Methylation and Expression of FTO and PLAG1 Genes in Childhood Obesity: Insight into Anthropometric Parameters and Glucose–Lipid Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Microarray Analysis
2.2. Methylation Analysis
2.2.1. DNA and Selection of Fragments
2.2.2. Restriction Enzyme Digestion
2.2.3. qPCR
2.3. Methylation Data Processing and Statistical Analysis
2.4. Anthropometric Measurements
2.5. Study Protocol
2.6. Laboratory Measurements
2.7. HOMA-IR
2.8. Blood Pressure
2.9. Statistical Analysis
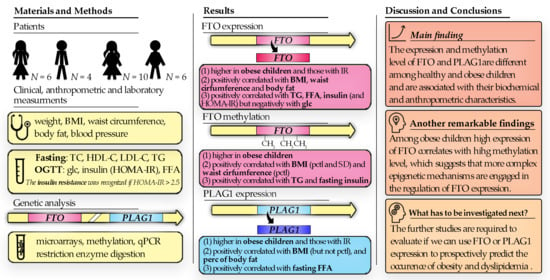
3. Results
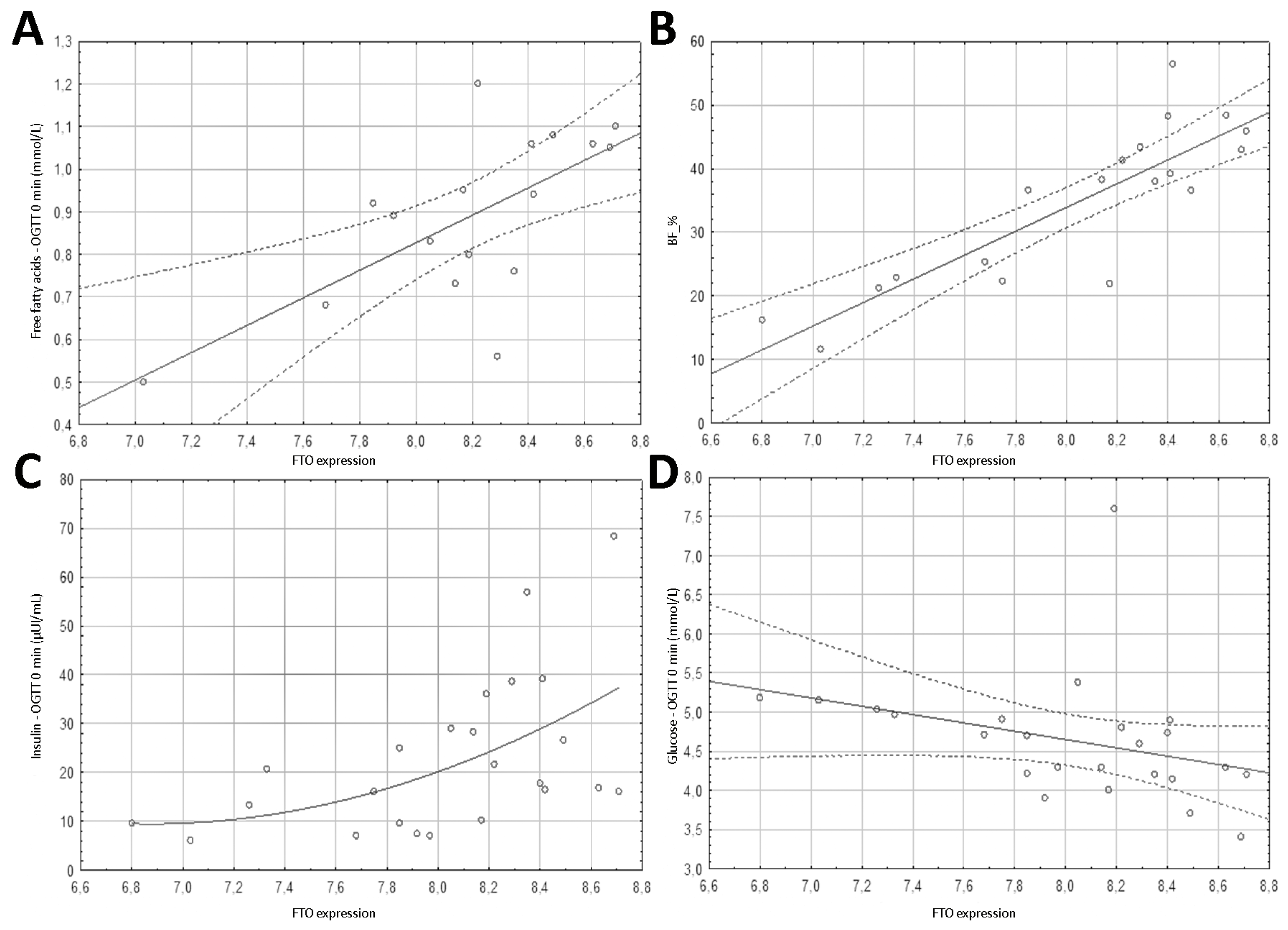
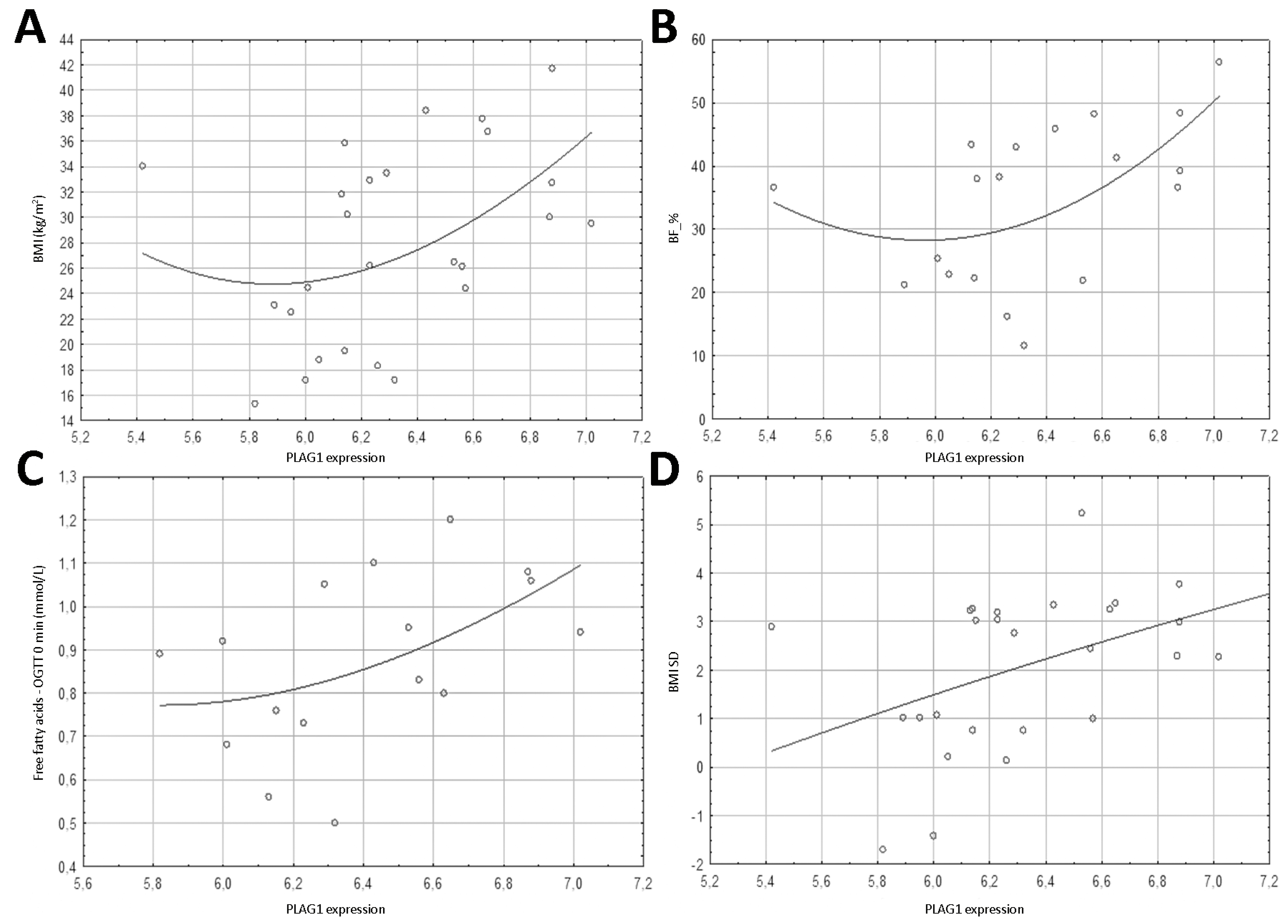
3.1. Genes Expression
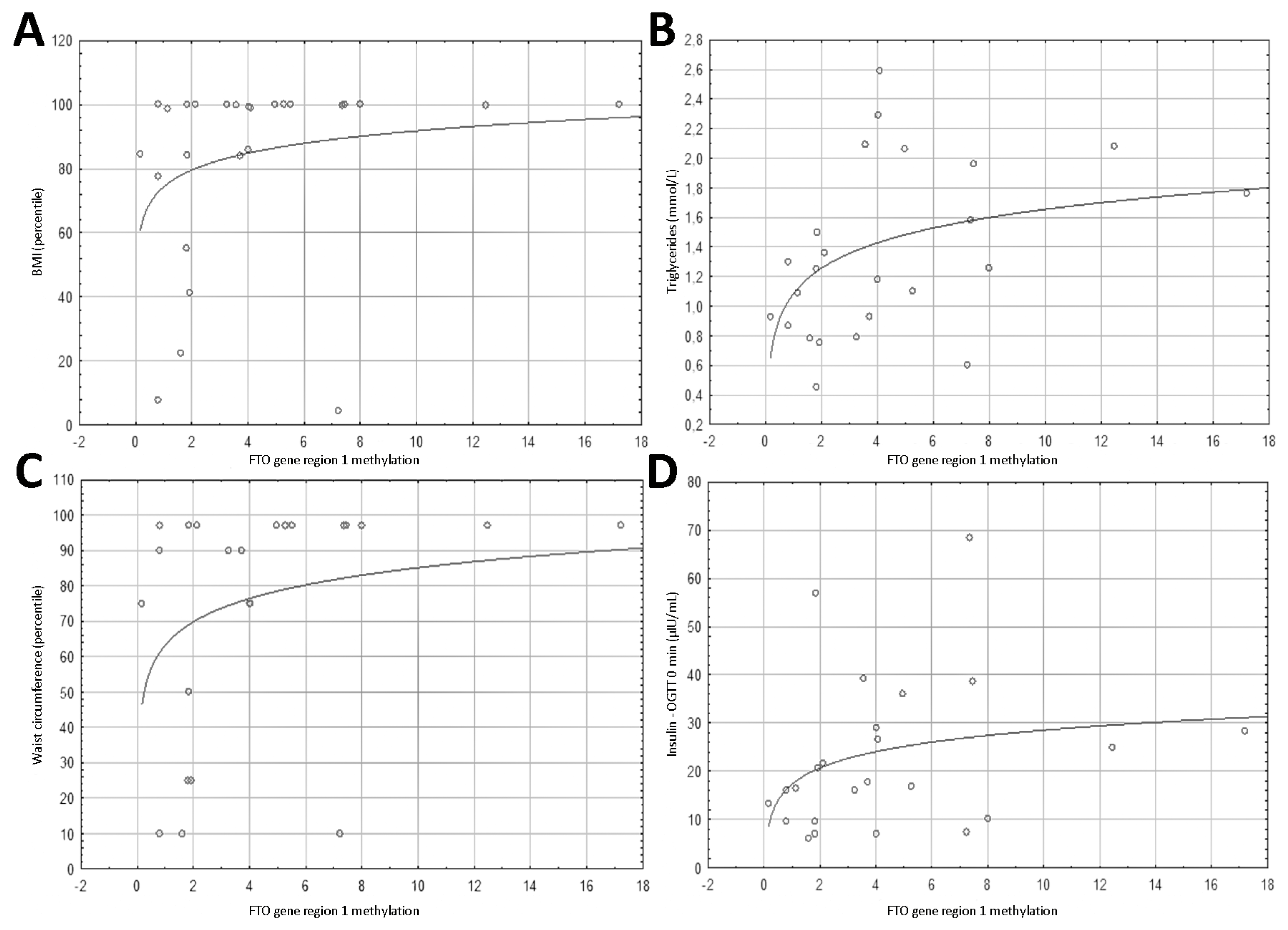
3.2. Methylation Results
3.3. Spearman’s Correlation Results of Methylation Level and Expression of FTO Gene and PLAG1 Gene
3.4. Spearman’s Correlation Results of Anthropometric Parameters with FTO Gene Methylation, Expression and PLAG1 Gene Expression
3.5. Spearman’s Correlation Results of Biochemical Parameters with FTO Gene Region 1 Methylation, FTO Expression, and PLAG1 Gene Expression
3.6. Effect of FTO Gene Region 1 Methylation, FTO Expression, and PLAG1 Expression on the Presence of Insulin Resistance
3.7. Multiple Regression Analysis
4. Discussion
4.1. Expression and Methylation of FTO Gene
4.2. Expression and Methylation of PLAG1 Gene
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danese, A.; Tan, M. Childhood Maltreatment and Obesity: Systematic Review and Meta-Analysis. Mol. Psychiatry 2014, 19, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Ogden, C.L.; Carroll, M.D.; Lawman, H.G.; Fryar, C.D.; Kruszon-Moran, D.; Kit, B.K.; Flegal, K.M. Trends in Obesity Prevalence Among Children and Adolescents in the United States, 1988–1994 through 2013–2014. JAMA 2016, 315, 2292. [Google Scholar] [CrossRef] [PubMed]
- Łuszczki, E.; Dereń, K.; Baran, J.; Weres, A.; Mazur, A. The Secular Trend of Overweight and Obesity in Preschool Children from Rzeszow Region. Pediatr. Endocrinol. 2015, 14, 29–34. [Google Scholar] [CrossRef][Green Version]
- Hill, J.O.; Peters, J.C. Environmental Contributions to the Obesity Epidemic. Science 1998, 280, 1371–1374. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Skoczen, S.; Tomasik, P.J.; Bik-Multanowski, M.; Surmiak, M.; Balwierz, W.; Pietrzyk, J.J.; Sztefko, K.; Gozdzik, J.; Galicka-Lataa, D.; Strojny, W. Plasma Levels of Leptin and Soluble Leptin Receptor and Polymorphisms of Leptin Gene -18G > A and Leptin Receptor Genes K109R and Q223R, in Survivors of Childhood Acute Lymphoblastic Leukemia. J. Exp. Clin. Cancer Res. 2011, 30. [Google Scholar] [CrossRef] [PubMed]
- Justice, A.E.; Chittoor, G.; Blanco, E.; Graff, M.; Wang, Y.; Albala, C.; Santos, J.L.; Angel, B.; Lozoff, B.; Voruganti, V.S.; et al. Genetic Determinants of BMI from Early Childhood to Adolescence: The Santiago Longitudinal Study. Pediatr. Obes. 2019, 14. [Google Scholar] [CrossRef]
- Molou, E.; Schulpis, K.H.; Birbilis, C.; Thodi, G.; Georgiou, V.; Dotsikas, Y.; Loukas, Y.L. Early Screening of FTO and MC4R Variants in Newborns of Greek Origin. J. Pediatr. Endocrinol. Metab. 2015, 28, 619–622. [Google Scholar] [CrossRef]
- Daoud, H.; Zhang, D.; McMurray, F.; Yu, A.; Luco, S.M.; Vanstone, J.; Jarinova, O.; Carson, N.; Wickens, J.; Shishodia, S.; et al. Identification of a Pathogenic FTO Mutation by Next-Generation Sequencing in a Newborn with Growth Retardation and Developmental Delay. J. Med. Genet. 2015, 53, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, L.; Marchegiani, F.; Piangerelli, M.; Napolioni, V.; Gabbianelli, R. Obesity-Related Genetic Polymorphisms and Adiposity Indices in a Young Italian Population. IUBMB Life 2017, 69, 98–105. [Google Scholar] [CrossRef]
- Almeida, S.M.; Furtado, J.M.; Mascarenhas, P.; Ferraz, M.E.; Ferreira, J.C.; Monteiro, M.P.; Vilanova, M.; Ferraz, F.P. Association between LEPR, FTO, MC4R, and PPARG-2 Polymorphisms with Obesity Traits and Metabolic Phenotypes in School-Aged Children. Endocrine 2018, 60, 466–478. [Google Scholar] [CrossRef]
- Da Silva, T.E.R.; Andrade, N.L.; de Oliveira Cunha, D.; Leão-Cordeiro, J.A.B.; Vilanova-Costa, C.A.S.T.; Silva, A.M.T.C. The FTO Rs9939609 Polymorphism and Obesity Risk in Teens: Evidence-Based Meta-Analysis. Obes. Res. Clin. Pract. 2018, 12, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Richards, M.; Wong, A.; Hardy, R. Fat Mass and Obesity-Associated (FTO) Rs9939609 Polymorphism Modifies the Relationship between Body Mass Index and Affective Symptoms through the Life Course: A Prospective Birth Cohort Study. Transl. Psychiatry 2018, 8. [Google Scholar] [CrossRef]
- Barseem, N.F.; El Ella, S.S.A.; Tawfik, M.A.; El-Nehrawy, R.R. The Potential Implication of FTO Rs17817449 Gene Polymorphism on BMI Mediated Risk for Type2 Diabetes Among Obese Egyptian Children And Adolescents. Endocr. Metab. Immune Disord. Drug Targets 2019, 19, 697–704. [Google Scholar] [CrossRef] [PubMed]
- González-Herrera, L.; Zavala-Castro, J.; Ayala-Cáceres, C.; Pérez-Mendoza, G.; López-González, M.J.; Pinto-Escalante, D.; Canto-Cetina, T.; García-Escalante, M.G.; Rubi-Castellanos, R.; Contreras-Capetillo, S.; et al. Genetic Variation of FTO: Rs1421085 T>C, Rs8057044 G>A, Rs9939609 T>A, and Copy Number (CNV) in Mexican Mayan School-Aged Children with Obesity/Overweight and with Normal Weight. Am. J. Hum. Biol. 2019, 31. [Google Scholar] [CrossRef]
- Warrington, N.M.; Beaumont, R.N.; Horikoshi, M.; Day, F.R.; Helgeland, Ø.; Laurin, C.; Bacelis, J.; Peng, S.; Hao, K.; Feenstra, B.; et al. Maternal and Fetal Genetic Effects on Birth Weight and Their Relevance to Cardio-Metabolic Risk Factors. Nat. Genet. 2019, 51, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Hübel, C.; Gaspar, H.A.; Coleman, J.R.I.; Finucane, H.; Purves, K.L.; Hanscombe, K.B.; Prokopenko, I.; Graff, M.; Ngwa, J.S.; Workalemahu, T.; et al. Genomics of Body Fat Percentage May Contribute to Sex Bias in Anorexia Nervosa. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2019, 180, 428–438. [Google Scholar] [CrossRef]
- Shungin, D.; Winkler, T.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New Genetic Loci Link Adipose and Insulin Biology to Body Fat Distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef]
- Gardner, K.R.; Sapienza, C.; Fisher, J.O. Genetic and Epigenetic Associations to Obesity-Related Appetite Phenotypes among African-American Children. Pediatr. Obes. 2015, 10, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Foraita, R.; Günther, F.; Gwozdz, W.; Reisch, L.A.; Russo, P.; Lauria, F.; Siani, A.; Veidebaum, T.; Tornaritis, M.; Iacoviello, L.; et al. Does the FTO Gene Interact with the Socioeconomic Status on the Obesity Development among Young European Children? Results from the IDEFICS Study. Int. J. Obes. 2015, 39, 1–6. [Google Scholar] [CrossRef]
- Sebert, S.; Salonurmi, T.; Keinänen-Kiukaanniemi, S.; Savolainen, M.; Herzig, K.-H.; Symonds, M.E.; Järvelin, M.-R. Programming Effects of FTO in the Development of Obesity. Acta Physiol. 2014, 210, 58–69. [Google Scholar] [CrossRef]
- Mǎrginean, C.O.; Mǎrginean, C.; Meliţ, L.E. New Insights Regarding Genetic Aspects of Childhood Obesity: A Minireview. Front. Pediatr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Kadakia, R.; Zheng, Y.; Zhang, Z.; Zhang, W.; Josefson, J.L.; Hou, L. Association of Cord Blood Methylation with Neonatal Leptin: An Epigenome Wide Association Study. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Keustermans, G.C.; Kofink, D.; Eikendal, A.; De Jager, W.; Meerding, J.; Nuboer, R.; Waltenberger, J.; Kraaijeveld, A.O.; Jukema, J.W.; Sels, J.W.; et al. Monocyte Gene Expression in Childhood Obesity Is Associated with Obesity and Complexity of Atherosclerosis in Adults. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Hashimoto, K.; Kokubun, S.; Itoi, E.; Roach, H.I. Improved Quantification of DNA Methylation Using Methylation-Sensitive Restriction Enzymes and Real-Time PCR. Epigenetics 2007, 2, 86–91. [Google Scholar] [CrossRef]
- Redshaw, N.; Huggett, J.F.; Taylor, M.S.; Foy, C.A.; Devonshire, A.S. Quantification of Epigenetic Biomarkers: An Evaluation of Established and Emerging Methods for DNA Methylation Analysis. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 13 May 2021).
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- WHO Anthro Survey Analyser and Other Tools. Available online: https://www.who.int/toolkits/child-growth-standards/software (accessed on 4 March 2021).
- Palczewska, I.; Niedzwiedzka, Z. Somatic development indices in children and youth of Warsaw. Med. Wieku Rozw. 2001, 5, 18–118. [Google Scholar]
- Chrzanowska, M.; Gołąb, S.; Żarów, R.; Sobiecki, J.; Brudecki, J. Dziecko Krakowskie 2000. Poziom Rozwoju Biologicznego Dzieci i Młodzieży Miasta Krakowa; Studia i Monografie; AWF Kraków: Kraków, Poland, 2002. [Google Scholar]
- Kushner, R.F.; Schoeller, D.A. Estimation of Total Body Water by Bioelectrical Impedance Analysis. Am. J. Clin. Nutr. 1986, 44, 417–424. [Google Scholar] [CrossRef]
- Blood Pressure Z-Score Application. Available online: https://www.bcm.edu/bodycomplab/BPappZjs/BPvAgeAPPz.html (accessed on 26 February 2021).
- GeneCards—Human Genes | Gene Database | Gene Search. Available online: https://www.genecards.org/ (accessed on 6 March 2021).
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinforma. 2016, 54, 3255. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Jia, G.; Yang, C.G.; Yang, S.; Jian, X.; Yi, C.; Zhou, Z.; He, C. Oxidative Demethylation of 3-Methylthymine and 3-Methyluracil in Single-Stranded DNA and RNA by Mouse and Human FTO. FEBS Lett. 2008, 582, 3313–3319. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Niu, T.; Chang, J.; Lei, X.; Zhao, M.; Wang, Q.; Cheng, W.; Wang, J.; Feng, Y.; Chai, J. Crystal Structure of the FTO Protein Reveals Basis for Its Substrate Specificity. Nature 2010, 464, 1205–1209. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.-H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, I.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef]
- Liu, Z.W.; Zhang, J.T.; Cai, Q.Y.; Zhang, H.X.; Wang, Y.H.; Yan, H.T.; Wu, H.M.; Yang, X.J. Birth Weight Is Associated with Placental Fat Mass- and Obesity-Associated Gene Expression and Promoter Methylation in a Chinese Population. J. Matern. Fetal Neonatal Med. 2016, 29, 106–111. [Google Scholar] [CrossRef]
- Doaei, S.; Kalantari, N.; Mohammadi, N.K.; Tabesh, G.A.; Gholamalizadeh, M. Macronutrients and the FTO Gene Expression in Hypothalamus; a Systematic Review of Experimental Studies. Indian Heart J. 2017, 69, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Dina, C.; Meyre, D.; Gallina, S.; Durand, E.; Körner, A.; Jacobson, P.; Carlsson, L.M.S.; Kiess, W.; Vatin, V.; Lecoeur, C.; et al. Variation in FTO Contributes to Childhood Obesity and Severe Adult Obesity. Nat. Genet. 2007, 39, 724–726. [Google Scholar] [CrossRef]
- Hinney, A.; Nguyen, T.T.; Scherag, A.; Friedel, S.; Brönner, G.; Müller, T.D.; Grallert, H.; Illig, T.; Wichmann, H.E.; Rief, W.; et al. Genome Wide Association (GWA) Study for Early Onset Extreme Obesity Supports the Role of Fat Mass and Obesity Associated Gene (FTO) Variants. PLoS ONE 2007, 2. [Google Scholar] [CrossRef]
- Scuteri, A.; Sanna, S.; Chen, W.M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orrú, M.; Usala, G.; et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007, 3, 1200–1210. [Google Scholar] [CrossRef]
- Gene: FTO (ENSG00000140718)—Summary—Homo_sapiens—Ensembl Genome Browser 103. Available online: https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000140718;r=16:53701692-54158512 (accessed on 6 March 2021).
- Liu, S.J.; Tang, H.L.; He, Q.; Lu, P.; Fu, T.; Xu, X.L.; Su, T.; Gao, M.M.; Duan, S.; Luo, Y.; et al. FTO Is a Transcriptional Repressor to Auto-Regulate Its Own Gene and Potentially Associated with Homeostasis of Body Weight. J. Mol. Cell Biol. 2018, 11, 118–132. [Google Scholar] [CrossRef]
- Zhou, Y.; Simmons, D.; Lai, D.; Hambly, B.D.; McLachlan, C.S. Rs9939609 FTO Genotype Associations with FTO Methylation Level Influences Body Mass and Telomere Length in an Australian Rural Population. Int. J. Obes. 2017, 41, 1427–1433. [Google Scholar] [CrossRef]
- Rauschert, S.; Melton, P.E.; Burdge, G.; Craig, J.; Godfrey, K.M.; Holbrook, J.D.; Lillycrop, K.; Mori, T.A.; Beilin, L.J.; Oddy, W.H.; et al. Maternal Smoking during Pregnancy Induces Persistent Epigenetic Changes into Adolescence, Independent of Postnatal Smoke Exposure and Is Associated with Cardiometabolic Risk. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Tehranifar, P.; Wu, H.C.; McDonald, J.A.; Jasmine, F.; Santella, R.M.; Gurvich, I.; Flom, J.D.; Terry, M.B. Maternal Cigarette Smoking during Pregnancy and Offspring DNA Methylation in Midlife. Epigenetics 2018, 13, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.K.; Shrestha, S.; Lillycrop, K.A.; Joglekar, C.V.; Pan, H.; Holbrook, J.D.; Fall, C.H.D.; Yajnik, C.S.; Chandak, G.R. Vitamin B12 Supplementation Influences Methylation of Genes Associated with Type 2 Diabetes and Its Intermediate Traits. Epigenomics 2018, 10, 71–90. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, S.A.; Mumtaz, A.; Shahid, S.U.; Shabana, N.A. Rs3751812, a Common Variant in Fat Mass and Obesity-Associated (FTO) Gene, Is Associated with Serum High- and Low-Density Lipoprotein Cholesterol in Pakistani Individuals. Nutrition 2017, 39–40, 92–95. [Google Scholar] [CrossRef]
- Nishida, H.; Onishi, K.; Kurose, S.; Tsutsumi, H.; Miyauchi, T.; Takao, N.; Yoshiuchi, S.; Fujii, A.; Kimura, Y. Changes in Body Composition and FTO Whole Blood Dna Methylation among Japanese Women: A Randomized Clinical Trial of Weight-Loss Program. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 2157–2167. [Google Scholar] [CrossRef]
- Yang, Q.; Xiao, T.; Guo, J.; Su, Z. Complex Relationship between Obesity and the Fat Mass and Obesity Locus. Int. J. Biol. Sci. 2017, 13, 615–629. [Google Scholar] [CrossRef]
- Khoshi, A.; Bajestani, M.K.; Shakeri, H.; Goodarzi, G.; Azizi, F. Association of Omentin Rs2274907 and FTO Rs9939609 Gene Polymorphisms with Insulin Resistance in Iranian Individuals with Newly Diagnosed Type 2 Diabetes. Lipids Health Dis. 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Mehrdad, M.; Doaei, S.; Gholamalizadeh, M.; Fardaei, M.; Fararouei, M.; Eftekhari, M.H. Association of FTO Rs9939609 Polymorphism with Serum Leptin, Insulin, Adiponectin, and Lipid Profile in Overweight Adults. Adipocyte 2020, 9, 51–56. [Google Scholar] [CrossRef]
- Czajkowski, P.; Adamska-Patruno, E.; Bauer, W.; Fiedorczuk, J.; Krasowska, U.; Moroz, M.; Gorska, M.; Kretowski, A. The Impact of FTO Genetic Variants on Obesity and Its Metabolic Consequences Is Dependent on Daily Macronutrient Intake. Nutrients 2020, 12, 3255. [Google Scholar] [CrossRef] [PubMed]
- Freathy, R.M.; Timpson, N.J.; Lawlor, D.A.; Pouta, A.; Ben-Shlomo, Y.; Ruokonen, A.; Ebrahim, S.; Shields, B.; Zeggini, E.; Weedon, M.N.; et al. Common Variation in the FTO Gene Alters Diabetes-Related Metabolic Traits to the Extent Expected given Its Effect on Bmi. Diabetes 2008, 57, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Xi, B.; Zhang, M.; Wang, C.; Shen, Y.; Zhao, X.; Wang, X.; Mi, J. The Common SNP (Rs9939609) in the FTO Gene Modifies the Association between Obesity and High Blood Pressure in Chinese Children. Mol. Biol. Rep. 2013, 40, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Sun, J.; Wu, J.; Yang, M. Combined Effects of FTO Rs9939609 and MC4R Rs17782313 on Elevated Nocturnal Blood Pressure in the Chinese Han Population. Cardiovasc. J. Afr. 2016, 27, 21–24. [Google Scholar] [CrossRef]
- PLAG1 Gene Symbol Report | HUGO Gene Nomenclature Committee. Available online: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:9045 (accessed on 6 March 2021).
- Braschi, B.; Denny, P.; Gray, K.; Jones, T.; Seal, R.; Tweedie, S.; Yates, B.; Bruford, E. Genenames.Org: The HGNC and VGNC Resources in 2019. Nucleic Acids Res. 2019, 47, D786–D792. [Google Scholar] [CrossRef] [PubMed]
- Van Dyck, F.; Declercq, J.; Braem, C.; Van de Ven, W. PLAG1, the Prototype of the PLAG Gene Family: Versatility in Tumour Development (Review). Int. J. Oncol. 2007, 30, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Zatkova, A.; Rouillard, J.M.; Hartmann, W.; Lamb, B.J.; Kuick, R.; Eckart, M.; Von Schweinitz, D.; Koch, A.; Fonatsch, C.; Pietsch, T.; et al. Amplification and Overexpression of the IGF2 Regulator PLAG1 in Hepatoblastoma. Genes Chromosom. Cancer 2004, 39, 126–137. [Google Scholar] [CrossRef]
- GWAS Catalog. Available online: https://www.ebi.ac.uk/gwas/home (accessed on 6 March 2021).
- Buniello, A.; Macarthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed]
- Kadakia, R.; Josefson, J. The Relationship of Insulin-like Growth Factor 2 to Fetal Growth and Adiposity. Horm. Res. Paediatr. 2016, 85, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Street, M.E.; Smerieri, A.; Montanini, L.; Predieri, B.; Iughetti, L.; Valenzise, M.; De Luca, F.; Vigone, M.; Weber, G.; Maghnie, M.; et al. Interactions among Pro-Inflammatory Cytokines, IGF System and Thyroid Function in Pre-Pubertal Obese Subjects. J. Biol. Regul. Homeost. Agents 2013, 27, 259–266. [Google Scholar] [PubMed]
- Ong, K.; Kratzsch, J.; Kiess, W.; Dunger, D. Circulating IGF-I Levels in Childhood Are Related to Both Current Body Composition and Early Postnatal Growth Rate. J. Clin. Endocrinol. Metab. 2002, 87, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Abi Habib, W.; Brioude, F.; Edouard, T.; Bennett, J.T.; Lienhardt-Roussie, A.; Tixier, F.; Salem, J.; Yuen, T.; Azzi, S.; Le Bouc, Y.; et al. Genetic Disruption of the Oncogenic HMGA2-PLAG1-IGF2 Pathway Causes Fetal Growth Restriction. Genet. Med. 2018, 20, 250–258. [Google Scholar] [CrossRef]
- EWAS Atlas. Available online: https://bigd.big.ac.cn/ewas/index (accessed on 6 March 2021).
- Li, M.; Zou, D.; Li, Z.; Gao, R.; Sang, J.; Zhang, Y.; Li, R.; Xia, L.; Zhang, T.; Niu, G.; et al. EWAS Atlas: A Curated Knowledgebase of Epigenome-Wide Association Studies. Nucleic Acids Res. 2019, 47, D983–D988. [Google Scholar] [CrossRef]
- MRC-IEU EWAS Catalog. Available online: http://www.ewascatalog.org/ (accessed on 6 March 2021).
- Battram, T.; Yousefi, P.; Crawford, G.; Prince, C.; Babaei, M.S.; Sharp, G.; Hatcher, C.; Vega-salas, M.J.; Khodabakhsh, S.; Whitehurst, O.; et al. The EWAS Catalog: A Database of Epigenome-Wide Association Studies. OSF Preprints 2021, 2–5. [Google Scholar] [CrossRef]
- Arpón, A.; Milagro, F.I.; Ramos-Lopez, O.; Mansego, M.L.; Riezu-Boj, J.-I.; Martínez, J.A. Methylome-Wide Association Study in Peripheral White Blood Cells Focusing on Central Obesity and Inflammation. Genes 2019, 10, 444. [Google Scholar] [CrossRef]
- Sharp, G.C.; Salas, L.A.; Monnereau, C.; Allard, C.; Yousefi, P.; Everson, T.M.; Bohlin, J.; Xu, Z.; Huang, R.C.; Reese, S.E.; et al. Maternal BMI at the Start of Pregnancy and Offspring Epigenome-Wide DNA Methylation: Findings from the Pregnancy and Childhood Epigenetics (PACE) Consortium. Hum. Mol. Genet. 2017, 26, 4067–4085. [Google Scholar] [CrossRef]
- Weng, X.; Liu, F.; Zhang, H.; Kan, M.; Wang, T.; Dong, M.; Liu, Y. Genome-Wide DNA Methylation Profiling in Infants Born to Gestational Diabetes Mellitus. Diabetes Res. Clin. Pract. 2018, 142, 10–18. [Google Scholar] [CrossRef]
- Declercq, J.; Kumar, A.; Van Diepen, J.A.; Vroegrijk, I.O.C.M.; Gysemans, C.; Di Pietro, C.; Voshol, P.J.; Mathieu, C.; Ectors, N.; Van De Ven, W.J.M.; et al. Increased β-Cell Mass by Islet Transplantation and PLAG1 Overexpression Causes Hyperinsulinemic Normoglycemia and Hepatic Insulin Resistance in Mice. Diabetes 2010, 59, 1957–1965. [Google Scholar] [CrossRef] [PubMed]
- Declercq, J.; Kumar, A.; Gysemans, C.; di Pietro, C.; Schraenen, A.; Chintinne, M.; Lemaire, K.; van Lomme, L.; van De Casteele, M.; Heimberg, H.; et al. Pdx1- and Ngn3-Cre-Mediated PLAG1 Expression in the Pancreas Leads to Endocrine Hormone Imbalances That Affect Glucose Metabolism. Cell Transplant. 2011, 20, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Home < Expression Atlas < EMBL-EBI. Available online: https://www.ebi.ac.uk/gxa/home (accessed on 6 March 2021).
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.-P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas Update: From Tissues to Single Cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef]
- Elo, L.L.; Mykkänen, J.; Nikula, T.; Järvenpää, H.; Simell, S.; Aittokallio, T.; Hyöty, H.; Ilonen, J.; Veijola, R.; Simell, T.; et al. Early Suppression of Immune Response Pathways Characterizes Children with Prediabetes in Genome-Wide Gene Expression Profiling. J. Autoimmun. 2010, 35, 70–76. [Google Scholar] [CrossRef]
- Kennedy, J.; Katsuta, H.; Jung, M.H.; Marselli, L.; Goldfine, A.B.; Balis, U.J.; Sgroi, D.; Bonner-Weir, S.; Weir, G.C. Protective Unfolded Protein Response in Human Pancreatic Beta Cells Transplanted into Mice. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Dynkevich, Y.; Rother, K.I.; Whitford, I.; Qureshi, S.; Galiveeti, S.; Szulc, A.L.; Danoff, A.; Breen, T.L.; Kaviani, N.; Shanik, M.H.; et al. Tumors, IGF-2, and Hypoglycemia: Insights From the Clinic, the Laboratory, and the Historical Archive. Endocr. Rev. 2013, 34, 798–826. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.M.; Laggai, S.; Van Wonterghem, E.; Gemperlein, K.; Müller, R.; Haybaeck, J.; Vandenbroucke, R.E.; Ogris, M.; Libert, C.; Kiemer, A.K. Transient Hepatic Overexpression of Insulin-like Growth Factor 2 Induces Free Cholesterol and Lipid Droplet Formation. Front. Physiol. 2016, 7. [Google Scholar] [CrossRef]
- Pouriamehr, S.; Barmaki, H.; Rastegary, M.; Lotfi, F.; Nabi Afjadi, M. Investigation of Insulin-like Growth Factors/Insulin-like Growth Factor Binding Proteins Regulation in Metabolic Syndrome Patients. BMC Res. Notes 2019, 12. [Google Scholar] [CrossRef]
- Cooley, S.M.; Donnelly, J.C.; Geary, M.P.; Rodeck, C.H.; Hindmarsh, P.C. Maternal Insulin-like Growth Factors 1 and 2 (IGF-1, IGF-2) and IGF BP-3 and the Hypertensive Disorders of Pregnancy. J. Matern. Fetal Neonatal Med. 2010, 23, 658–661. [Google Scholar] [CrossRef]
- Faienza, M.F.; Santoro, N.; Lauciello, R.; Calabró, R.; Giordani, L.; Di Salvo, G.; Ventura, A.; Delvecchio, M.; Perrone, L.; Del Giudice, E.M.; et al. IGF2 Gene Variants and Risk of Hypertension in Obese Children and Adolescents. Pediatr. Res. 2010, 67, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.; Guardiola, M.; Ferré, R.; Coll, B.; Alonso-Villaverde, C.; Winklhofer-Roob, B.M.; Rock, E.; Fernández-Ballart, J.D.; Civeira, F.; Pocoví, M.; et al. Association of a Polymorphism in the Promoter of the Cellular Retinoic Acid-Binding Protein II Gene (CRABP2) with Increased Circulating Low-Density Lipoprotein Cholesterol. Clin. Chem. Lab. Med. 2007, 45, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Boden, G. Obesity, Insulin Resistance and Free Fatty Acids. Curr. Opin. Endocrinol. Diabetes Obes. 2011, 18, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and Characterization of the Promoter Region of Perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in Bovine Adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef] [PubMed]
| Gene | Localization | Fragment Size | No. of CCGG Sites | Forward | Reverse |
|---|---|---|---|---|---|
| FTO | upstream | 216 bp | 2 | CAACTCCAGGGCCTTCTC | GGAGCCTGCCATGTTTCT |
| FTO | 3′UTR | 214 bp | 1 | GGAAGGAGAGAATAGAGCCAAG | TCCTCAGCACTTTAGCCTTAC |
| PLAG1 | exon1 | 202 bp | 2 | ACAATGGCTGCTGGAAAGA | CCCTGATATTTCTCCCGCTAAA |
| Baseline Characteristic | Obesity Group n = 16 | Control Group n = 10 | p Value |
|---|---|---|---|
| boys/girls n (%) | 6/10 (37.5%/62.5%) | 4/6 (40%/60%) | 0.609 |
| Age (years) | 13.78 ± 2.98 | 14.32 ± 1.92 | 0.938 |
| Height (cm) | 162.88 ± 15.77 | 168.01 ± 10.22 | 0.324 |
| Weight (kg) | 88.43 ± 22.48 | 57.72 ± 15.69 | <0.001 |
| BMI (kg/m2) | 32.74 ± 4.59 | 20.08 ± 3.3 | <0.001 |
| BMI (percentile) | 99.78 ± 0.41 | 54.77 ± 33.45 | 0.002 |
| Waist circumference (cm) | 97.46 ± 14.23 | 72.18 ± 9.24 | <0.001 |
| Waist circumference (percentile) | 94.77 ± 6.25 | 46 ± 33.9 | 0.001 |
| BMI SD | 3.15 ± 0.69 | 0.27 ± 1.03 | <0.001 |
| BF_kg | 37.89 ± 11.28 | 15.23 ± 9.95 | <0.001 |
| BF_% | 40.72 ± 8.25 | 23.96 ± 11.65 | 0.008 |
| Total cholesterol (mmol/L) | 4.41 ± 1.04 | 3.9 ± 0.65 | 0.159 |
| LDL cholesterol (mmol/L) | 2.6 ± 0.92 | 2.07 ± 0.48 | 0.084 |
| HDL cholesterol (mmol/L) | 1.04 ± 0.23 | 1.42 ± 0.55 | 0.138 |
| Triglycerides (mmol/L) | 1.68 ± 0.52 | 0.9 ± 0.28 | <0.001 |
| Glucose—OGTT 0 min (mmol/L) | 4.59 ± 1 | 4.71 ± 0.43 | 0.154 |
| Glucose—OGTT 60 min (mmol/L) | 7.37 ± 1.7 | 6.68 ± 2.05 | 0.395 |
| Glucose—OGTT 120 min (mmol/L) | 6.24 ± 1.67 | 5.36 ± 1.23 | 0.152 |
| Insulin—OGTT 0 min (µIU/mL) | 30.67 ± 16.25 | 11.49 ± 5.19 | <0.001 |
| Insulin—OGTT 60 min (µIU/mL) | 174.29 ± 93.12 | 72.32 ± 53.52 | 0.002 |
| Insulin—OGTT 120 min (µIU/mL) | 159.91 ± 83.56 | 52.41 ± 26.94 | 0.001 |
| HOMA-IR | 6.23 ± 3.25 | 2.43 ± 1.19 | 0.001 |
| Insulin resistance, n (%) | 13(92.86%) | 4(40%) | 0.009 |
| Free fatty acids—OGTT 0 min (mmol/L) | 0.93 ± 0.18 | 0.75 ± 0.2 | 0.159 |
| Free fatty acids—OGTT 60 min (mmol/L) | 0.41 ± 0.15 | 0.32 ± 0.08 | 0.093 |
| Free fatty acids—OGTT 120 min (mmol/L) | 0.34 ± 0.12 | 0.25 ± 0.1 | 0.103 |
| Mean systolic blood pressure (mmHg) | 118 ± 9.23 | 108.9 ± 5.43 | 0.006 |
| Mean systolic blood pressure (percentile) | 73.14 ± 8.195 | 44.7 ± 16.97 | 0.001 |
| Mean diastolic blood pressure (mmHg) | 66.64 ± 7.7 | 63.5 ± 3.89 | 0.205 |
| Mean diastolic blood pressure (percentile) | 53.86 ± 21.96 | 41.8 ± 17.03 | 0.145 |
| Mean heart rate | 78.29 ± 8.68 | 73.9 ± 5.3 | 0.14 |
| FTO Gene Region 1 Methylation | FTO Expression | PLAG1 Expression | ||||
|---|---|---|---|---|---|---|
| Anthropometric Parameters | Spearman’s Correlation Coefficient r | p Value | Spearman’s Correlation Coefficient r | p Value | Spearman’s Correlation Coefficient r | p Value |
| BMI (percentile) | 0.428 | 0.029 | 0.449 | 0.021 | 0.363 | 0.068 |
| BMI (kg/m2) | 0.376 | 0.059 | 0.608 | 0.001 | 0.445 | 0.023 |
| BMI SD | 0.415 | 0.035 | 0.473 | 0.015 | 0.430 | 0.028 |
| Waist circumference (cm) | 0.370 | 0.062 | 0.445 | 0.023 | 0.267 | 0.187 |
| Waist circumference (percentile) | 0.504 | 0.014 | 0.509 | 0.013 | 0.369 | 0.083 |
| BF_kg | 0.437 | 0.061 | 0.704 | <0.001 | 0.246 | 0.311 |
| BF_% | 0.289 | 0.229 | 0.837 | <0.001 | 0.523 | 0.022 |
| FTO Gene Region 1 Methylation | FTO Expression | PLAG1 Expression | ||||
|---|---|---|---|---|---|---|
| Biochemical Parameters | Spearman’s Correlation Coefficient r | p Value | Spearman’s Correlation Coefficient r | p Value | Spearman’s Correlation Coefficient r | p Value |
| Triglycerides (mmol/L) | 0.474 | 0.019 | 0.314 | 0.135 | 0.352 | 0.092 |
| Free fatty acids—OGTT 0 min (mmol/L) | −0.094 | 0.719 | 0.661 | 0.004 | 0.563 | 0.019 |
| Glucose—OGTT 0 min (mmol/L) | −0.320 | 0.127 | −0.566 | 0.004 | −0.002 | 0.994 |
| Insulin—OGTT 0 min (µIU/mL) | 0.428 | 0.037 | 0.566 | 0.004 | 0.348 | 0.1 |
| Insulin—OGTT 60 min (µIU/mL) | 0.264 | 0.213 | 0.444 | 0.03 | 0.177 | 0.408 |
| Insulin—OGTT 120 min (µIU/mL) | 0.269 | 0.215 | 0.458 | 0.028 | 0.221 | 0.311 |
| HOMA-IR | 0.344 | 0.1 | 0.442 | 0.03 | 0.322 | 0.125 |
| FTO Gene Region 1 Methylation | FTO Expression | PLAG1 Expression | |||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | padj Value | Mean ± SD | padj Value | Mean ± SD | padj Value | ||
| Insulin resistance | Present, n = 17 | 4.79 ± 4.38 | 0.348 | 302.63 ± 78.95 | 0.037 | 87.68 ± 24.04 | 0.125 |
| Absent, n = 7 | 3.62 ± 2.91 | 208.41 ± 64.84 | 70.81 ± 12.62 | ||||
| FTO Gene Region 1 Methylation | FTO Expression | PLAG1 Expression | ||||
|---|---|---|---|---|---|---|
| OR (95%CI) | p Value | OR (95%CI) | p Value | OR (95%CI) | p Value | |
| Obesity | 1.52 (0.97–2.39) | 0.066 | 4.33 (1.38–13.64) | 0.012 | - | 0.129 |
| Insulin resistance | 0.041 (0.01–7.26) | 0.227 | 2.33 (1.01–5.40) | 0.048 | 2.67 (0.67–10.62) | 0.165 |
| FTO Region 1 Methylation | FTO Expression | PLAG1 Expression | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Standardized Regression Coefficient ± SEM, p-Value) | R2adj | p-Value for R2 | Standardized Regression Coefficient ± SEM, p-Value) | R2adj | p-Value for R2 | Standardized Regression Coefficient ± SEM, p-Value) | R2adj | p-Value for R2 | |
| BMI (kg/m2) | 0.32 ± 0.20, 0.117 | 0.057 | 0.242 | 0.68 ± 0.17, <0.001 | 0.386 | 0.003 | 0.46 ± 0.19, 0.021 | 0.174 | 0.067 |
| BMI percentile | 0.34 ± 0.20, 0.113 | <0.01 | 0.411 | 0.72 ± 0.17, <0.001 Sex: −0.35 ± 0.17, 0.049 | 0.368 | 0.004 | 0.47 ± 0.19, 0.025 | 0.112 | 0.137 |
| BMI SD | 0.47 ± 0.20, 0.031 | 0.11 | 0.156 | 0.79 ± 0.17, <0.001 | 0.445 | 0.002 | 0.37 ± 0.22, 0.103 | 0.013 | 0.371 |
| Waist circumference (cm) | 0.44 ± 0.17, 0.022 | 0.230 | 0.032 | 0.46 ± 0.19, 0.026 | 0.21 | 0.039 | 0.25 ± 0.20, 0.212 | 0.082 | 0.187 |
| Waist circumference percentile | 0.45 ± 0.21, 0.045 | 0.073 | 0.227 | 0.65 ± 0.20, 0.005 | 0.25 | 0.038 | 0.36 ± 0.22, 0.120 | >−0.01 | 0.444 |
| BF_kg | 0.46 ± 0.20, 0.032 Age: 0.47 ± 0.18, 0.022 | 0.423 | 0.01 | 0.75 ± 0.11, <0.001; Age: 0.42 ± 0.11, 0.001 | 0.80 | <0.001 | 0.36 ± 0.20, 0.101 Age: 0.55 ± 0.20, 0.013 | 0.325 | 0.031 |
| BF_% | 0.21 ± 0.22, 0.354 Sex: 0.49 ± 0.22, 0.041 | 0.257 | 0.060 | 0.76 ± 0.12, <0.001 Age: 0.29 ± 0.11, 0.021 | 0.785 | <0.001 | 0.54 ± 0.17, 0.006 Age: 0.42 ± 0.16, 0.022 | 0.531 | 0.002 |
| Triglycerides (mmol/L) | 0.41 ± 0.22, 0.073 | 0.031 | 0.321 | 0.37 ± 0.22, 0.107 | >−0.01 | 0.415 | 0.35 ± 0.22, 0.115 | >−0.01 | 0.434 |
| Free fatty acids—OGTT 0 min (mmol/L) | −0.11 ± 0.26, 0.677 | <0.01 | 0.418 | 0.63 ± 0.23, 0.016 | 0.386 | 0.025 | 0.50 ± 0.21, 0.033 | 0.311 | 0.050 |
| Glucose—OGTT 0 min (mmol/L) | −0.23 ± 0.23, 0.299 | −0.06 | 0.648 | −0.41 ± 0.22, 0.080 | 0.037 | 0.305 | 0.10 ± 0.23, 0.646 | −0.123 | 0.920 |
| Insulin—OGTT 0 min (μIU/mL) | 0.37 ± 0.21, 0.101 | 0.053 | 0.264 | 0.52 ± 0.20, 0.016 | 0.191 | 0.065 | 0.09 ± 0.22, 0.700 | −0.088 | 0.768 |
| Insulin—OGTT 60 min (μIU/mL) | 0.30 ± 0.22, 0.20 | −0.03 | 0.530 | 0.48 ± 0.21, 0.034 | 0.106 | 0.161 | −0.22 ± 0.21, 0.314 | >−0.01 | 0.428 |
| Insulin—OGTT 120 min (μIU/mL) | 0.30 ± 0.23, 0.195 | −0.03 | 0.52 | 0.43 ± 0.22, 0.068 | 0.056 | 0.264 | 0.08 ± 0.23, 0.747 | −0.123 | 0.898 |
| HOMA-IR | 0.30 ± 0.22, 0.184 | >−0.01 | 0.429 | 0.42 ± 0.22, 0.069 | 0.072 | 0.222 | 0.18 ± 0.22, 0.427 | −0.065 | 0.664 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czogała, W.; Czogała, M.; Strojny, W.; Wątor, G.; Wołkow, P.; Wójcik, M.; Bik Multanowski, M.; Tomasik, P.; Wędrychowicz, A.; Kowalczyk, W.; et al. Methylation and Expression of FTO and PLAG1 Genes in Childhood Obesity: Insight into Anthropometric Parameters and Glucose–Lipid Metabolism. Nutrients 2021, 13, 1683. https://doi.org/10.3390/nu13051683
Czogała W, Czogała M, Strojny W, Wątor G, Wołkow P, Wójcik M, Bik Multanowski M, Tomasik P, Wędrychowicz A, Kowalczyk W, et al. Methylation and Expression of FTO and PLAG1 Genes in Childhood Obesity: Insight into Anthropometric Parameters and Glucose–Lipid Metabolism. Nutrients. 2021; 13(5):1683. https://doi.org/10.3390/nu13051683
Chicago/Turabian StyleCzogała, Wojciech, Małgorzata Czogała, Wojciech Strojny, Gracjan Wątor, Paweł Wołkow, Małgorzata Wójcik, Mirosław Bik Multanowski, Przemysław Tomasik, Andrzej Wędrychowicz, Wojciech Kowalczyk, and et al. 2021. "Methylation and Expression of FTO and PLAG1 Genes in Childhood Obesity: Insight into Anthropometric Parameters and Glucose–Lipid Metabolism" Nutrients 13, no. 5: 1683. https://doi.org/10.3390/nu13051683
APA StyleCzogała, W., Czogała, M., Strojny, W., Wątor, G., Wołkow, P., Wójcik, M., Bik Multanowski, M., Tomasik, P., Wędrychowicz, A., Kowalczyk, W., Miklusiak, K., Łazarczyk, A., Hałubiec, P., & Skoczeń, S. (2021). Methylation and Expression of FTO and PLAG1 Genes in Childhood Obesity: Insight into Anthropometric Parameters and Glucose–Lipid Metabolism. Nutrients, 13(5), 1683. https://doi.org/10.3390/nu13051683