Inhibitory Effects of Breast Milk-Derived Lactobacillus rhamnosus Probio-M9 on Colitis-Associated Carcinogenesis by Restoration of the Gut Microbiota in a Mouse Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animal Experiments
2.3. Histopathological Evaluation
2.4. Immunohistochemistry
2.5. Real-Time Polymerase Chain Reaction (PCR) Analysis
2.6. Metagenomic Sequencing and Quality Control
2.7. Taxonomy and Functional Annotation
2.8. Statistical Analyses
2.9. Datasets Generated by the Whole Genome Metagenomics
3. Results
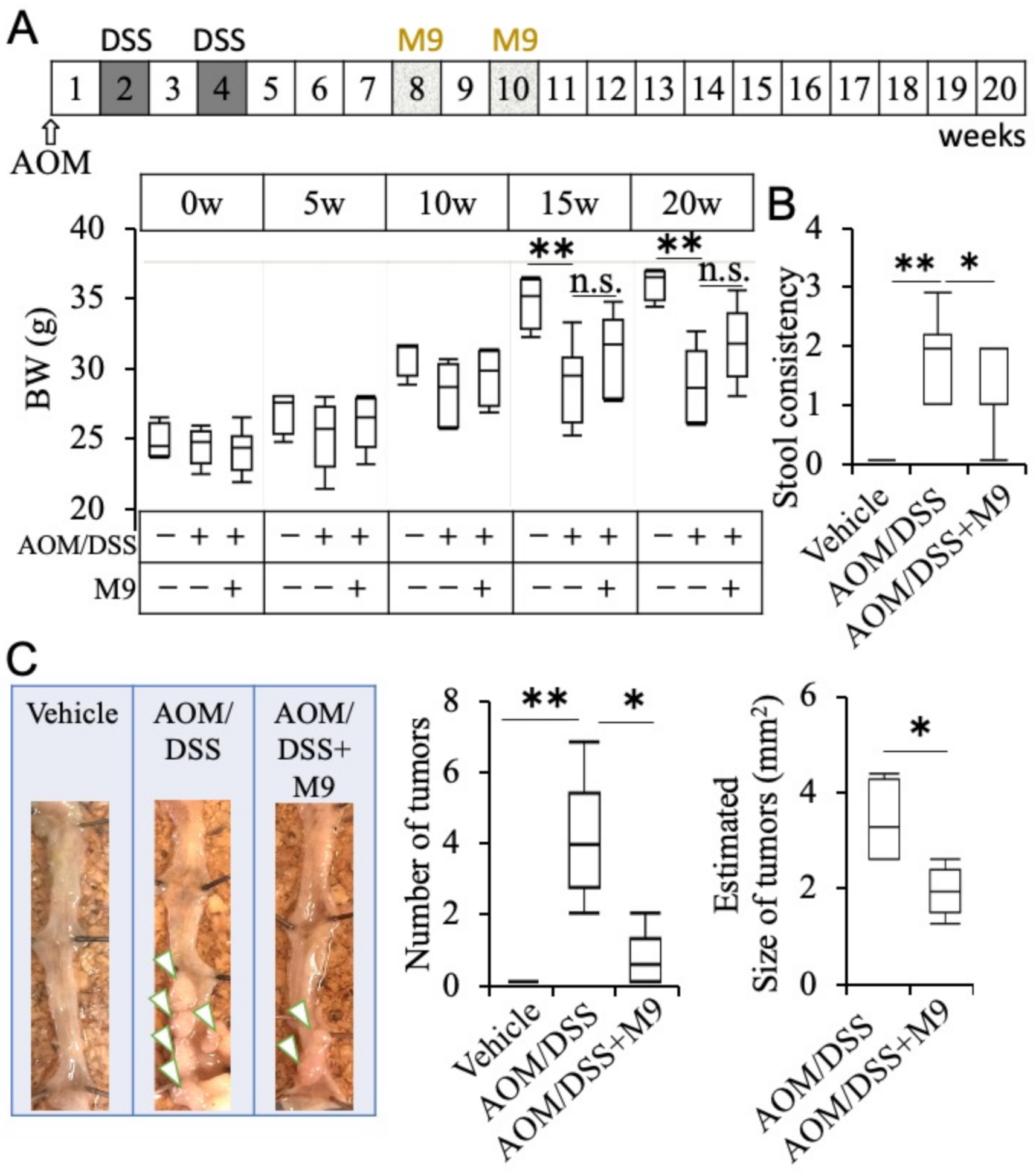
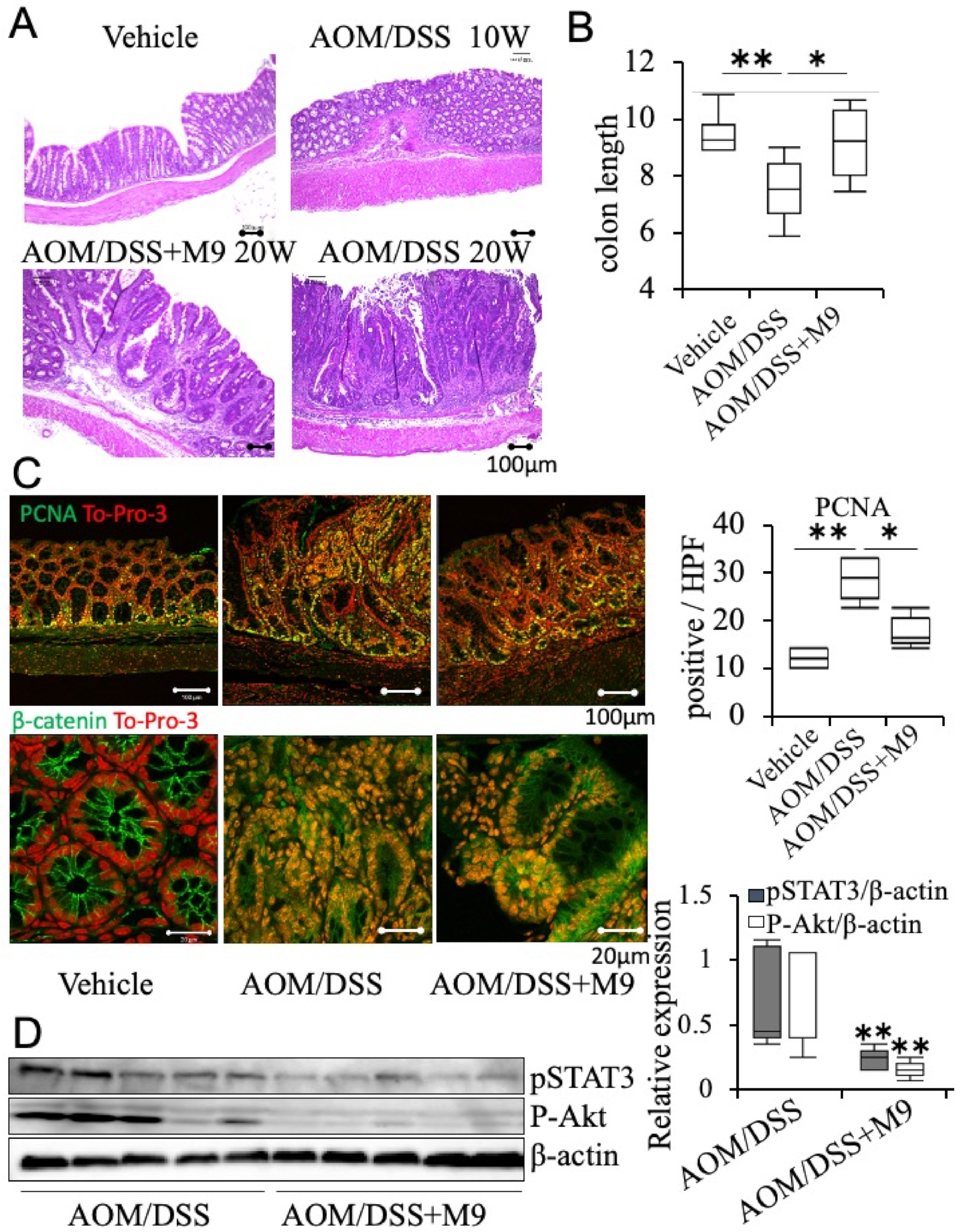
3.1. Probio-M9 Administration Suppresses Inflammatory Carcinogenesis
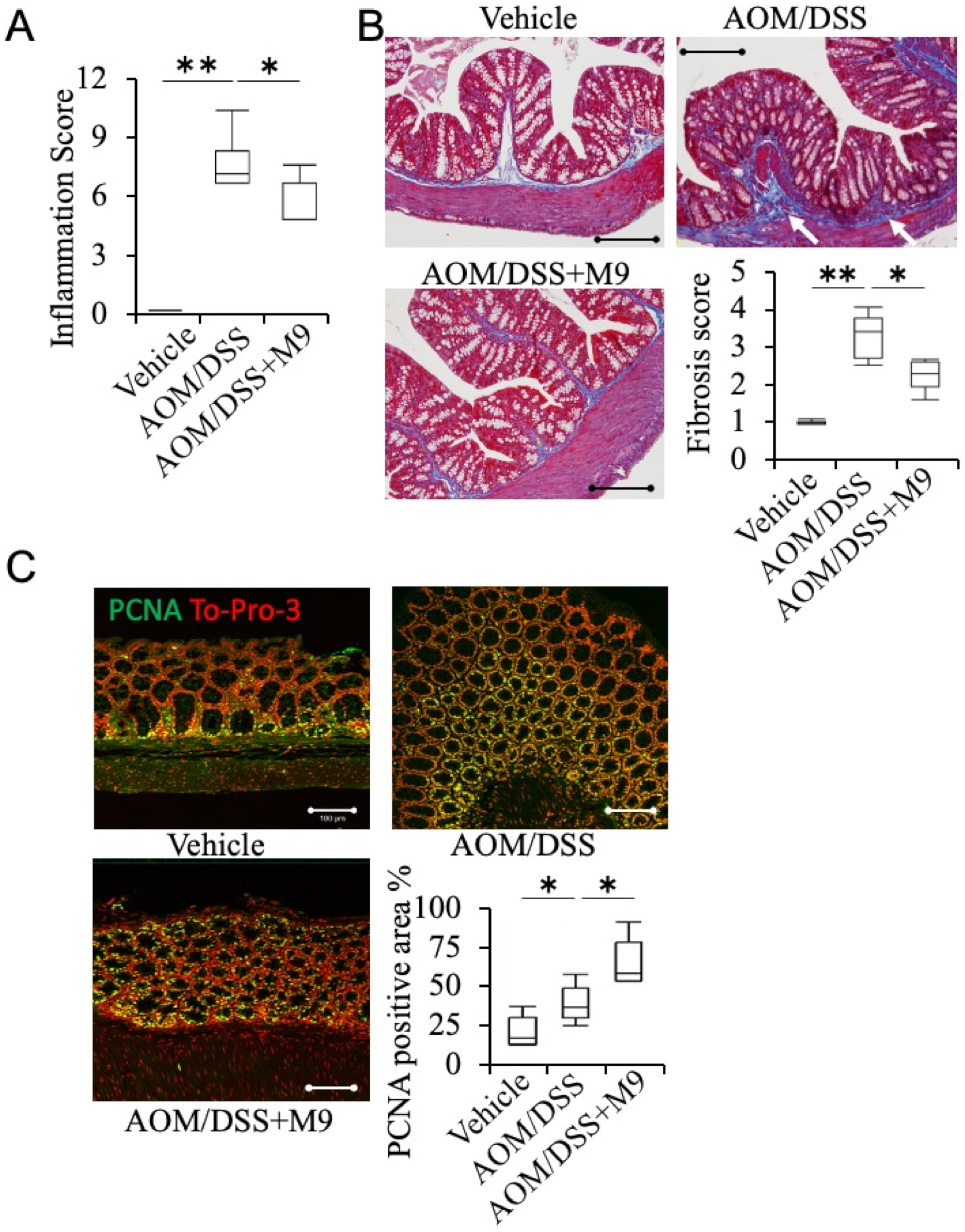
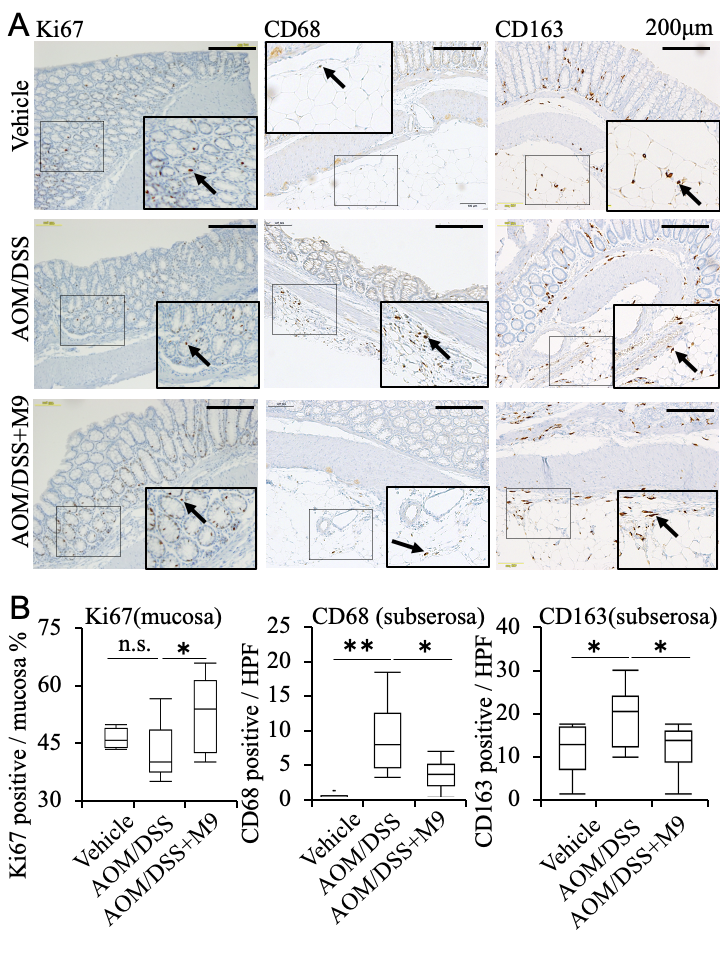
3.2. Analysis of the Therapeutic Effect of Probio-M9 by Pathological Immunostaining in Non-Tumor Areas
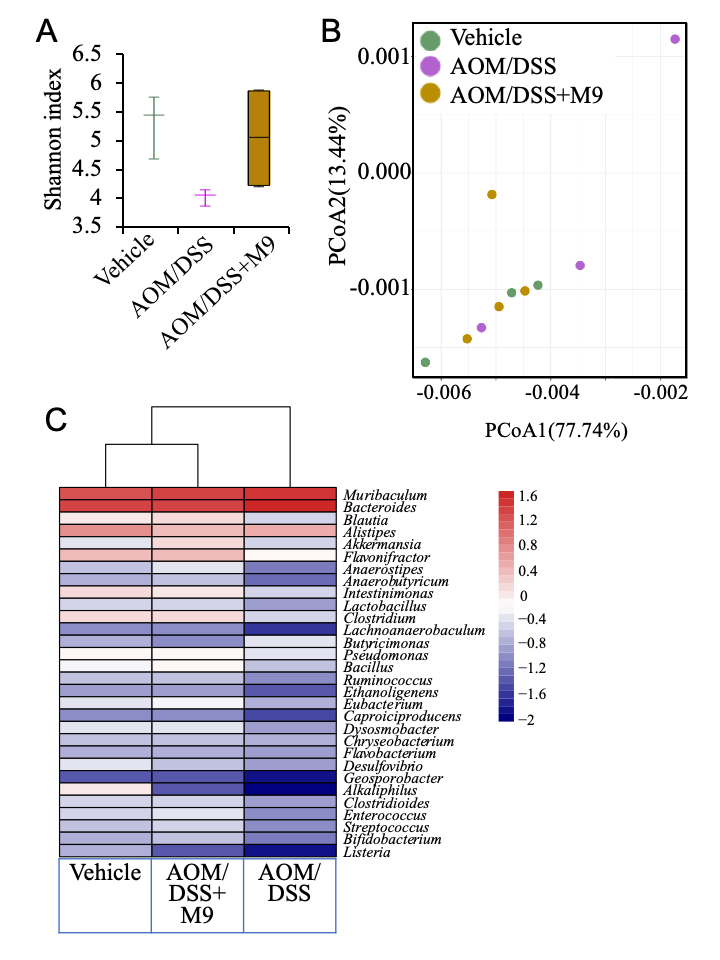
3.3. Changes in the Fecal Microbiota after Treatment
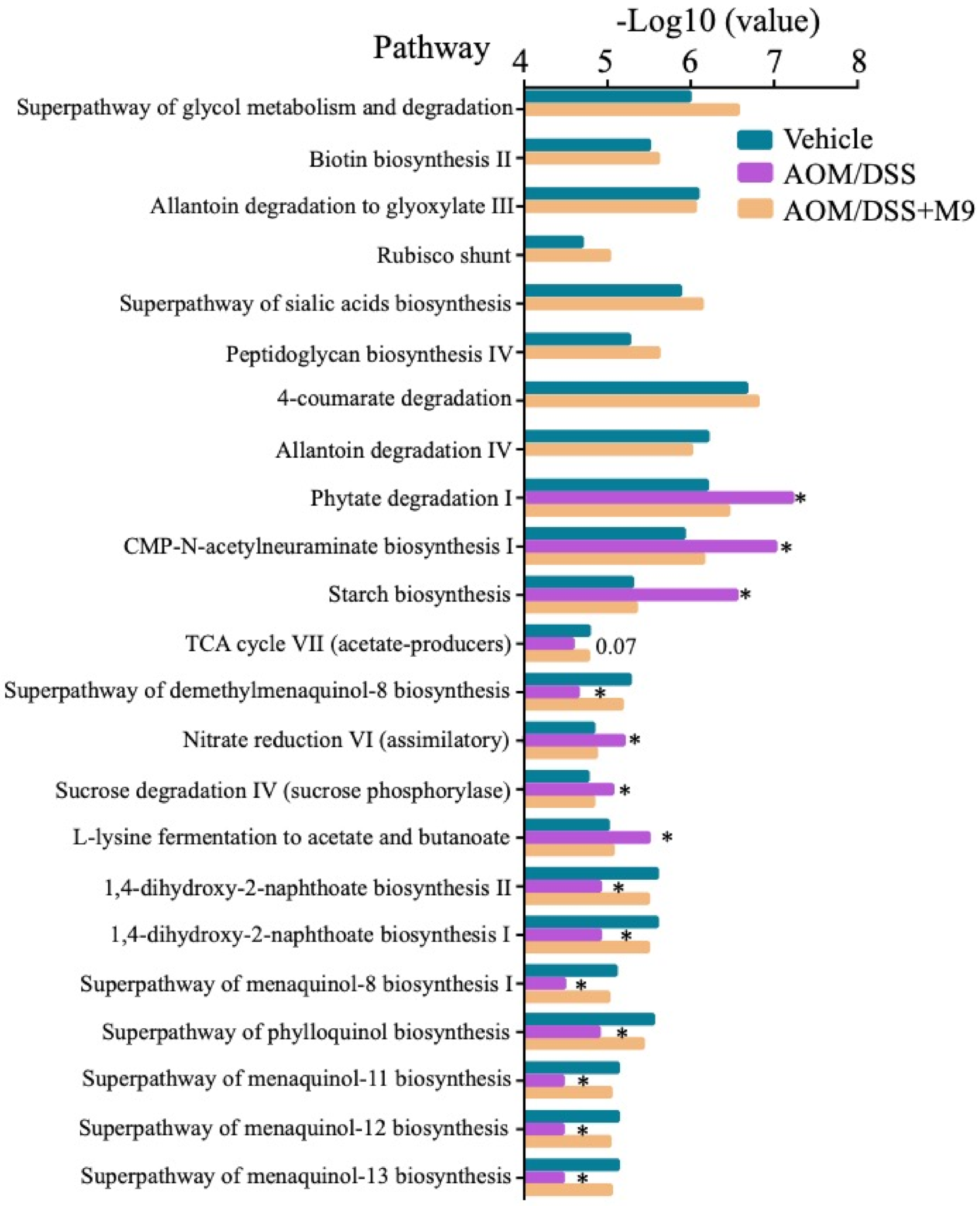
3.4. Changes in the Fecal Microbial Function after Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Beaugerie, L.; Itzkowitz, S.H. Cancers Complicating Inflammatory Bowel Disease. N. Engl. J. Med. 2015, 373, 195. [Google Scholar] [CrossRef] [PubMed]
- Maresso, K.C.; Tsai, K.Y.; Brown, P.H.; Szabo, E.; Lippman, S.; Hawk, E.T. Molecular cancer prevention: Current status and future directions. CA Cancer J. Clin. 2015, 65, 345–383. [Google Scholar] [CrossRef] [PubMed]
- Omar Al-Hassi, H.; Ng, O.; Brookes, M. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2018, 67, 395. [Google Scholar] [CrossRef]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Gagliani, N.; Hu, B.; Huber, S.; Elinav, E.; Flavell, R.A. The fire within: Microbes inflame tumors. Cell 2014, 157, 776–783. [Google Scholar] [CrossRef]
- Appleyard, C.B.; Cruz, M.L.; Isidro, A.A.; Arthur, J.C.; Jobin, C.; De Simone, C. Pretreatment with the probiotic VSL#3 delays transition from inflammation to dysplasia in a rat model of colitis-associated cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G1004–G1013. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Xu, L.; Lochhead, P.; Ko, Y.; Claggett, B.; Leong, R.W.; Ananthakrishnan, A.N. Systematic review with meta-analysis: Breastfeeding and the risk of Crohn’s disease and ulcerative colitis. Aliment. Pharmacol. Ther. 2017, 46, 780–789. [Google Scholar] [CrossRef]
- Liu, W.; Chen, M.; Duo, L.; Wang, J.; Guo, S.; Sun, H.; Menghe, B.; Zhang, H. Characterization of potentially probiotic lactic acid bacteria and bifidobacteria isolated from human colostrum. J. Dairy Sci. 2020, 103, 4013–4025. [Google Scholar] [CrossRef]
- Bromberg, J.; Wang, T.C. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 2009, 15, 79–80. [Google Scholar] [CrossRef]
- Song, G.; Lu, Y.; Yu, Z.; Xu, L.; Liu, J.; Chen, K.; Zhang, P. The inhibitory effect of polysaccharide from Rhizopus nigricans on colitis-associated colorectal cancer. Biomed. Pharmacother. 2019, 112, 108593. [Google Scholar] [CrossRef]
- Yang, C.; Wei, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Dou, R.; Xiong, B. Elevated CD163(+)/CD68(+) Ratio at Tumor Invasive Front is Closely Associated with Aggressive Phenotype and Poor Prognosis in Colorectal Cancer. Int. J. Biol. Sci. 2019, 15, 984–998. [Google Scholar] [CrossRef]
- Johnson, C.H.; Spilker, M.E.; Goetz, L.; Peterson, S.N.; Siuzdak, G. Metabolite and Microbiome Interplay in Cancer Immunotherapy. Cancer Res. 2016, 76, 6146–6152. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Luo, Z.; Guan, L.; Zhu, W. The Colonic Microbiome and Epithelial Transcriptome Are Altered in Rats Fed a High-Protein Diet Compared with a Normal-Protein Diet. J. Nutr. 2016, 146, 474–483. [Google Scholar] [CrossRef]
- Delcenserie, V.; Martel, D.; Lamoureux, M.; Amiot, J.; Boutin, Y.; Roy, D. Immunomodulatory effects of probiotics in the intestinal tract. Curr. Issues Mol. Biol. 2008, 10, 37–54. [Google Scholar]
- Burns, A.J.; Rowland, I.R. Antigenotoxicity of probiotics and prebiotics on faecal water-induced DNA damage in human colon adenocarcinoma cells. Mutat. Res. 2004, 551, 233–243. [Google Scholar] [CrossRef]
- Zhu, J.; Zhu, C.; Ge, S.; Zhang, M.; Jiang, L.; Cui, J.; Ren, F. Lactobacillus salivarius Ren prevent the early colorectal carcinogenesis in 1, 2-dimethylhydrazine-induced rat model. J. Appl. Microbiol. 2014, 117, 208–216. [Google Scholar] [CrossRef]
- Takedatsu, H.; Mitsuyama, K.; Mochizuki, S.; Kobayashi, T.; Sakurai, K.; Takeda, H.; Fujiyama, Y.; Koyama, Y.; Nishihira, J.; Sata, M. A new therapeutic approach using a schizophyllan-based drug delivery system for inflammatory bowel disease. Mol. Ther. 2012, 20, 1234–1241. [Google Scholar] [CrossRef]
- Nakano-Narusawa, Y.; Yokohira, M.; Yamakawa, K.; Saoo, K.; Imaida, K.; Matsuda, Y. Single Intratracheal Quartz Instillation Induced Chronic Inflammation and Tumourigenesis in Rat Lungs. Sci. Rep. 2020, 10, 6647. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Bassaganya-Riera, J.; Viladomiu, M.; Pedragosa, M.; De Simone, C.; Hontecillas, R. Immunoregulatory mechanisms underlying prevention of colitis-associated colorectal cancer by probiotic bacteria. PLoS ONE 2012, 7, e34676. [Google Scholar] [CrossRef]
- Quince, C.; Ijaz, U.Z.; Loman, N.; Eren, A.M.; Saulnier, D.; Russell, J.; Haig, S.J.; Calus, S.T.; Quick, J.; Barclay, A.; et al. Extensive Modulation of the Fecal Metagenome in Children With Crohn’s Disease During Exclusive Enteral Nutrition. Am. J. Gastroenterol. 2015, 110, 1718–1729, quiz 1730. [Google Scholar] [CrossRef]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Shreiner, A.B.; Gillilland, M.G., 3rd; Kitamoto, S.; Ishii, C.; Hirayama, A.; Kuffa, P.; El-Zaatari, M.; Grasberger, H.; Seekatz, A.M.; et al. Functional Characterization of Inflammatory Bowel Disease-Associated Gut Dysbiosis in Gnotobiotic Mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 468–481. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef]
- Udayappan, S.; Manneras-Holm, L.; Chaplin-Scott, A.; Belzer, C.; Herrema, H.; Dallinga-Thie, G.M.; Duncan, S.H.; Stroes, E.S.G.; Groen, A.K.; Flint, H.J.; et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. NPJ Biofilms Microbiomes 2016, 2, 16009. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, X.; Li, M.; Yu, M.; Ping, F.; Zheng, J.; Wang, T.; Wang, X. Vildagliptin increases butyrate-producing bacteria in the gut of diabetic rats. PLoS ONE 2017, 12, e0184735. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Reinoso Webb, C.; den Bakker, H.; Koboziev, I.; Jones-Hall, Y.; Rao Kottapalli, K.; Ostanin, D.; Furr, K.L.; Mu, Q.; Luo, X.M.; Grisham, M.B. Differential Susceptibility to T Cell-Induced Colitis in Mice: Role of the Intestinal Microbiota. Inflamm. Bowel Dis. 2018, 24, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Dobranowski, P.A.; Tang, C.; Sauve, J.P.; Menzies, S.C.; Sly, L.M. Compositional changes to the ileal microbiome precede the onset of spontaneous ileitis in SHIP deficient mice. Gut Microbes 2019, 10, 578–598. [Google Scholar] [CrossRef] [PubMed]
- Eichenlaub, T.; Villadsen, R.; Freitas, F.C.P.; Andrejeva, D.; Aldana, B.I.; Nguyen, H.T.; Petersen, O.W.; Gorodkin, J.; Herranz, H.; Cohen, S.M. Warburg Effect Metabolism Drives Neoplasia in a Drosophila Genetic Model of Epithelial Cancer. Curr. Biol. 2018, 28, 3220–3228 e3226. [Google Scholar] [CrossRef] [PubMed]
| Score | 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|---|
| I | Inflammation | none | mild | moderate | severe | --- |
| E | Extend | none | mucosa and submucosa | transmural | --- | --- |
| R | Regeneration | complete regeneration | almost complete regeneration | regeneration with crypt deletion | surface epithelium not intact | no tissue repair |
| C | Crypt damage | none | 1/3 of basal damaged | 2/3 of basal damaged | only surface epithelium intact | entire crypt and epithelium lost |
| P | Percent involvement | none | 1–25% | 26–50% | 51–75% | 76–100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Hiraishi, K.; Kurahara, L.-H.; Nakano-Narusawa, Y.; Li, X.; Hu, Y.; Matsuda, Y.; Zhang, H.; Hirano, K. Inhibitory Effects of Breast Milk-Derived Lactobacillus rhamnosus Probio-M9 on Colitis-Associated Carcinogenesis by Restoration of the Gut Microbiota in a Mouse Model. Nutrients 2021, 13, 1143. https://doi.org/10.3390/nu13041143
Xu H, Hiraishi K, Kurahara L-H, Nakano-Narusawa Y, Li X, Hu Y, Matsuda Y, Zhang H, Hirano K. Inhibitory Effects of Breast Milk-Derived Lactobacillus rhamnosus Probio-M9 on Colitis-Associated Carcinogenesis by Restoration of the Gut Microbiota in a Mouse Model. Nutrients. 2021; 13(4):1143. https://doi.org/10.3390/nu13041143
Chicago/Turabian StyleXu, Haiyan, Keizo Hiraishi, Lin-Hai Kurahara, Yuko Nakano-Narusawa, Xiaodong Li, Yaopeng Hu, Yoko Matsuda, Heping Zhang, and Katsuya Hirano. 2021. "Inhibitory Effects of Breast Milk-Derived Lactobacillus rhamnosus Probio-M9 on Colitis-Associated Carcinogenesis by Restoration of the Gut Microbiota in a Mouse Model" Nutrients 13, no. 4: 1143. https://doi.org/10.3390/nu13041143
APA StyleXu, H., Hiraishi, K., Kurahara, L.-H., Nakano-Narusawa, Y., Li, X., Hu, Y., Matsuda, Y., Zhang, H., & Hirano, K. (2021). Inhibitory Effects of Breast Milk-Derived Lactobacillus rhamnosus Probio-M9 on Colitis-Associated Carcinogenesis by Restoration of the Gut Microbiota in a Mouse Model. Nutrients, 13(4), 1143. https://doi.org/10.3390/nu13041143







