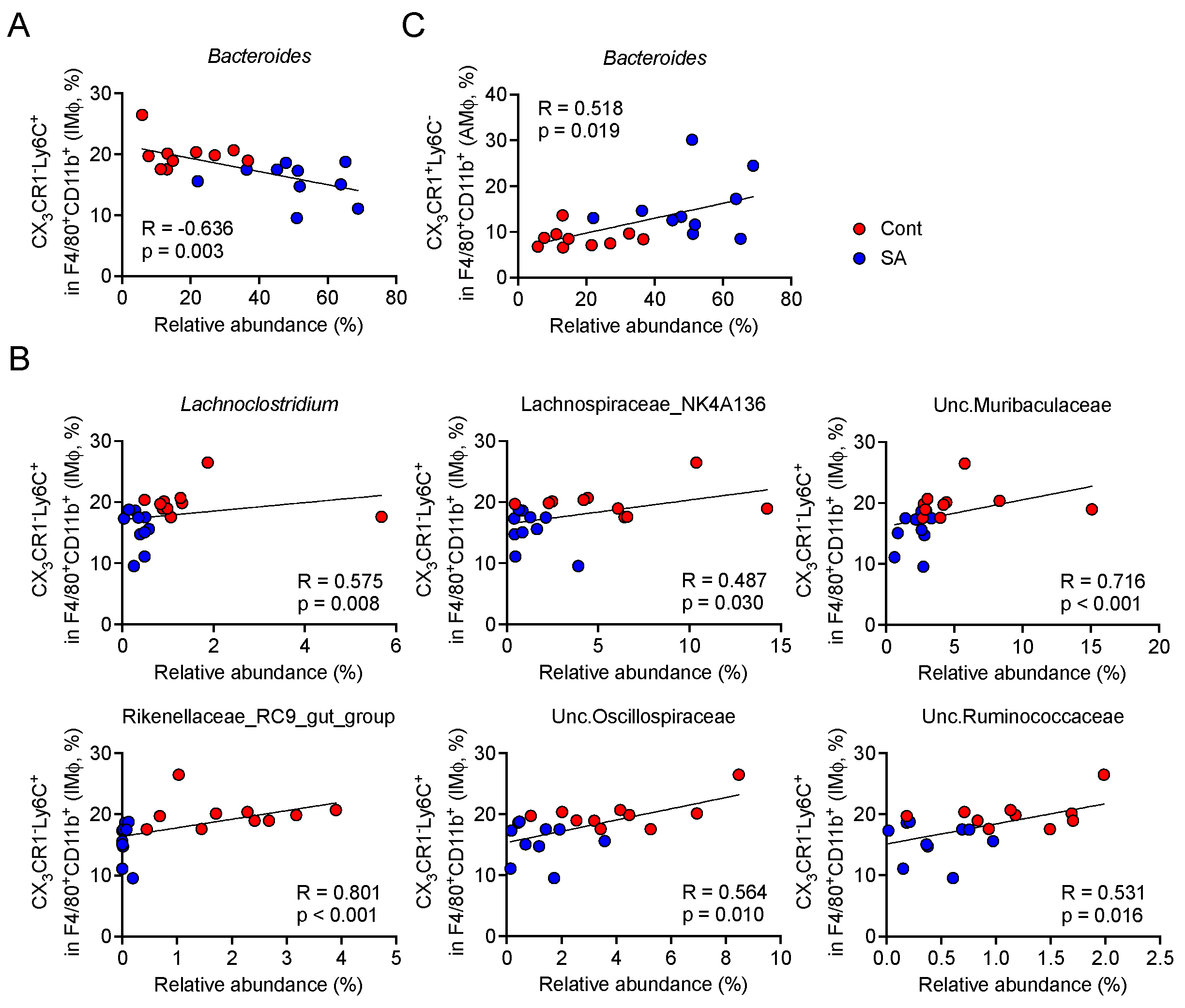
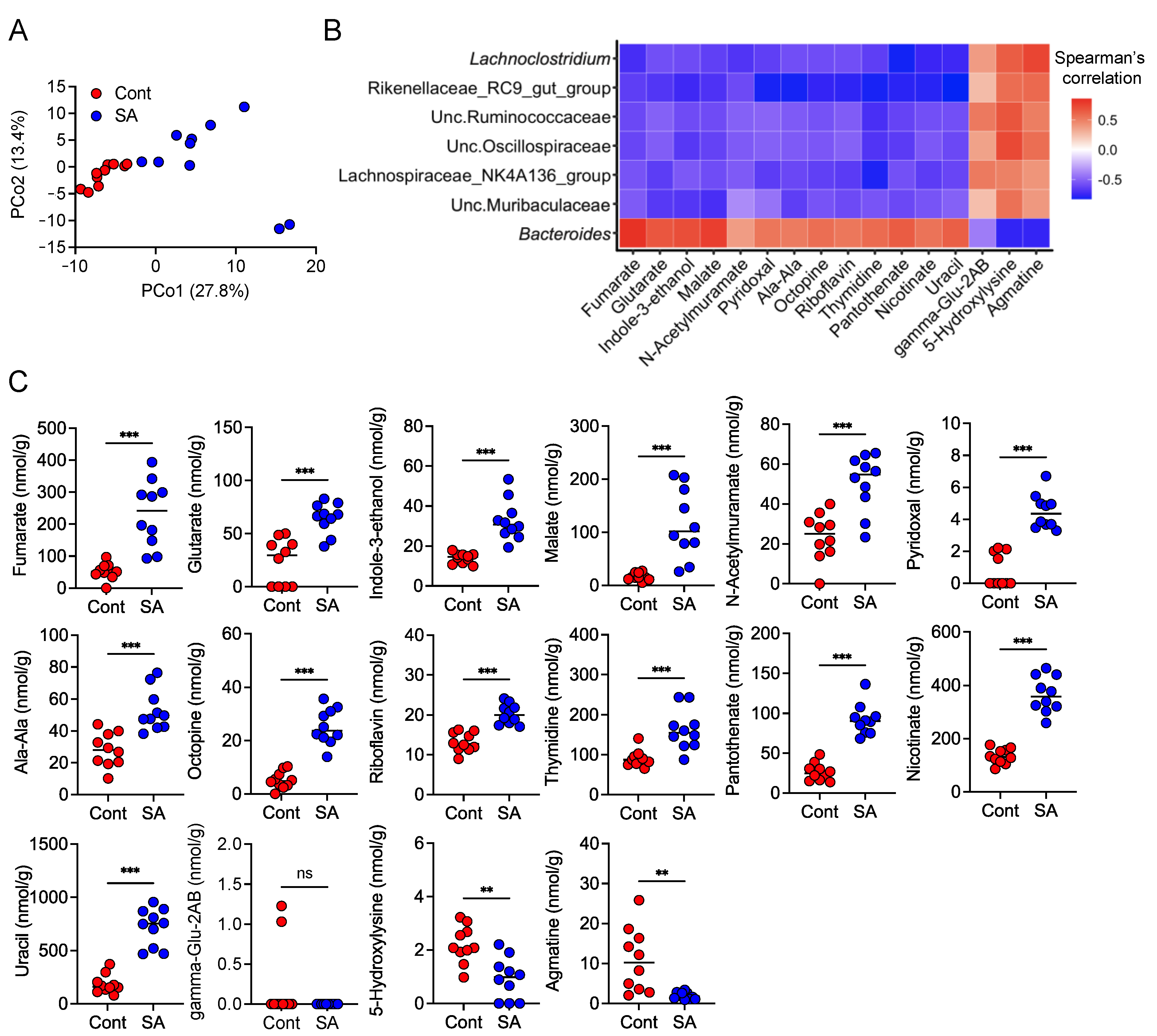
| Product # | D12492 | D18102802 | ||
| g% | kcal% | g% | kcal% | |
| Protein | 26 | 20 | 26 | 20 |
| Carbohydrate | 26 | 20 | 26 | 20 |
| Fat | 35 | 60 | 35 | 60 |
| Total | 100 | 100 | ||
| kcal/g | 5.2 | 5.2 | ||
| Ingredient | g | kcal | g | kcal |
| Casein | 200 | 800 | 200 | 800 |
| L-cystine | 3 | 12 | 3 | 12 |
| Maltodextrin 10 | 125 | 500 | 125 | 500 |
| Sucrose | 68.8 | 275 | 68.8 | 275 |
| Cellulose | 50 | 0 | 11.3 | 0 |
| Soybean oil | 25 | 225 | 25 | 225 |
| Lard | 245 | 2205 | 245 | 2205 |
| Mineral mix, S10026 | 10 | 0 | 10 | 0 |
| Dicalcium phosphate | 13 | 0 | 13 | 0 |
| Calcium carbonate | 5.5 | 0 | 5.5 | 0 |
| Potassium citrate, 1 H2O | 16.5 | 0 | 16.5 | 0 |
| Vitamin mix, V10001 | 10 | 40 | 10 | 40 |
| Choline bitartrate | 2 | 0 | 2 | 0 |
| Sodium alginate | 0 | 0 | 38.7 | 0 |
| FD&C Yellow Dye #5 | 0 | 0 | 0.025 | 0 |
| FD&C Red Dye #40 | 0 | 0 | 0 | 0 |
| FD&C Blue Dye #1 | 0.05 | 0 | 0.025 | 0 |
| Total | 773.85 | 4057 | 773.85 | 4057 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ejima, R.; Akiyama, M.; Sato, H.; Tomioka, S.; Yakabe, K.; Kimizuka, T.; Seki, N.; Fujimura, Y.; Hirayama, A.; Fukuda, S.; et al. Seaweed Dietary Fiber Sodium Alginate Suppresses the Migration of Colonic Inflammatory Monocytes and Diet-Induced Metabolic Syndrome via the Gut Microbiota. Nutrients 2021, 13, 2812. https://doi.org/10.3390/nu13082812
Ejima R, Akiyama M, Sato H, Tomioka S, Yakabe K, Kimizuka T, Seki N, Fujimura Y, Hirayama A, Fukuda S, et al. Seaweed Dietary Fiber Sodium Alginate Suppresses the Migration of Colonic Inflammatory Monocytes and Diet-Induced Metabolic Syndrome via the Gut Microbiota. Nutrients. 2021; 13(8):2812. https://doi.org/10.3390/nu13082812
Chicago/Turabian StyleEjima, Ryuta, Masahiro Akiyama, Hiroki Sato, Sawako Tomioka, Kyosuke Yakabe, Tatsuki Kimizuka, Natsumi Seki, Yumiko Fujimura, Akiyoshi Hirayama, Shinji Fukuda, and et al. 2021. "Seaweed Dietary Fiber Sodium Alginate Suppresses the Migration of Colonic Inflammatory Monocytes and Diet-Induced Metabolic Syndrome via the Gut Microbiota" Nutrients 13, no. 8: 2812. https://doi.org/10.3390/nu13082812
APA StyleEjima, R., Akiyama, M., Sato, H., Tomioka, S., Yakabe, K., Kimizuka, T., Seki, N., Fujimura, Y., Hirayama, A., Fukuda, S., Hase, K., & Kim, Y.-G. (2021). Seaweed Dietary Fiber Sodium Alginate Suppresses the Migration of Colonic Inflammatory Monocytes and Diet-Induced Metabolic Syndrome via the Gut Microbiota. Nutrients, 13(8), 2812. https://doi.org/10.3390/nu13082812