Microbiota and Metabolomic Patterns in the Breast Milk of Subjects with Celiac Disease on a Gluten-Free Diet
Abstract
:1. Introduction
2. Materials and Methods
3. Results
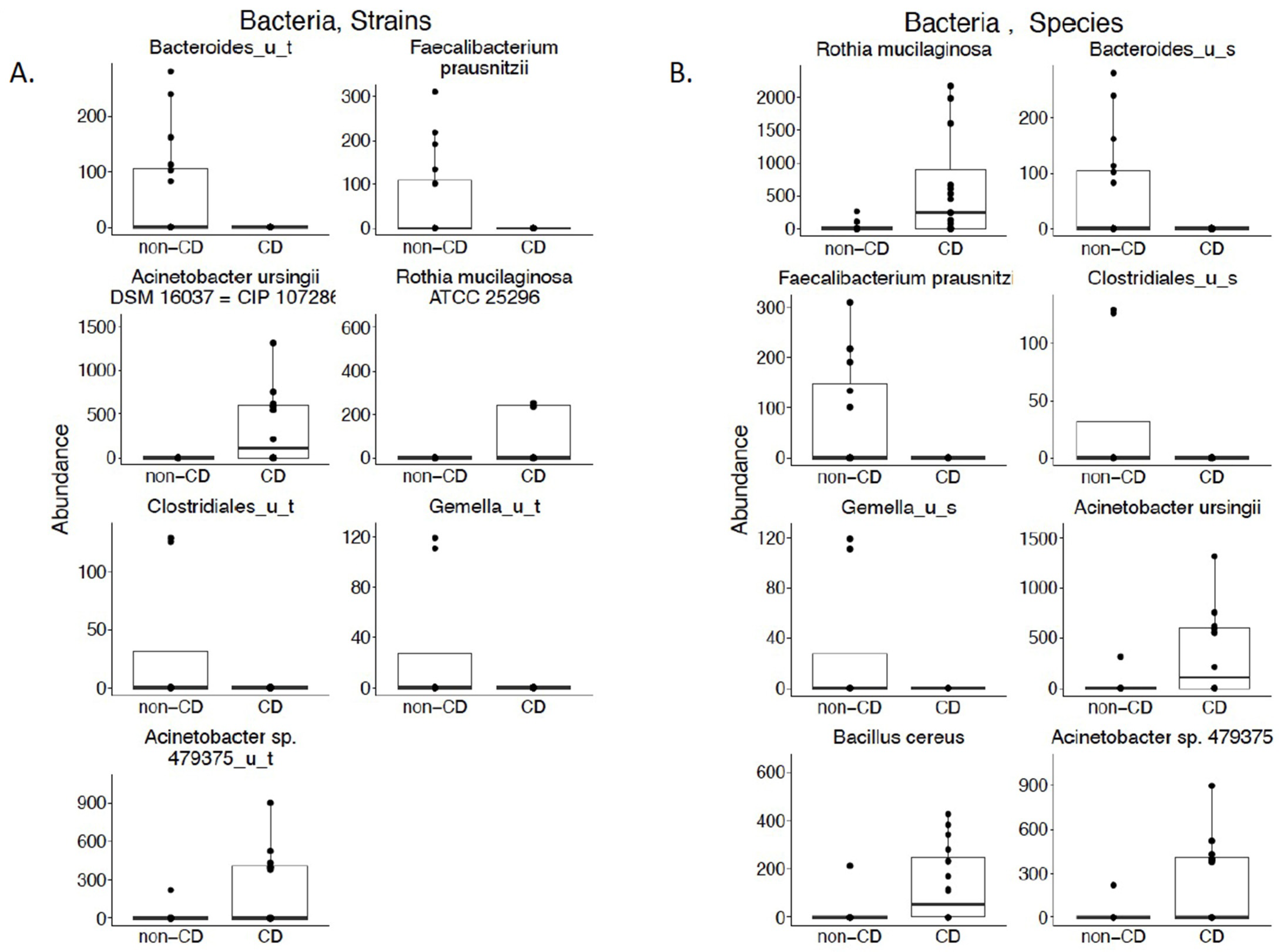
3.1. Bacterial Composition
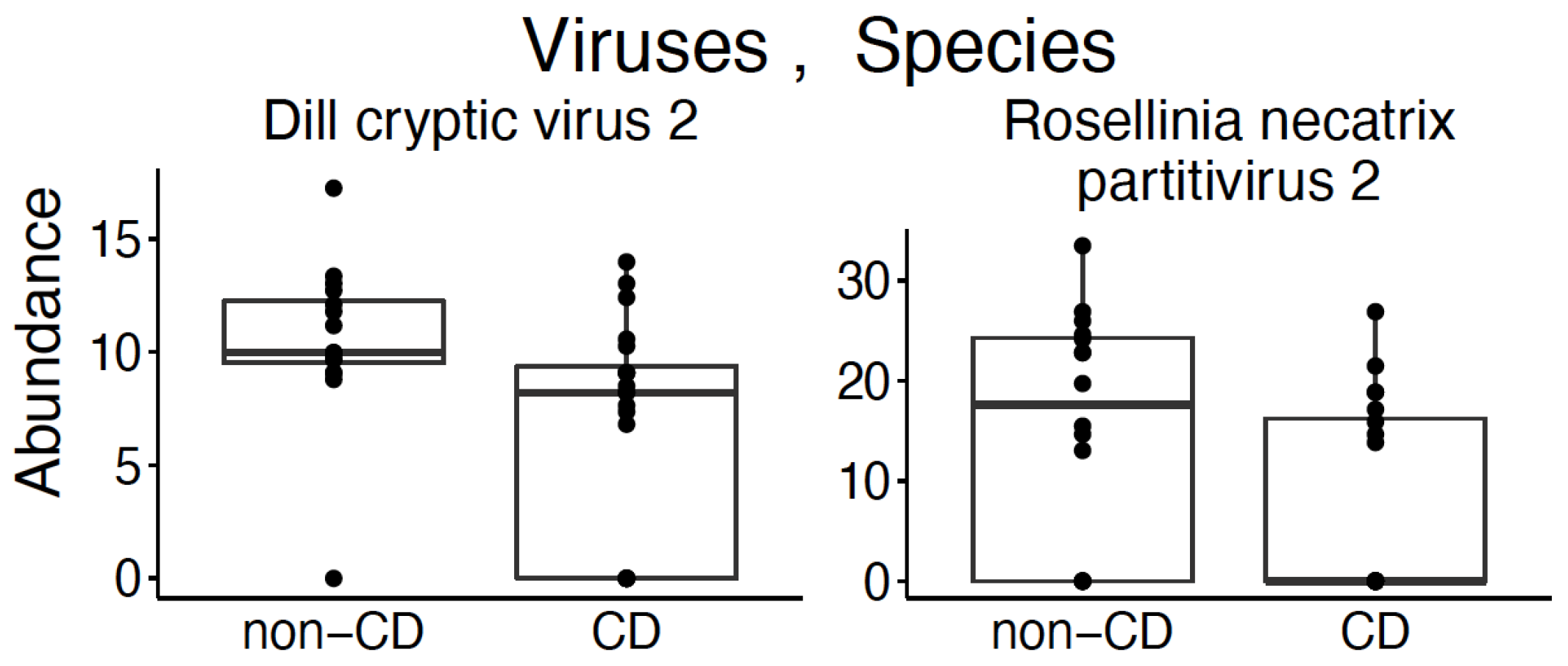
3.2. Virome
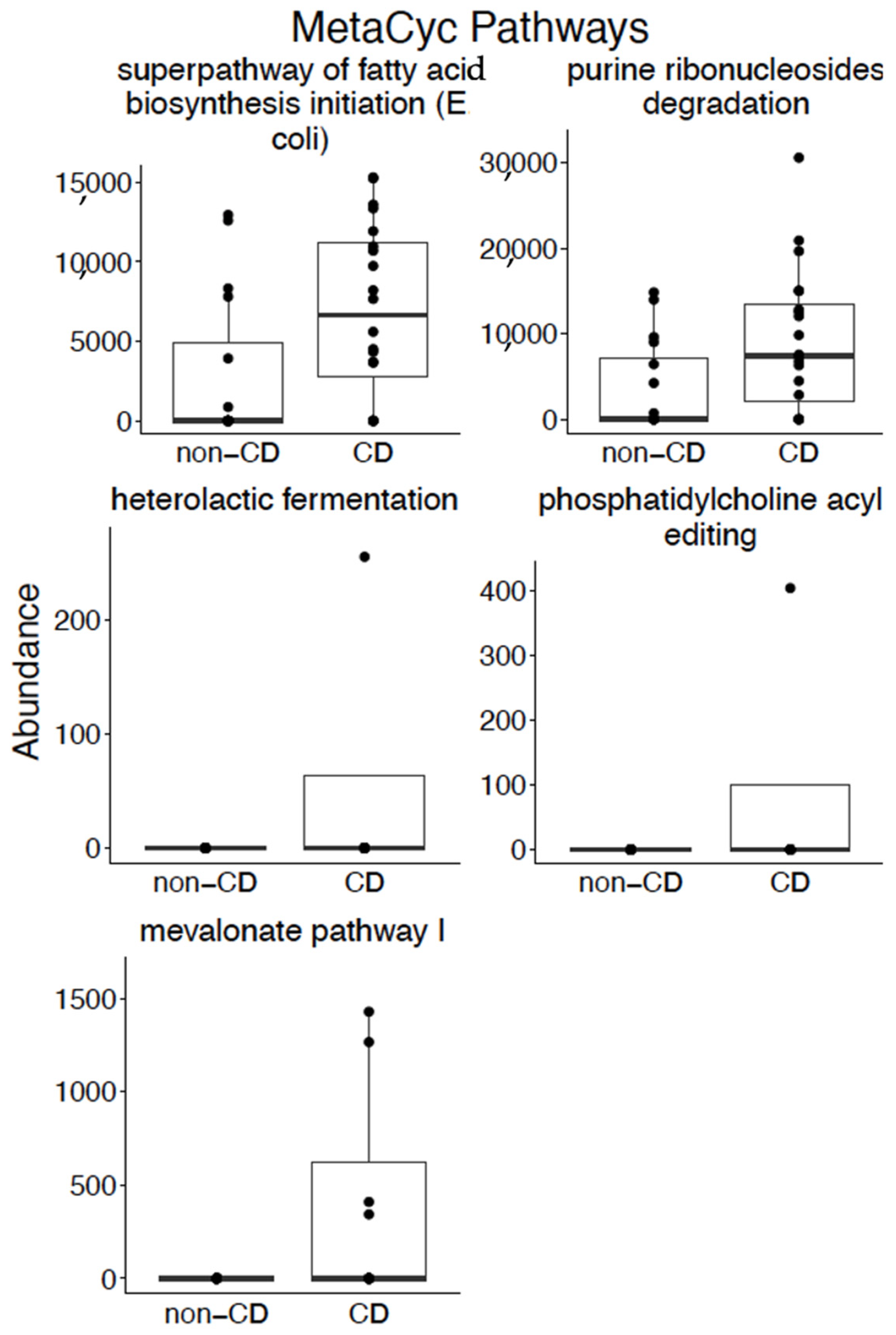
3.3. Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | celiac disease |
| HLA | human leukocyte antigen |
| PCR | polymerase chain reaction |
| GC-MS | gas chromatography–mass spectrometry |
| BMI | body mass index |
References
- Schuppan, D. Current concepts of celiac disease pathogenesis. Gastroenterology 2000, 119, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Ricano-Ponce, I.; Wijmenga, C.; Gutierrez-Achury, J. Genetics of celiac disease. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 399–412. [Google Scholar] [CrossRef]
- Lionetti, E.; Castellaneta, S.; Francavilla, R.; Pulvirenti, A.; Tonutti, E.; Amarri, S.; Barbato, M.; Barbera, C.; Barera, G.; Bellantoni, A.; et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 2014, 371, 1295–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vriezinga, S.; Auricchio, R.; Bravi, E.; Castillejo, G.; Chmielewska, A.; Crespo Escobar, P.; Kolacek, S.; Koletzko, S.; Korponay-Szabo, I.; Mummert, E.; et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 2014, 371, 1304–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blazquez, A.B.; Berin, M.C. Microbiome and food allergy. Transl. Res. 2017, 179, 199–203. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallus, S.; Brandt, L. The Intestinal Microbiota and Obesity. J. Clin. Gastroenterol. 2012, 46, 16–24. [Google Scholar] [CrossRef]
- Akobeng, A.; Ramanan, A.; Buchan, I.; Heller, R. Effect of breast feeding on risk of coeliac disease: A systematic review and meta-analysis of observational studies. Arch. Dis. Child. 2006, 91, 39–43. [Google Scholar] [CrossRef] [Green Version]
- Szajewska, H.; Chmielewska, A.; Piescik-Lech, M.; Ivarsson, A.; Kolacek, S.; Koletzko, S.; Mearin, M.; Shamir, R.; Auricchio, R.; Troncone, R.; et al. Systematic review: Early infant feeding and the prevention of coeliac disease. Aliment. Pharmacol. Ther. 2012, 36, 607–618. [Google Scholar] [CrossRef] [Green Version]
- Korpela, K.S.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; de Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef]
- Maurice, C.F.; Haiser, H.J.; Turnbaugh, P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013, 152, 39–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, M.A.G.; Goodrich, J.K.; Maxan, M.E.; Freedberg, D.E.; Abrams, J.A.; Poole, A.C.; Sutter, J.L.; Welter, D.; Ley, R.E.; Bell, J.T.; et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut 2016, 65, 749–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host. Microbe. 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmsen, H.W.V.; Wildeboer–Veloo, A.C.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Capilla, A.; Nova, E.; Castillejo, G.; Varea, V.; Pozo, T.; Garrote, J.; Polanco, I.; Lopez, A.; Ribes-Koninckx, C.; et al. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: The PROFICEL study. PLoS ONE 2012, 7, e30791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivares, M.; Benitez-Paez, A.; de Palma, G.; Capilla, A.; Nova, E.; Castillejo, G.; Varea, V.; Marcos, A.; Garrote, J.; Polanco, I.; et al. Increased prevalence of pathogenic bacteria in the gut microbiota of infants at risk of developing celiac disease: The PROFICEL study. Gut Microbes. 2018, 9, 551–558. [Google Scholar] [CrossRef] [Green Version]
- Leonard, M.K.; Pujolassos, M.; Troisi, J.; Valitutti, F.; Subramanian, P.; Camhi, S.; Kenyon, V.; Colucci, A.; Serena, G.; Cucchiara, S.; et al. Multi-omics analysis reveals the influence of genetic and environmental risk factors on developing gut microbiota in infants at risk of celiac disease. Microbiome 2020. [Google Scholar] [CrossRef]
- Fernandez, L.L.; Martin, V.; Maldonado, A.; Jimenez, E.; Martin, R.; Rodriguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef]
- Martin, R.J.; Heilig, H.; Fernandez, L.; Marin, M.L.; Zoetendal, E.G.; Rodriguez, J.M. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl. Environ. Microbiol. 2009, 75, 965–969. [Google Scholar] [CrossRef] [Green Version]
- Grönlund, M.M.; Gueimonde, M.; Laitinen, K.; Kociubinski, G.; Gronroos, T.; Salminen, S.; Isolauri, E. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin. Exp. Allergy 2007, 37, 1764–1772. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.D.; Maldonado, A.; Rodriguez, J.M. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 2009, 48, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Heikkila, M.P.; Saris, P.E.J. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 2003, 95, 471–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Olivares, M.; Marin, M.L.; Fernandez, L.; Xaus, J.; Rodriguez, J.M. Probiotic potential of 3 Lactobacilli strains isolated from breast milk. J. Hum. Lact. 2005, 21, 8–17. [Google Scholar] [CrossRef]
- Olivares, M.; Albrecht, S.; De Palma, G.; Ferrer, M.D.; Castillejo, G.; Schols, H.A.; Sanz, Y. Human milk composition differs in healthy mothers and mothers with celiac disease. Eur. J. Nutr. 2015, 54, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Páez, A.; Olivares, M.; Szajewska, H.; Pieścik-Lech, M.; Polanco, I.; Castillejo, G.; Nuñez, M.; Ribes-Koninckx, C.; Korponay-Szabó, I.; Koletzko, S.; et al. Breast-Milk Microbiota Linked to Celiac Disease Development in Children: A Pilot Study from the PreventCD Cohort. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Leonard, M.; Camhi, S.; Huedo-Medina, T.; Fasano, A. Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease. Nutrients 2015, 7, 9325–9336. [Google Scholar] [CrossRef] [Green Version]
- Lackey, K.A.W.; Meehan, C.L.; Zachek, J.A.; Benda, E.D.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; Mbugua, S.; et al. What’s Normal? Microbiomes in Human Milk and Infant Feces Are Related to Each Other but Vary Geographically: The INSPIRE Study. Front. Nutr. 2019, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Aparicio, M.; Alba, C.; Proctocolitis Study Group Of Cam Public Health Area; Rodriguez, J.M.; Fernandez, L. Microbiological and Immunological Markers in Milk and Infant Feces for Common Gastrointestinal Disorders: A Pilot Study. Nutrients 2020, 12, 634. [Google Scholar] [CrossRef] [Green Version]
- Pace, R.M.; Williams, J.E.; Robertson, B.; Lackey, K.A.; Meehan, C.L.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. Variation in Human Milk Composition Is Related to Differences in Milk and Infant Fecal Microbial Communities. Microorganisms 2021, 9, 1153. [Google Scholar] [CrossRef]
- Hasan, N.A.Y.; Minard-Smith, A.T.; Saeed, K.; Li, H.; Heizer, E.M.; McMillan, N.J.; Isom, R.; Abdullah, A.S.; Bornman, D.M.; Faith, S.A.; et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS ONE 2014, 9, e97699. [Google Scholar] [CrossRef]
- Ponnusamy, D.K.; Sha, J.; Erova, T.E.; Azar, S.R.; Fitts, E.C.; Kirtley, M.L.; Tiner, B.L.; Andersson, J.A.; Grim, C.J.; Isom, R.P.; et al. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci. USA 2016, 113, 722–727. [Google Scholar] [CrossRef] [Green Version]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2019. Available online: https://cran.r-project.org/web/packages/vegan/ (accessed on 5 May 2020).
- Goslee, S.C.; Urban, D.L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 2007, 22, 1–19. [Google Scholar] [CrossRef]
- Spiegelhauer, M.R.; Reza, C.; Iversen, A.K.S.; Andersen, L.P. Infection with Rothia mucilaginosa: Review of Findings at a Danish Tertiary Hospital and Reports in Literature. EC Microbiol. 2019, 15, 50–63. [Google Scholar]
- Biagi, E.; Aceti, A.; Quercia, S.; Beghetti, I.; Rampelli, S.; Turroni, S.; Soverini, M.; Zambrini, A.V.; Faldella, G.; Candela, M.; et al. Microbial Community Dynamics in Mother’s Milk and Infant’s Mouth and Gut in Moderately Preterm Infants. Front. Microbiol. 2018, 9, 2512. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.W.; Lin, Y.L.; Huang, M.S. Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J. Food Drug Anal. 2018, 26, 1235–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stinson, L.F.; Sindi, A.S.M.; Cheema, A.S.; Lai, C.T.; Muhlhausler, B.S.; Wlodek, M.E.; Payne, M.S.; Geddes, D.T. The human milk microbiome: Who, what, when, where, why, and how? Nutr. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 2013, 110, 1253–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, P.; Curtis, N. Breast milk microbiota: A review of the factors that influence composition. J. Infect. 2020, 81, 17–47. [Google Scholar] [CrossRef]
- Kim, S.Y.; Yi, D.Y. Analysis of the human breast milk microbiome and bacterial extracellular vesicles in healthy mothers. Exp. Mol. Med. 2020, 52, 1288–1297. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Qi, C.; Yang, Z.; Jiang, S.; Bi, Y.; Lai, J.; Sun, J. Geographical location specific composition of cultured microbiota and Lactobacillus occurrence in human breast milk in China. Food Funct. 2019, 10, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Bajer, L.; Kverka, M.; Kostovcik, M.; Macinga, P.; Dvorak, J.; Stehlikova, Z.; Brezina, J.; Wohl, P.; Spicak, J.; Drastich, P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 2017, 23, 4548–4558. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Dore, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.P.; Riordan, S.M.; Castano-Rodriguez, N.; Wilkins, M.R.; Kaakoush, N.O. Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome 2018, 6, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Healthy Controls (n = 16) | Subjects with CD on a Gluten-Free Diet (n = 20) | |
|---|---|---|
| Maternal Characteristics | ||
| Age (years) * | 33.4 | 32.5 |
| Duration gestation (weeks) * | 39.9 | 39.4 |
| Pregnancy (%) | ||
| Nulliparous | 3 (18.8) | 6 (30) |
| Multiparous | 13 (81.3) | 14 (70) |
| Mode of delivery (%) | ||
| Vaginal | 13 (81.3) | 15 (75) |
| Cesarean | 3 (18.8) | 5 (25) |
| Antibiotics during pregnancy (%) | 5 (31.3) | 5 (25) |
| Antibiotics during delivery (%) | 3 (18.8) | 6 (30) |
| Probiotics during pregnancy (%) | 7 (43.8) | 5 (25) |
| Probiotics while breastfeeding (%) | 5 (31.3) | 4 (20) |
| Delivery setting (%) | ||
| Hospital | 15 (93.8) | 19 (95) |
| Other (birthing center, home) | 1 (6.2) | 1 (5) |
| Pre-pregnancy body mass index (BMI) (kg/m2) * | 22.4 | 23 |
| Infant Characteristics | ||
| Sex (%) | ||
| Male | 10 (62.5) | 13 (65) |
| Female | 6 (37.5) | 7 (35) |
| Antibiotics at birth (%) | 0 (0) | 1 (5) |
| Duration of hospitalization (days) * | 2.03 | 1.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olshan, K.L.; Zomorrodi, A.R.; Pujolassos, M.; Troisi, J.; Khan, N.; Fanelli, B.; Kenyon, V.; Fasano, A.; Leonard, M.M. Microbiota and Metabolomic Patterns in the Breast Milk of Subjects with Celiac Disease on a Gluten-Free Diet. Nutrients 2021, 13, 2243. https://doi.org/10.3390/nu13072243
Olshan KL, Zomorrodi AR, Pujolassos M, Troisi J, Khan N, Fanelli B, Kenyon V, Fasano A, Leonard MM. Microbiota and Metabolomic Patterns in the Breast Milk of Subjects with Celiac Disease on a Gluten-Free Diet. Nutrients. 2021; 13(7):2243. https://doi.org/10.3390/nu13072243
Chicago/Turabian StyleOlshan, Katherine L., Ali R. Zomorrodi, Meritxell Pujolassos, Jacopo Troisi, Nayeim Khan, Brian Fanelli, Victoria Kenyon, Alessio Fasano, and Maureen M. Leonard. 2021. "Microbiota and Metabolomic Patterns in the Breast Milk of Subjects with Celiac Disease on a Gluten-Free Diet" Nutrients 13, no. 7: 2243. https://doi.org/10.3390/nu13072243
APA StyleOlshan, K. L., Zomorrodi, A. R., Pujolassos, M., Troisi, J., Khan, N., Fanelli, B., Kenyon, V., Fasano, A., & Leonard, M. M. (2021). Microbiota and Metabolomic Patterns in the Breast Milk of Subjects with Celiac Disease on a Gluten-Free Diet. Nutrients, 13(7), 2243. https://doi.org/10.3390/nu13072243








