Human Milk Microbiota and Oligosaccharides: A Glimpse into Benefits, Diversity, and Correlations
Abstract
1. Introduction to Human Milk and Importance of Breastfeeding
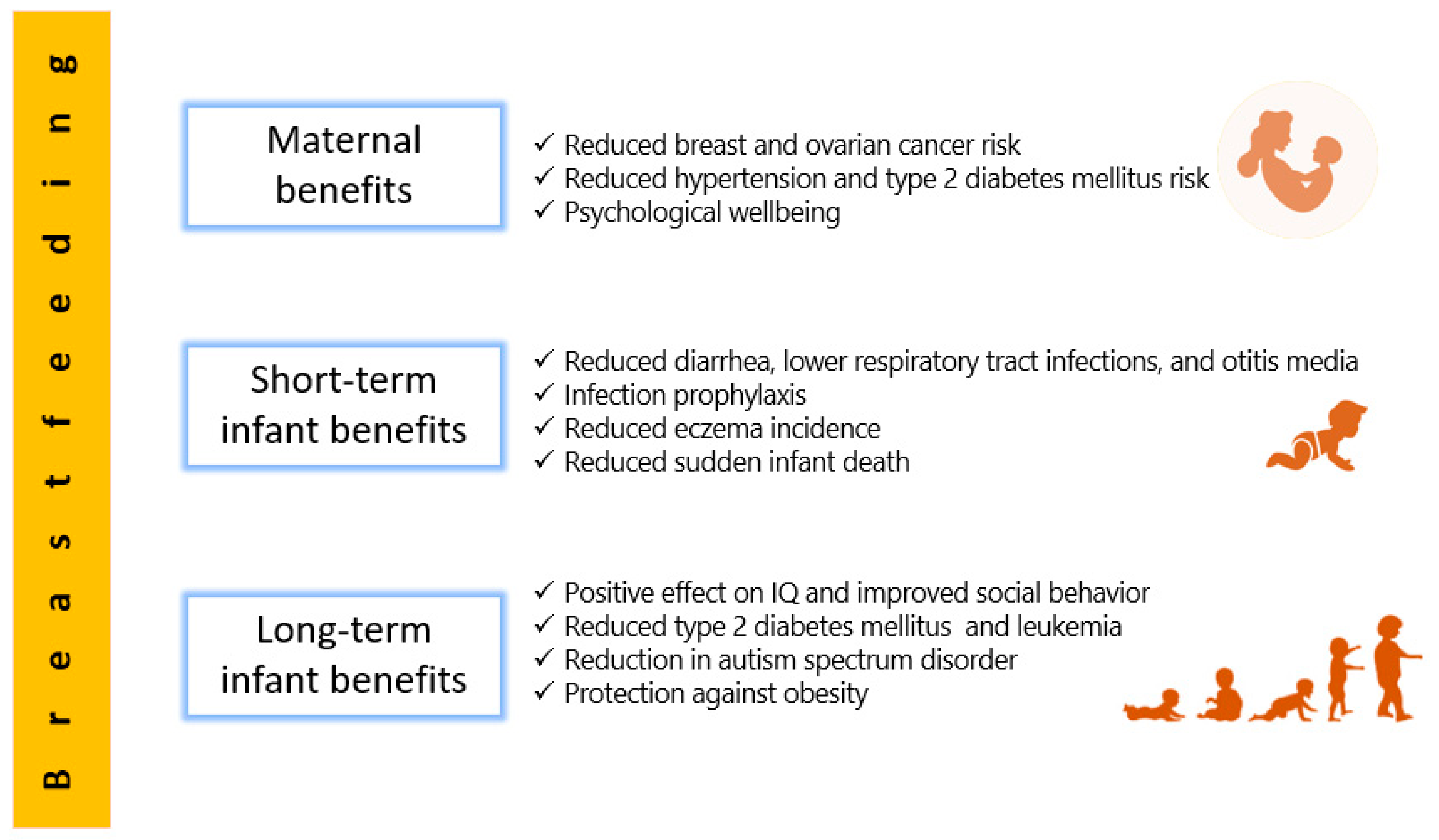
2. Human Milk Benefits
2.1. Short-Term Benefits for Infants
2.2. Long-Term Benefits for Infants
2.3. Maternal Benefits
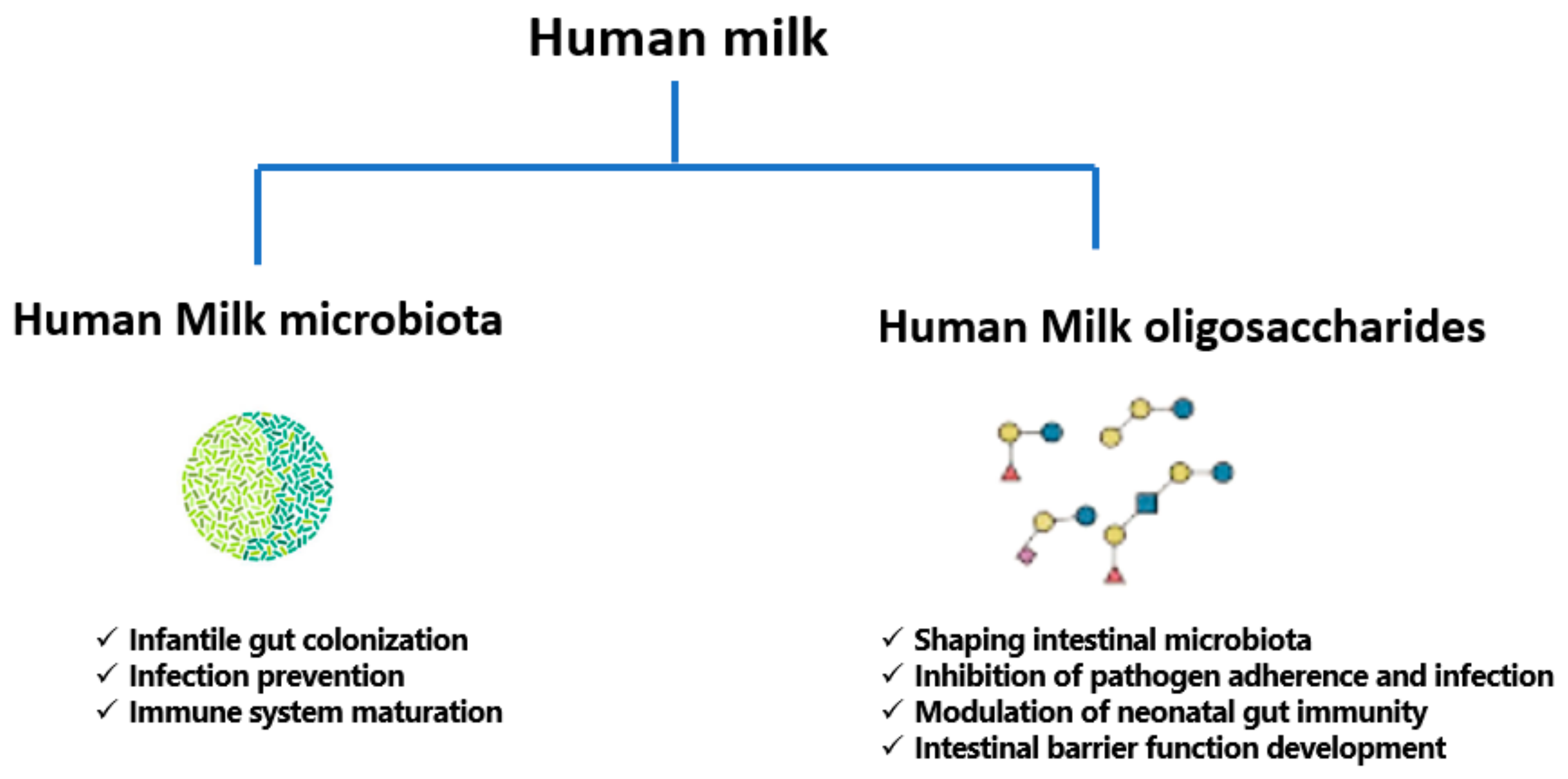
3. A Focused Insight into Human Milk Microbiota
3.1. The Origin of Human Milk Microbiota
3.1.1. The Entero-Mammary Pathway
3.1.2. Retrograde Origin
3.1.3. Transfer from Maternal Skin
3.1.4. Mammary Tissue Origin
3.2. Types of Microbes in Human Milk
3.2.1. Bacteria in Human Milk
3.2.2. Fungi in Human Milk
3.2.3. Human Milk Virome
3.3. Milk Microbiota Diversity and Associated Factors
3.4. The Effect of Maternal Diet on Human Milk Microbiota and on Intestinal Microbiota of Infants
3.5. Beneficial Effects of Milk Microbiota on Infant Health
3.6. Probiotics of Human Milk
4. A Review of Human Milk Oligosaccharides (HMOs)
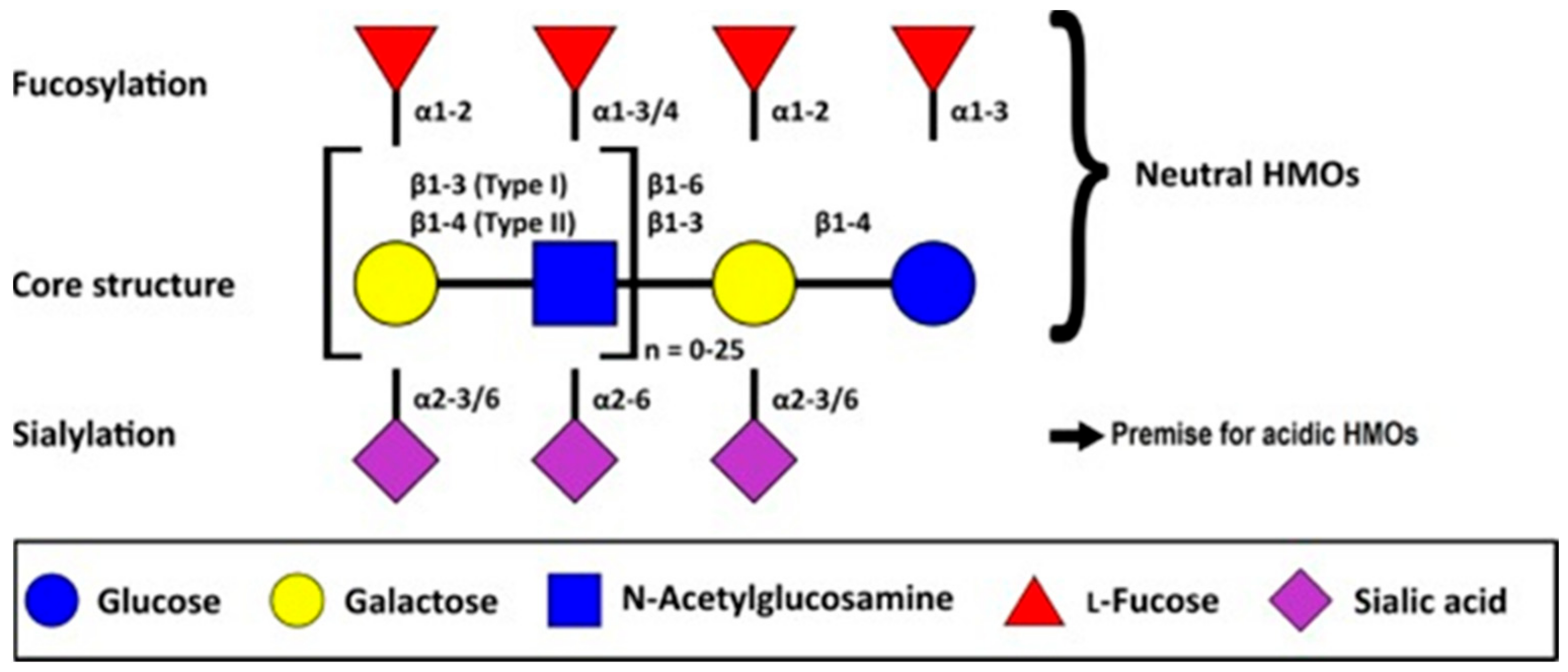
4.1. Overview and Chemical Composition of HMOs
- (a)
- A fraction of 35–50% of the total HMO content; these are neutral and contain fucose at the terminal position. These are called neutral (fucosylated) HMOs.
- (b)
- A fraction of 42–55% of the total HMO content; these are neutral N-containing HMOs and contain N-acetylglucosamine at the terminal position. These are called neutral (nonfucosylated) HMOs.
- (c)
4.2. Milk Groups Related to Lewis Blood Group-Dependent HMOs: Definition and Relevance to HMOs Research
4.3. Physiological Importance and Benefits of HMOs for Infant Health
4.3.1. HMOs and Gut Microbiota Development
4.3.2. HMOs and Infection Prevention
4.3.3. HMOs and Immunomodulatory Effect
4.3.4. HMOs and Intestinal Barrier Function
4.4. HMOs and Prebiotics
5. Milk Microbiota and HMOs: Are There Any Correlations?
6. Future Implications in Milk Microbiota and HMOs Research
7. Conclusions
Funding
Conflicts of Interest
References
- Mosca, F.; Giannì, M.L. Human Milk: Composition and Health Benefits. Pediatr. Med. Chir. 2017, 39, 155. [Google Scholar] [CrossRef]
- Johnson, A.; Kirk, R.; Rosenblum, K.L.; Muzik, M. Enhancing Breastfeeding Rates among African American Women: A Systematic Review of Current Psychosocial Interventions. Breastfeed Med. 2015, 10, 45–62. [Google Scholar] [CrossRef]
- Westerfield, K.L.; Koenig, K.; Oh, R. Breastfeeding: Common Questions and Answers. Am. Fam. Phys. 2018, 98, 368–373. [Google Scholar]
- Bagci Bosi, A.T.; Eriksen, K.G.; Sobko, T.; Wijnhoven, T.M.A.; Breda, J. Breastfeeding Practices and Policies in WHO European Region Member States. Public Health Nutr. 2016, 19, 753–764. [Google Scholar] [CrossRef]
- World Breastfeeding Week 2020 Message. Available online: https://www.who.int/news-room/detail/31-07-2020-world-breastfeeding-week-2020-message (accessed on 19 September 2020).
- Section on Breastfeeding. Breastfeeding and the Use of Human Milk. Pediatrics 2012, 129, e827–e841. [Google Scholar] [CrossRef]
- Ojo-Okunola, A.; Cacciatore, S.; Nicol, M.P.; du Toit, E. The Determinants of the Human Milk Metabolome and Its Role in Infant Health. Metabolites 2020, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human Milk Composition: Nutrients and Bioactive Factors. Pediatr. Clin. 2013, 60, 49–74. [Google Scholar] [CrossRef]
- Gidrewicz, D.A.; Fenton, T.R. A Systematic Review and Meta-Analysis of the Nutrient Content of Preterm and Term Breast Milk. BMC Pediatr. 2014, 14, 216. [Google Scholar] [CrossRef]
- Dror, D.K.; Allen, L.H. Overview of Nutrients in Human Milk. Adv. Nutr. 2018, 9, 278S–294S. [Google Scholar] [CrossRef] [PubMed]
- Picciano, M.F. Representative Values for Constituents of Human Milk. Pediatr. Clin. N. Am. 2001, 48, 263–264. [Google Scholar] [CrossRef]
- Gay, M.C.L.; Koleva, P.T.; Slupsky, C.M.; Toit, E.D.; Eggesbo, M.; Johnson, C.C.; Wegienka, G.; Shimojo, N.; Campbell, D.E.; Prescott, S.L.; et al. Worldwide Variation in Human Milk Metabolome: Indicators of Breast Physiology and Maternal Lifestyle? Nutrients 2018, 10, 1151. [Google Scholar] [CrossRef] [PubMed]
- Musilova, S.; Rada, V.; Vlkova, E.; Bunesova, V. Beneficial Effects of Human Milk Oligosaccharides on Gut Microbiota. Benef. Microbes 2014, 5, 273–283. [Google Scholar] [CrossRef]
- Bhandari, N.; Prajapati, R. Prevalence of Exclusive Breast Feeding and Its Associated Factors among Mothers. Kathmandu Univ. Med. J. KUMJ 2018, 16, 166–170. [Google Scholar]
- Gertosio, C.; Meazza, C.; Pagani, S.; Bozzola, M. Breastfeeding and Its Gamut of Benefits. Minerva. Pediatr. 2016, 68, 201–212. [Google Scholar]
- Binns, C.; Lee, M.; Low, W.Y. The Long-Term Public Health Benefits of Breastfeeding. Asia Pac. J. Public Health 2016, 28, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Horta, B.L.; Victora, C.G. Short-Term Effects of Breastfeeding. A Systematic Review on the Benefits of Breastfeeding on Diarrhoea and Pneumonia Mortality; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Sankar, M.J.; Sinha, B.; Chowdhury, R.; Bhandari, N.; Taneja, S.; Martines, J.; Bahl, R. Optimal Breastfeeding Practices and Infant and Child Mortality: A Systematic Review and Meta-Analysis. Acta Paediatr. 2015, 104, 3–13. [Google Scholar] [CrossRef]
- Lamberti, L.M.; Fischer Walker, C.L.; Noiman, A.; Victora, C.; Black, R.E. Breastfeeding and the Risk for Diarrhea Morbidity and Mortality. BMC Public Health 2011, 11 (Suppl. 3), S15. [Google Scholar] [CrossRef]
- Santos, F.S.; Santos, F.C.S.; dos Santos, L.H.; Leite, A.M.; de Mello, D.F. Breastfeeding and Protection against Diarrhea: An Integrative Review of Literature. Einstein Sao Paulo 2015, 13, 435–440. [Google Scholar] [CrossRef]
- Rouw, E.; von Gartzen, A.; Weißenborn, A. The importance of breastfeeding for the infant. Bundesgesundheitsblatt Gesundh. Gesundh. 2018, 61, 945–951. [Google Scholar] [CrossRef]
- Lamberti, L.M.; Zakarija-Grković, I.; Fischer Walker, C.L.; Theodoratou, E.; Nair, H.; Campbell, H.; Black, R.E. Breastfeeding for Reducing the Risk of Pneumonia Morbidity and Mortality in Children under Two: A Systematic Literature Review and Meta-Analysis. BMC Public Health 2013, 13 (Suppl. 3), S18. [Google Scholar] [CrossRef]
- Quigley, M.A.; Carson, C.; Sacker, A.; Kelly, Y. Exclusive Breastfeeding Duration and Infant Infection. Eur. J. Clin. Nutr. 2016, 70, 1420–1427. [Google Scholar] [CrossRef] [PubMed]
- Bowatte, G.; Tham, R.; Allen, K.J.; Tan, D.J.; Lau, M.; Dai, X.; Lodge, C.J. Breastfeeding and Childhood Acute Otitis Media: A Systematic Review and Meta-Analysis. Acta Paediatr. 2015, 104, 85–95. [Google Scholar] [CrossRef]
- Christensen, N.; Bruun, S.; Søndergaard, J.; Christesen, H.T.; Fisker, N.; Zachariassen, G.; Sangild, P.T.; Husby, S. Breastfeeding and Infections in Early Childhood: A Cohort Study. Pediatrics 2020, 146. [Google Scholar] [CrossRef]
- Zivich, P.; Lapika, B.; Behets, F.; Yotebieng, M. Implementation of Steps 1–9 to Successful Breastfeeding Reduces the Frequency of Mild and Severe Episodes of Diarrhea and Respiratory Tract Infection Among 0–6 Month Infants in Democratic Republic of Congo. Matern. Child Health J. 2018, 22, 762–771. [Google Scholar] [CrossRef]
- Davanzo, R.; Moro, G.; Sandri, F.; Agosti, M.; Moretti, C.; Mosca, F. Breastfeeding and Coronavirus Disease-2019: Ad Interim Indications of the Italian Society of Neonatology Endorsed by the Union of European Neonatal & Perinatal Societies. Matern. Child Nutr. 2020, 16, e13010. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Breastfeeding and COVID-19. Available online: https://www.who.int/news-room/commentaries/detail/breastfeeding-and-covid-19 (accessed on 30 January 2021).
- CDC Coronavirus Disease (COVID-19) and Breastfeeding. Available online: https://www.cdc.gov/breastfeeding/breastfeeding-special-circumstances/maternal-or-infant-illnesses/covid-19-and-breastfeeding.html (accessed on 30 January 2021).
- Fernández-Carrasco, F.J.; Vázquez-Lara, J.M.; González-Mey, U.; Gómez-Salgado, J.; Parrón-Carreño, T.; Rodríguez-Díaz, L. Coronavirus Covid-19 Infection and Breastfeeding: An Exploratory Review. Available online: https://pubmed.ncbi.nlm.nih.gov/32458823/ (accessed on 20 September 2020).
- Greer, F.R.; Sicherer, S.H.; Burks, A.W.; Committee on Nutrition; Section on Allergy and Immunology. The Effects of Early Nutritional Interventions on the Development of Atopic Disease in Infants and Children: The Role of Maternal Dietary Restriction, Breastfeeding, Hydrolyzed Formulas, and Timing of Introduction of Allergenic Complementary Foods. Pediatrics 2019, 143. [Google Scholar] [CrossRef]
- Matsumoto, N.; Yorifuji, T.; Nakamura, K.; Ikeda, M.; Tsukahara, H.; Doi, H. Breastfeeding and Risk of Food Allergy: A Nationwide Birth Cohort in Japan. Allergol. Int. 2020, 69, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Carlin, R.F.; Moon, R.Y. Risk Factors, Protective Factors, and Current Recommendations to Reduce Sudden Infant Death Syndrome: A Review. JAMA Pediatr. 2017, 171, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Alm, B.; Wennergren, G.; Möllborg, P.; Lagercrantz, H. Breastfeeding and Dummy Use Have a Protective Effect on Sudden Infant Death Syndrome. Acta Paediatr. 2016, 105, 31–38. [Google Scholar] [CrossRef]
- Horne, R.S.C. Sudden Infant Death Syndrome: Current Perspectives. Intern. Med. J. 2019, 49, 433–438. [Google Scholar] [CrossRef]
- Adams, S.M.; Ward, C.E.; Garcia, K.L. Sudden Infant Death Syndrome. Am. Fam. Phys. 2015, 91, 778–783. [Google Scholar]
- Hauck, F.R.; Thompson, J.M.D.; Tanabe, K.O.; Moon, R.Y.; Vennemann, M.M. Breastfeeding and Reduced Risk of Sudden Infant Death Syndrome: A Meta-Analysis. Pediatrics 2011, 128, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Franco, P.; Scaillet, S.; Wermenbol, V.; Valente, F.; Groswasser, J.; Kahn, A. The Influence of a Pacifier on Infants’ Arousals from Sleep. J. Pediatr. 2000, 136, 775–779. [Google Scholar]
- Horne, R.S.C.; Parslow, P.M.; Ferens, D.; Watts, A.-M.; Adamson, T.M. Comparison of Evoked Arousability in Breast and Formula Fed Infants. Arch. Dis. Child. 2004, 89, 22–25. [Google Scholar]
- Horta, B.L.; Loret de Mola, C.; Victora, C.G. Long-Term Consequences of Breastfeeding on Cholesterol, Obesity, Systolic Blood Pressure and Type 2 Diabetes: A Systematic Review and Meta-Analysis. Acta Paediatr. 2015, 104, 30–37. [Google Scholar] [CrossRef]
- Marseglia, L.; Manti, S.; D’Angelo, G.; Cuppari, C.; Salpietro, V.; Filippelli, M.; Trovato, A.; Gitto, E.; Salpietro, C.; Arrigo, T. Obesity and Breastfeeding: The Strength of Association. Women Birth 2015, 28, 81–86. [Google Scholar] [CrossRef]
- Yan, J.; Liu, L.; Zhu, Y.; Huang, G.; Wang, P.P. The Association between Breastfeeding and Childhood Obesity: A Meta-Analysis. BMC Public Health 2014, 14, 1267. [Google Scholar] [CrossRef]
- Ortega-García, J.A.; Kloosterman, N.; Alvarez, L.; Tobarra-Sánchez, E.; Cárceles-Álvarez, A.; Pastor-Valero, R.; López-Hernández, F.A.; Sánchez-Solis, M.; Claudio, L. Full Breastfeeding and Obesity in Children: A Prospective Study from Birth to 6 Years. Child Obes. 2018, 14, 327–337. [Google Scholar] [CrossRef]
- Ak, V. Does Breastfeeding Shape Food Preferences? Links to Obesity. Available online: https://pubmed.ncbi.nlm.nih.gov/28903109/ (accessed on 20 September 2020).
- Dahiya, D.K.; Puniya, M.; Shandilya, U.K.; Dhewa, T.; Kumar, N.; Kumar, S.; Puniya, A.K.; Shukla, P. Gut Microbiota Modulation and Its Relationship with Obesity Using Prebiotic Fibers and Probiotics: A Review. Front. Microbiol. 2017, 8, 563. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.L.; Kim, J.H. Human Milk and Necrotizing Enterocolitis. Semin. Pediatr. Surg. 2018, 27, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Cotten, C.M. Modifiable Risk Factors in Necrotizing Enterocolitis. Clin. Perinatol. 2019, 46, 129–143. [Google Scholar] [CrossRef]
- Maffei, D.; Schanler, R.J. Human Milk Is the Feeding Strategy to Prevent Necrotizing Enterocolitis! Semin. Perinatol. 2017, 41, 36–40. [Google Scholar] [CrossRef]
- Lindoso, L.; Mondal, K.; Venkateswaran, S.; Somineni, H.K.; Ballengee, C.; Walters, T.D.; Griffiths, A.; Noe, J.D.; Crandall, W.; Snapper, S.; et al. The Effect of Early-Life Environmental Exposures on Disease Phenotype and Clinical Course of Crohn’s Disease in Children. Am. J. Gastroenterol. 2018, 113, 1524–1529. [Google Scholar] [CrossRef]
- Silano, M.; Agostoni, C.; Sanz, Y.; Guandalini, S. Infant Feeding and Risk of Developing Celiac Disease: A Systematic Review. BMJ Open 2016, 6, e009163. [Google Scholar] [CrossRef]
- Murff, H.J.; Edwards, T.L. Endogenous Production of Long-Chain Polyunsaturated Fatty Acids and Metabolic Disease Risk. Curr. Cardiovasc. Risk Rep. 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Amitay, E.L.; Keinan-Boker, L. Breastfeeding and Childhood Leukemia Incidence: A Meta-Analysis and Systematic Review. JAMA Pediatr. 2015, 169, e151025. [Google Scholar] [CrossRef]
- Küçükçongar, A.; Oğuz, A.; Pınarlı, F.G.; Karadeniz, C.; Okur, A.; Kaya, Z.; Çelik, B. Breastfeeding and Childhood Cancer: Is Breastfeeding Preventative to Childhood Cancer? Pediatric Hematol. Oncol. 2015, 32, 374–381. [Google Scholar]
- Bar, S.; Milanaik, R.; Adesman, A. Long-Term Neurodevelopmental Benefits of Breastfeeding. Curr. Opin. Pediatr. 2016, 28, 559–566. [Google Scholar] [CrossRef]
- Kramer, M.S.; Aboud, F.; Mironova, E.; Vanilovich, I.; Platt, R.W.; Matush, L.; Igumnov, S.; Fombonne, E.; Bogdanovich, N.; Ducruet, T.; et al. Breastfeeding and Child Cognitive Development: New Evidence from a Large Randomized Trial. Arch. Gen. Psychiatry 2008, 65, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Holme, A.; MacArthur, C.; Lancashire, R. The Effects of Breastfeeding on Cognitive and Neurological Development of Children at 9 Years. Child Care Health Dev. 2010, 36, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; Rodriguez-Palmero, M. Polyunsaturated Fatty Acids in Human Milk and Their Role in Early Infant Development. J. Mammary Gland Biol. Neoplasia 1999, 4, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Bernard, J.Y.; Armand, M.; Peyre, H.; Garcia, C.; Forhan, A.; De Agostini, M.; Charles, M.-A.; Heude, B.; EDEN Mother-Child Cohort Study Group (Etude des Déterminants pré- et postnatals précoces du développement et de la santé de l’Enfant). Breastfeeding, Polyunsaturated Fatty Acid Levels in Colostrum and Child Intelligence Quotient at Age 5–6 Years. J. Pediatr. 2017, 183, 43–50.e3. [Google Scholar] [CrossRef] [PubMed]
- Hadley, K.B.; Ryan, A.S.; Forsyth, S.; Gautier, S.; Salem, N. The Essentiality of Arachidonic Acid in Infant Development. Nutrients 2016, 8, 216. [Google Scholar] [CrossRef] [PubMed]
- Ghozy, S.; Tran, L.; Naveed, S.; Quynh, T.T.H.; Helmy Zayan, A.; Waqas, A.; Sayed, A.K.H.; Karimzadeh, S.; Hirayama, K.; Huy, N.T. Association of Breastfeeding Status with Risk of Autism Spectrum Disorder: A Systematic Review, Dose-Response Analysis and Meta-Analysis. Asian J. Psychiatr. 2020, 48, 101916. [Google Scholar] [CrossRef] [PubMed]
- Kielbratowska, B.; Kazmierczak, M.; Michalek, J.; Preis, K. Temperament and the Mother-Infant Dyad: Associations with Breastfeeding and Formula Feeding with a Bottle. Infant Ment. Health J. 2015, 36, 243–250. [Google Scholar] [CrossRef]
- Shelton, K.H.; Collishaw, S.; Rice, F.J.; Harold, G.T.; Thapar, A. Using a Genetically Informative Design to Examine the Relationship between Breastfeeding and Childhood Conduct Problems. Eur. Child Adolesc. Psychiatry 2011, 20, 571–579. [Google Scholar] [CrossRef]
- Schwarz, E.B.; Nothnagle, M. The Maternal Health Benefits of Breastfeeding. Am. Fam. Phys. 2015, 91, 603–604. [Google Scholar]
- Victora, C.G.; Bahl, R.; Barros, A.J.D.; França, G.V.A.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st Century: Epidemiology, Mechanisms, and Lifelong Effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef]
- Unar-Munguía, M.; Torres-Mejía, G.; Colchero, M.A.; González de Cosío, T. Breastfeeding Mode and Risk of Breast Cancer: A Dose-Response Meta-Analysis. J. Hum. Lact. 2017, 33, 422–434. [Google Scholar] [CrossRef]
- Pan, H.; He, Z.; Ling, L.; Ding, Q.; Chen, L.; Zha, X.; Zhou, W.; Liu, X.; Wang, S. Reproductive Factors and Breast Cancer Risk among BRCA1 or BRCA2 Mutation Carriers: Results from Ten Studies. Cancer Epidemiol. 2014, 38, 1–8. [Google Scholar] [CrossRef]
- Manrique Tejedor, J.; Figuerol Calderó, M.I.; Cuéllar De Frutos, A. Breastfeeding as a method of breast cancer prevention. Rev. Enferm. 2015, 38, 32–38. [Google Scholar]
- Modugno, F.; Goughnour, S.L.; Wallack, D.; Edwards, R.P.; Odunsi, K.; Kelley, J.L.; Moysich, K.; Ness, R.B.; Brooks, M.M. Breastfeeding Factors and Risk of Epithelial Ovarian Cancer. Gynecol. Oncol. 2019, 153, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.K.; Ma, S.H.; Choi, J.-Y.; Hwang, Y.; Ahn, C.; Kim, B.-G.; Kim, Y.-M.; Kim, J.W.; Kang, S.; Kim, J.; et al. The Effect of Breastfeeding Duration and Parity on the Risk of Epithelial Ovarian Cancer: A Systematic Review and Meta-Analysis. J. Prev. Med. Public Health 2016, 49, 349–366. [Google Scholar] [CrossRef]
- Rajaei, S.; Rigdon, J.; Crowe, S.; Tremmel, J.; Tsai, S.; Assimes, T.L. Breastfeeding Duration and the Risk of Coronary Artery Disease. J. Womens Health Larchmt 2019, 28, 30–36. [Google Scholar] [CrossRef]
- Peters, S.A.; van der Schouw, Y.T.; Wood, A.M.; Sweeting, M.J.; Moons, K.G.; Weiderpass, E.; Arriola, L.; Benetou, V.; Boeing, H.; Bonnet, F.; et al. Parity, Breastfeeding and Risk of Coronary Heart Disease: A Pan-European Case-Cohort Study. Eur. J. Prev. Cardiol. 2016, 23, 1755–1765. [Google Scholar] [CrossRef] [PubMed]
- Stuebe, A.M.; Michels, K.B.; Willett, W.C.; Manson, J.E.; Rexrode, K.; Rich-Edwards, J.W. Duration of Lactation and Incidence of Myocardial Infarction in Middle-to-Late Adulthood. Am. J. Obstet. Gynecol. 2009, 200, 138.e1–138.e8. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.R.; Shrestha, D. Breastfeeding for Diabetes Prevention. J. Pak. Med. Assoc. 2016, 66, S88–S90. [Google Scholar]
- Park, S.; Choi, N.-K. Breastfeeding and Maternal Hypertension. Am. J. Hypertens. 2018, 31, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Qu, G.; Wang, L.; Tang, X.; Wu, W.; Sun, Y. Association Between Duration of Breastfeeding and Maternal Hypertension: A Systematic Review and Meta-Analysis. Breastfeed Med. 2018, 13, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, H.; Bliddal, M.; Støvring, H.; Rasmussen, K.M.; Gunderson, E.P.; Køber, L.; Sørensen, T.I.A.; Nohr, E.A. Breastfeeding and Later Maternal Risk of Hypertension and Cardiovascular Disease—The Role of Overall and Abdominal Obesity. Prev. Med. 2018, 114, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Jacobs, S.; Kröger, J.; Fritsche, A.; Schienkiewitz, A.; Rubin, D.; Boeing, H.; Schulze, M.B. Breast-Feeding and Maternal Risk of Type 2 Diabetes: A Prospective Study and Meta-Analysis. Diabetologia 2014, 57, 1355–1365. [Google Scholar] [CrossRef]
- Luo, J.; Hendryx, M.; LeBlanc, E.S.; Shadyab, A.H.; Qi, L.; Sealy-Jefferson, S.; Manson, J.E. Associations Between Parity, Breastfeeding, and Risk of Maternal Type 2 Diabetes Among Postmenopausal Women. Obstet. Gynecol. 2019, 134, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Stuebe, A.M.; Rich-Edwards, J.W. The Reset Hypothesis: Lactation and Maternal Metabolism. Am. J. Perinatol. 2009, 26, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, Y.; Xu, M.; Ning, G.; Lu, J.; Dai, M.; Xu, B.; Sun, J.; Sun, W.; Lai, S.; et al. Circulating Prolactin and Risk of Type 2 Diabetes: A Prospective Study. Am. J. Epidemiol. 2016, 184, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Bobrow, K.L.; Quigley, M.A.; Green, J.; Reeves, G.K.; Beral, V.; Million Women Study Collaborators. Persistent Effects of Women’s Parity and Breastfeeding Patterns on Their Body Mass Index: Results from the Million Women Study. Int. J. Obes. 2013, 37, 712–717. [Google Scholar] [CrossRef]
- Groër, M.W. Differences between Exclusive Breastfeeders, Formula-Feeders, and Controls: A Study of Stress, Mood, and Endocrine Variables. Biol. Res. Nurs. 2005, 7, 106–117. [Google Scholar] [CrossRef]
- Doan, T.; Gardiner, A.; Gay, C.L.; Lee, K.A. Breast-Feeding Increases Sleep Duration of New Parents. J. Perinat. Neonatal. Nurs. 2007, 21, 200–206. [Google Scholar] [CrossRef]
- Figueiredo, B.; Dias, C.C.; Brandão, S.; Canário, C.; Nunes-Costa, R. Breastfeeding and Postpartum Depression: State of the Art Review. J. Pediatr. Rio J. 2013, 89, 332–338. [Google Scholar] [CrossRef]
- Da Silva Tanganhito, D.; Bick, D.; Chang, Y.-S. Breastfeeding Experiences and Perspectives among Women with Postnatal Depression: A Qualitative Evidence Synthesis. Women Birth 2020, 33, 231–239. [Google Scholar] [CrossRef]
- Lara-Cinisomo, S.; McKenney, K.; Di Florio, A.; Meltzer-Brody, S. Associations Between Postpartum Depression, Breastfeeding, and Oxytocin Levels in Latina Mothers. Breastfeed Med. 2017, 12, 436–442. [Google Scholar] [CrossRef]
- Diez-Sampedro, A.; Flowers, M.; Olenick, M.; Maltseva, T.; Valdes, G. Women’s Choice Regarding Breastfeeding and Its Effect on Well-Being. Nurs. Womens Health 2019, 23, 383–389. [Google Scholar] [CrossRef]
- Silva, C.S.; Lima, M.C.; Sequeira-de-Andrade, L.A.S.; Oliveira, J.S.; Monteiro, J.S.; Lima, N.M.S.; Santos, R.M.A.B.; Lira, P.I.C. Association between Postpartum Depression and the Practice of Exclusive Breastfeeding in the First Three Months of Life. J. Pediatr. Rio J. 2017, 93, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Cooklin, A.R.; Amir, L.H.; Nguyen, C.D.; Buck, M.L.; Cullinane, M.; Fisher, J.R.W.; Donath, S.M.; CASTLE Study Team. Physical Health, Breastfeeding Problems and Maternal Mood in the Early Postpartum: A Prospective Cohort Study. Arch. Womens Ment. Health 2018, 21, 365–374. [Google Scholar] [CrossRef]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Rantasalo, I.; Kauppinen, M.A. The Occurrence of Staphylococcus Aureus in Mother’s Milk. Ann. Chir. Gynaecol. Fenn. 1959, 48, 246–258. [Google Scholar] [PubMed]
- Ojo-Okunola, A.; Nicol, M.; du Toit, E. Human Breast Milk Bacteriome in Health and Disease. Nutrients 2018, 10, 1643. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human Milk Is a Source of Lactic Acid Bacteria for the Infant Gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef]
- Civardi, E.; Garofoli, F.; Tzialla, C.; Paolillo, P.; Bollani, L.; Stronati, M. Microorganisms in Human Milk: Lights and Shadows. J. Matern. Fetal. Neonatal. Med. 2013, 26 (Suppl. 2), 30–34. [Google Scholar] [CrossRef]
- Ruiz, L.; García-Carral, C.; Rodriguez, J.M. Unfolding the Human Milk Microbiome Landscape in the Omics Era. Front. Microbiol. 2019, 10, 1378. [Google Scholar] [CrossRef]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 25, 324–335.e4. [Google Scholar] [CrossRef] [PubMed]
- Damaceno, Q.S.; Souza, J.P.; Nicoli, J.R.; Paula, R.L.; Assis, G.B.; Figueiredo, H.C.; Azevedo, V.; Martins, F.S. Evaluation of Potential Probiotics Isolated from Human Milk and Colostrum. Probiotics Antimicrob. Proteins 2017, 9, 371–379. [Google Scholar] [CrossRef]
- Rodríguez, J.M. The Origin of Human Milk Bacteria: Is There a Bacterial Entero-Mammary Pathway during Late Pregnancy and Lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef]
- Perez, P.F.; Doré, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J.; Donnet-Hughes, A. Bacterial Imprinting of the Neonatal Immune System: Lessons from Maternal Cells? Pediatrics 2007, 119, e724–e732. [Google Scholar] [CrossRef]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.P.; Ricciardi-Castagnoli, P. Dendritic Cells Express Tight Junction Proteins and Penetrate Gut Epithelial Monolayers to Sample Bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef]
- Geddes, D.T. The Use of Ultrasound to Identify Milk Ejection in Women—Tips and Pitfalls. Int. Breastfeed J. 2009, 4, 5. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the Diversity and Temporal Stability of Bacterial Communities in Human Milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef]
- Demmelmair, H.; Jiménez, E.; Collado, M.C.; Salminen, S.; McGuire, M.K. Maternal and Perinatal Factors Associated with the Human Milk Microbiome. Curr. Dev. Nutr. 2020, 4, nzaa027. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of Human Breast Tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [PubMed]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the Microbiome of Nipple Aspirate Fluid of Breast Cancer Survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef]
- Going, J.J.; Moffat, D.F. Escaping from Flatland: Clinical and Biological Aspects of Human Mammary Duct Anatomy in Three Dimensions. J. Pathol. 2004, 203, 538–544. [Google Scholar] [CrossRef]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef] [PubMed]
- Jeurink, P.V.; van Bergenhenegouwen, J.; Jiménez, E.; Knippels, L.M.J.; Fernández, L.; Garssen, J.; Knol, J.; Rodríguez, J.M.; Martín, R. Human Milk: A Source of More Life than We Imagine. Benef. Microbes 2013, 4, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical Mother-Neonate Transfer of Maternal Gut Bacteria via Breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Togo, A.; Dufour, J.-C.; Lagier, J.-C.; Dubourg, G.; Raoult, D.; Million, M. Repertoire of Human Breast and Milk Microbiota: A Systematic Review. Future Microbiol. 2019, 14, 623–641. [Google Scholar] [CrossRef] [PubMed]
- Latuga, M.S.; Stuebe, A.; Seed, P.C. A Review of the Source and Function of Microbiota in Breast Milk. Semin. Reprod. Med. 2014, 32, 68–73. [Google Scholar] [CrossRef]
- Fitzstevens, J.L.; Smith, K.C.; Hagadorn, J.I.; Caimano, M.J.; Matson, A.P.; Brownell, E.A. Systematic Review of the Human Milk Microbiota. Nutr. Clin. Pract. 2017, 32, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The Human Milk Microbiome Changes over Lactation and Is Shaped by Maternal Weight and Mode of Delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef]
- Amir, L.H.; Donath, S.M.; Garland, S.M.; Tabrizi, S.N.; Bennett, C.M.; Cullinane, M.; Payne, M.S. Does Candida and/or Staphylococcus Play a Role in Nipple and Breast Pain in Lactation? A Cohort Study in Melbourne, Australia. BMJ Open 2013, 3. [Google Scholar] [CrossRef]
- Jiménez, E.; de Andrés, J.; Manrique, M.; Pareja-Tobes, P.; Tobes, R.; Martínez-Blanch, J.F.; Codoñer, F.M.; Ramón, D.; Fernández, L.; Rodríguez, J.M. Metagenomic Analysis of Milk of Healthy and Mastitis-Suffering Women. J. Hum. Lact. 2015, 31, 406–415. [Google Scholar] [CrossRef]
- Moossavi, S.; Fehr, K.; Derakhshani, H.; Sbihi, H.; Robertson, B.; Bode, L.; Brook, J.; Turvey, S.E.; Moraes, T.J.; Becker, A.B.; et al. Human Milk Fungi: Environmental Determinants and Inter-Kingdom Associations with Milk Bacteria in the CHILD Cohort Study. BMC Microbiol. 2020, 20, 146. [Google Scholar] [CrossRef]
- Dinleyici, M.; Pérez-Brocal, V.; Arslanoglu, S.; Aydemir, O.; Ozumut, S.S.; Tekin, N.; Vandenplas, Y.; Moya, A.; Dinleyici, E.C. Human Milk Mycobiota Composition: Relationship with Gestational Age, Delivery Mode, and Birth Weight. Benef. Microbes 2020, 11, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhao, C.; Zhang, H.; Mattei, L.; Sherrill-Mix, S.; Bittinger, K.; Kessler, L.R.; Wu, G.D.; Baldassano, R.N.; DeRusso, P.; et al. The Stepwise Assembly of the Neonatal Virome Is Modulated by Breastfeeding. Nature 2020, 581, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Pannaraj, P.S.; Ly, M.; Cerini, C.; Saavedra, M.; Aldrovandi, G.M.; Saboory, A.A.; Johnson, K.M.; Pride, D.T. Shared and Distinct Features of Human Milk and Infant Stool Viromes. Front. Microbiol. 2018, 9, 1162. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, S.; Pannaraj, P.S. Beyond the Bacterial Microbiome: Virome of Human Milk and Effects on the Developing Infant. Milk Mucosal Immun. Microbiome Impact Neonate 2020, 94, 86–93. [Google Scholar] [CrossRef]
- Khodayar-Pardo, P.; Mira-Pascual, L.; Collado, M.C.; Martínez-Costa, C. Impact of Lactation Stage, Gestational Age and Mode of Delivery on Breast Milk Microbiota. J. Perinatol. 2014, 34, 599–605. [Google Scholar] [CrossRef]
- Decker, E.; Engelmann, G.; Findeisen, A.; Gerner, P.; Laass, M.; Ney, D.; Posovszky, C.; Hoy, L.; Hornef, M.W. Cesarean Delivery Is Associated with Celiac Disease but Not Inflammatory Bowel Disease in Children. Pediatrics 2010, 125, e1433–e1440. [Google Scholar] [CrossRef]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human Milk Microbiota Profiles in Relation to Birthing Method, Gestation and Infant Gender. Microbiome 2016, 4, 1. [Google Scholar] [CrossRef]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the Infant Microbiome Community Structure and Function across Multiple Body Sites and in Relation to Mode of Delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef]
- Gérard, P. Gut Microbiota and Obesity. Cell. Mol. Life Sci. 2016, 73, 147–162. [Google Scholar] [CrossRef]
- Olivares, M.; Albrecht, S.; De Palma, G.; Ferrer, M.D.; Castillejo, G.; Schols, H.A.; Sanz, Y. Human Milk Composition Differs in Healthy Mothers and Mothers with Celiac Disease. Eur. J. Nutr. 2015, 54, 119–128. [Google Scholar] [CrossRef]
- Valitutti, F.; Cucchiara, S.; Fasano, A. Celiac Disease and the Microbiome. Nutrients 2019, 11, 2403. [Google Scholar] [CrossRef]
- Hermansson, H.; Kumar, H.; Collado, M.C.; Salminen, S.; Isolauri, E.; Rautava, S. Breast Milk Microbiota Is Shaped by Mode of Delivery and Intrapartum Antibiotic Exposure. Front. Nutr. 2019, 6, 4. [Google Scholar] [CrossRef]
- Browne, P.D.; Aparicio, M.; Alba, C.; Hechler, C.; Beijers, R.; Rodríguez, J.M.; Fernández, L.; de Weerth, C. Human Milk Microbiome and Maternal Postnatal Psychosocial Distress. Front. Microbiol. 2019, 10, 2333. [Google Scholar] [CrossRef]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct Patterns in Human Milk Microbiota and Fatty Acid Profiles Across Specific Geographic Locations. Front. Microbiol. 2016, 7, 1619. [Google Scholar] [CrossRef]
- Boix-Amorós, A.; Puente-Sánchez, F.; du Toit, E.; Linderborg, K.M.; Zhang, Y.; Yang, B.; Salminen, S.; Isolauri, E.; Tamames, J.; Mira, A.; et al. Mycobiome Profiles in Breast Milk from Healthy Women Depend on Mode of Delivery, Geographic Location, and Interaction with Bacteria. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Ayoub Moubareck, C.; Lootah, M.; Tahlak, M.; Venema, K. Profiles of Human Milk Oligosaccharides and Their Relations to the Milk Microbiota of Breastfeeding Mothers in Dubai. Nutrients 2020, 12, 1727. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.; Martín, V.; Jiménez, E.; Mader, I.; Rodríguez, J.M.; Fernández, L. Lactobacilli and Bifidobacteria in Human Breast Milk: Influence of Antibiotherapy and Other Host and Clinical Factors. J. Pediatric Gastroenterol. Nutr. 2014, 59, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Breast Milk Microbiota: A Review of the Factors That Influence Composition. J. Infect. 2020, 81, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.E.; Carrothers, J.M.; Lackey, K.A.; Beatty, N.F.; York, M.A.; Brooker, S.L.; Shafii, B.; Price, W.J.; Settles, M.L.; McGuire, M.A.; et al. Human Milk Microbial Community Structure Is Relatively Stable and Related to Variations in Macronutrient and Micronutrient Intakes in Healthy Lactating Women. J. Nutr. 2017, 147, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Carrothers, J.M.; York, M.A.; Brooker, S.L.; Lackey, K.A.; Williams, J.E.; Shafii, B.; Price, W.J.; Settles, M.L.; McGuire, M.A.; McGuire, M.K. Fecal Microbial Community Structure Is Stable over Time and Related to Variation in Macronutrient and Micronutrient Intakes in Lactating Women123. J. Nutr. 2015, 145, 2379–2388. [Google Scholar] [CrossRef]
- Padilha, M.; Danneskiold-Samsøe, N.B.; Brejnrod, A.; Hoffmann, C.; Cabral, V.P.; Iaucci, J.d.M.; Sales, C.H.; Fisberg, R.M.; Cortez, R.V.; Brix, S.; et al. The Human Milk Microbiota Is Modulated by Maternal Diet. Microorganisms 2019, 7, 502. [Google Scholar] [CrossRef]
- Miyake, Y.; Sasaki, S.; Tanaka, K.; Hirota, Y. Consumption of Vegetables, Fruit, and Antioxidants during Pregnancy and Wheeze and Eczema in Infants. Allergy 2010, 65, 758–765. [Google Scholar] [CrossRef]
- Coakley, M.; Ross, R.P.; Nordgren, M.; Fitzgerald, G.; Devery, R.; Stanton, C. Conjugated Linoleic Acid Biosynthesis by Human-Derived Bifidobacterium Species. J. Appl. Microbiol. 2003, 94, 138–145. [Google Scholar] [CrossRef]
- Shively, C.A.; Register, T.C.; Appt, S.E.; Clarkson, T.B.; Uberseder, B.; Clear, K.Y.J.; Wilson, A.S.; Chiba, A.; Tooze, J.A.; Cook, K.L. Consumption of Mediterranean versus Western Diet Leads to Distinct Mammary Gland Microbiome Populations. Cell Rep. 2018, 25, 47–56.e3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lv, J.; Pan, L.; Zhang, Y. Roles and Applications of Probiotic Lactobacillus Strains. Appl. Microbiol. Biotechnol. 2018, 102, 8135–8143. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Babakobi, M.D.; Reshef, L.; Gihaz, S.; Belgorodsky, B.; Fishman, A.; Bujanover, Y.; Gophna, U. Effect of Maternal Diet and Milk Lipid Composition on the Infant Gut and Maternal Milk Microbiomes. Nutrients 2020, 12, 2539. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Viappiani, A.; Lugli, G.A.; Ferrario, C.; Gioiosa, L.; Ferrarini, A.; et al. Deciphering Bifidobacterial-Mediated Metabolic Interactions and Their Impact on Gut Microbiota by a Multi-Omics Approach. ISME J. 2016, 10, 1656–1668. [Google Scholar] [CrossRef]
- Simpson, M.R.; Avershina, E.; Storrø, O.; Johnsen, R.; Rudi, K.; Øien, T. Breastfeeding-Associated Microbiota in Human Milk Following Supplementation with Lactobacillus Rhamnosus GG, Lactobacillus Acidophilus La-5, and Bifidobacterium Animalis Ssp. Lactis Bb-12. J. Dairy Sci. 2018, 101, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Rautava, S. Early Microbial Contact, the Breast Milk Microbiome and Child Health. J. Dev. Orig. Health Dis. 2016, 7, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Gueimonde, M.; Laitinen, K.; Salminen, S.; Isolauri, E. Breast Milk: A Source of Bifidobacteria for Infant Gut Development and Maturation? Neonatology 2007, 92, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.R.; Barile, D.; Underwood, M.A.; Mills, D.A. The Impact of the Milk Glycobiome on the Neonate Gut Microbiota. Annu. Rev. Anim. Biosci. 2015, 3, 419–445. [Google Scholar] [CrossRef]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of Bifidobacteria within the Infant Gut Microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Foroni, E.; Pizzetti, P.; Giubellini, V.; Ribbera, A.; Merusi, P.; Cagnasso, P.; Bizzarri, B.; de’Angelis, G.L.; Shanahan, F.; et al. Exploring the Diversity of the Bifidobacterial Population in the Human Intestinal Tract. Appl. Environ. Microbiol. 2009, 75, 1534–1545. [Google Scholar] [CrossRef]
- Chong, C.Y.L.; Bloomfield, F.H.; O’Sullivan, J.M. Factors Affecting Gastrointestinal Microbiome Development in Neonates. Nutrients 2018, 10, 274. [Google Scholar] [CrossRef]
- Solís, G.; de Los Reyes-Gavilan, C.G.; Fernández, N.; Margolles, A.; Gueimonde, M. Establishment and Development of Lactic Acid Bacteria and Bifidobacteria Microbiota in Breast-Milk and the Infant Gut. Anaerobe 2010, 16, 307–310. [Google Scholar] [CrossRef]
- Bergmann, H.; Rodríguez, J.M.; Salminen, S.; Szajewska, H. Probiotics in Human Milk and Probiotic Supplementation in Infant Nutrition: A Workshop Report. Br. J. Nutr. 2014, 112, 1119–1128. [Google Scholar] [CrossRef]
- Jiao, X.; Fu, M.-D.; Wang, Y.-Y.; Xue, J.; Zhang, Y. Bifidobacterium and Lactobacillus for Preventing Necrotizing Enterocolitis in Very-Low-Birth-Weight Preterm Infants: A Systematic Review and Meta-Analysis. World J. Pediatr. 2020, 16, 135–142. [Google Scholar] [CrossRef]
- Jiménez, E.; Delgado, S.; Maldonado, A.; Arroyo, R.; Albújar, M.; García, N.; Jariod, M.; Fernández, L.; Gómez, A.; Rodríguez, J.M. Staphylococcus Epidermidis: A Differential Trait of the Fecal Microbiota of Breast-Fed Infants. BMC Microbiol. 2008, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Peirotén, Á.; Medina, M.; Arqués, J.L.; Rodríguez-Mínguez, E. Virulence and Antibiotic Resistance of Enterococci Isolated from Healthy Breastfed Infants. Microb. Drug Resist. 2018, 24, 63–69. [Google Scholar] [CrossRef]
- Martín, V.; Maldonado, A.; Fernández, L.; Rodríguez, J.M.; Connor, R.I. Inhibition of Human Immunodeficiency Virus Type 1 by Lactic Acid Bacteria from Human Breastmilk. Breastfeed Med. 2010, 5, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO. Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. In Proceedings of the Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria, Cordoba, Argentina, 1–4 October 2001. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Bozzi Cionci, N.; Baffoni, L.; Gaggìa, F.; Di Gioia, D. Therapeutic Microbiology: The Role of Bifidobacterium Breve as Food Supplement for the Prevention/Treatment of Paediatric Diseases. Nutrients 2018, 10, 1723. [Google Scholar] [CrossRef]
- Halloran, K.; Underwood, M.A. Probiotic Mechanisms of Action. Early Hum. Dev. 2019, 135, 58–65. [Google Scholar] [CrossRef]
- Bermudez-Brito, M.; Plaza-Díaz, J.; Muñoz-Quezada, S.; Gómez-Llorente, C.; Gil, A. Probiotic Mechanisms of Action. Ann. Nutr. Metab. 2012, 61, 160–174. [Google Scholar] [CrossRef]
- Montalban-Arques, A.; De Schryver, P.; Bossier, P.; Gorkiewicz, G.; Mulero, V.; Gatlin, D.M.; Galindo-Villegas, J. Selective Manipulation of the Gut Microbiota Improves Immune Status in Vertebrates. Front. Immunol. 2015, 6, 512. [Google Scholar] [CrossRef]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus Reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Li, L.; Fang, Z.; Liu, X.; Hu, W.; Lu, W.; Lee, Y.-K.; Zhao, J.; Zhang, H.; Chen, W. Lactobacillus Reuteri Attenuated Allergic Inflammation Induced by HDM in the Mouse and Modulated Gut Microbes. PLoS ONE 2020, 15, e0231865. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qi, C.; Zhu, H.; Yu, R.; Xie, C.; Peng, Y.; Yin, S.-W.; Fan, J.; Zhao, S.; Sun, J. Lactobacillus Reuteri Improves Gut Barrier Function and Affects Diurnal Variation of the Gut Microbiota in Mice Fed a High-Fat Diet. Food Funct. 2019, 10, 4705–4715. [Google Scholar] [CrossRef]
- Savino, F.; Ceratto, S.; Poggi, E.; Cartosio, M.E.; Cordero di Montezemolo, L.; Giannattasio, A. Preventive Effects of Oral Probiotic on Infantile Colic: A Prospective, Randomised, Blinded, Controlled Trial Using Lactobacillus Reuteri DSM 17938. Benef. Microbes 2015, 6, 245–251. [Google Scholar] [CrossRef] [PubMed]
- López-Huertas, E. Safety and Efficacy of Human Breast Milk Lactobacillus Fermentum CECT 5716. A Mini-Review of Studies with Infant Formulae. Benef. Microbes 2015, 6, 219–224. [Google Scholar] [CrossRef]
- Asan-Ozusaglam, M.; Gunyakti, A. Lactobacillus Fermentum Strains from Human Breast Milk with Probiotic Properties and Cholesterol-Lowering Effects. Food Sci. Biotechnol. 2019, 28, 501–509. [Google Scholar] [CrossRef]
- Pérez-Cano, F.J.; Dong, H.; Yaqoob, P. In Vitro Immunomodulatory Activity of Lactobacillus Fermentum CECT5716 and Lactobacillus Salivarius CECT5713: Two Probiotic Strains Isolated from Human Breast Milk. Immunobiology 2010, 215, 996–1004. [Google Scholar] [CrossRef]
- Riaz Rajoka, M.S.; Zhao, H.; Lu, Y.; Lian, Z.; Li, N.; Hussain, N.; Shao, D.; Jin, M.; Li, Q.; Shi, J. Anticancer Potential against Cervix Cancer (HeLa) Cell Line of Probiotic Lactobacillus Casei and Lactobacillus Paracasei Strains Isolated from Human Breast Milk. Food Funct. 2018, 9, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Riaz Rajoka, M.S.; Zhao, H.; Mehwish, H.M.; Li, N.; Lu, Y.; Lian, Z.; Shao, D.; Jin, M.; Li, Q.; Zhao, L.; et al. Anti-Tumor Potential of Cell Free Culture Supernatant of Lactobacillus Rhamnosus Strains Isolated from Human Breast Milk. Food Res. Int. 2019, 123, 286–297. [Google Scholar] [CrossRef]
- Guarino, A.; Guandalini, S.; Lo Vecchio, A. Probiotics for Prevention and Treatment of Diarrhea. J. Clin. Gastroenterol. 2015, 49 (Suppl. 1), S37–S45. [Google Scholar] [CrossRef]
- Maldonado, J.; Gil-Campos, M.; Maldonado-Lobón, J.A.; Benavides, M.R.; Flores-Rojas, K.; Jaldo, R.; Jiménez Del Barco, I.; Bolívar, V.; Valero, A.D.; Prados, E.; et al. Evaluation of the Safety, Tolerance and Efficacy of 1-Year Consumption of Infant Formula Supplemented with Lactobacillus Fermentum CECT5716 Lc40 or Bifidobacterium Breve CECT7263: A Randomized Controlled Trial. BMC Pediatr. 2019, 19, 361. [Google Scholar] [CrossRef]
- Fahmy, C.A.; Gamal-Eldeen, A.M.; El-Hussieny, E.A.; Raafat, B.M.; Mehanna, N.S.; Talaat, R.M.; Shaaban, M.T. Bifidobacterium Longum Suppresses Murine Colorectal Cancer through the Modulation of OncomiRs and Tumor Suppressor MiRNAs. Nutr. Cancer 2019, 71, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Kwon, B.; Ku, S.; Ji, G.E. The Efficacy of Bifidobacterium Longum BORI and Lactobacillus Acidophilus AD031 Probiotic Treatment in Infants with Rotavirus Infection. Nutrients 2017, 9, 887. [Google Scholar] [CrossRef] [PubMed]
- Khalkhali, S.; Mojgani, N. In Vitro and in Vivo Safety Analysis of Enterococcus Faecium 2C Isolated from Human Breast Milk. Microb. Pathog. 2018, 116, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Khalkhali, S.; Mojgani, N. Bacteriocinogenic Potential and Virulence Traits of Enterococcus Faecium and E. Faecalis Isolated from Human Milk. Iran. J. Microbiol. 2017, 9, 224–233. [Google Scholar]
- Hassan, Z.; Mustafa, S.; Rahim, R.A.; Isa, N.M. Anti-Breast Cancer Effects of Live, Heat-Killed and Cytoplasmic Fractions of Enterococcus Faecalis and Staphylococcus Hominis Isolated from Human Breast Milk. Cell. Dev. Biol. Anim. 2016, 52, 337–348. [Google Scholar] [CrossRef]
- Wiciński, M.; Sawicka, E.; Gębalski, J.; Kubiak, K.; Malinowski, B. Human Milk Oligosaccharides: Health Benefits, Potential Applications in Infant Formulas, and Pharmacology. Nutrients 2020, 12, 266. [Google Scholar] [CrossRef]
- Bode, L. Human Milk Oligosaccharides: Every Baby Needs a Sugar Mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef]
- Cheng, L.; Akkerman, R.; Kong, C.; Walvoort, M.T.C.; de Vos, P. More than Sugar in the Milk: Human Milk Oligosaccharides as Essential Bioactive Molecules in Breast Milk and Current Insight in Beneficial Effects. Crit. Rev. Food Sci. Nutr. 2020, 1–17. [Google Scholar] [CrossRef]
- Thurl, S.; Munzert, M.; Boehm, G.; Matthews, C.; Stahl, B. Systematic Review of the Concentrations of Oligosaccharides in Human Milk. Nutr. Rev. 2017, 75, 920–933. [Google Scholar] [CrossRef]
- Gabrielli, O.; Zampini, L.; Galeazzi, T.; Padella, L.; Santoro, L.; Peila, C.; Giuliani, F.; Bertino, E.; Fabris, C.; Coppa, G.V. Preterm Milk Oligosaccharides during the First Month of Lactation. Pediatrics 2011, 128, e1520–e1531. [Google Scholar] [CrossRef]
- Nakhla, T.; Fu, D.; Zopf, D.; Brodsky, N.L.; Hurt, H. Neutral Oligosaccharide Content of Preterm Human Milk. Br. J. Nutr. 1999, 82, 361–367. [Google Scholar] [CrossRef]
- Bering, S.B. Human Milk Oligosaccharides to Prevent Gut Dysfunction and Necrotizing Enterocolitis in Preterm Neonates. Nutrients 2018, 10, 1461. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; Lebrilla, C.B.; Mills, D.A.; German, J.B.; Freeman, S.L. Breast Milk Oligosaccharides: Structure-Function Relationships in the Neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A.; Baumgärtner, F.; Albermann, C. Production of Human Milk Oligosaccharides by Enzymatic and Whole-Cell Microbial Biotransformations. J. Biotechnol. 2017, 258, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Díaz, J.; Fontana, L.; Gil, A. Human Milk Oligosaccharides and Immune System Development. Nutrients 2018, 10, 1038. [Google Scholar] [CrossRef]
- Bode, L.; Jantscher-Krenn, E. Structure-Function Relationships of Human Milk Oligosaccharides. Adv. Nutr. 2012, 3, 383S–391S. [Google Scholar] [CrossRef]
- Totten, S.M.; Zivkovic, A.M.; Wu, S.; Ngyuen, U.; Freeman, S.L.; Ruhaak, L.R.; Darboe, M.K.; German, J.B.; Prentice, A.M.; Lebrilla, C.B. Comprehensive Profiles of Human Milk Oligosaccharides Yield Highly Sensitive and Specific Markers for Determining Secretor Status in Lactating Mothers. J. Proteome Res. 2012, 11, 6124–6133. [Google Scholar] [CrossRef]
- McGuire, M.K.; Meehan, C.L.; McGuire, M.A.; Williams, J.E.; Foster, J.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; Mbugua, S.; Moore, S.E.; et al. What’s Normal? Oligosaccharide Concentrations and Profiles in Milk Produced by Healthy Women Vary Geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100. [Google Scholar] [CrossRef] [PubMed]
- Denomme, G.A. Molecular Basis of Blood Group Expression. Transfus. Apher. Sci. 2011, 44, 53–63. [Google Scholar] [CrossRef]
- Ewald, D.R.; Sumner, S.C.J. Blood Type Biochemistry and Human Disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 517–535. [Google Scholar] [CrossRef] [PubMed]
- Wacklin, P.; Mäkivuokko, H.; Alakulppi, N.; Nikkilä, J.; Tenkanen, H.; Räbinä, J.; Partanen, J.; Aranko, K.; Mättö, J. Secretor Genotype (FUT2 Gene) Is Strongly Associated with the Composition of Bifidobacteria in the Human Intestine. PLoS ONE 2011, 6, e20113. [Google Scholar] [CrossRef]
- Tonon, K.M.; de Morais, M.B.; Abrão, A.C.F.V.; Miranda, A.; Morais, T.B. Maternal and Infant Factors Associated with Human Milk Oligosaccharides Concentrations According to Secretor and Lewis Phenotypes. Nutrients 2019, 11, 1358. [Google Scholar] [CrossRef]
- Kobata, A. Structures and Application of Oligosaccharides in Human Milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 731–747. [Google Scholar] [CrossRef]
- Thurl, S.; Henker, J.; Siegel, M.; Tovar, K.; Sawatzki, G. Detection of Four Human Milk Groups with Respect to Lewis Blood Group Dependent Oligosaccharides. Glycoconj. J. 1997, 14, 795–799. [Google Scholar] [CrossRef]
- Stahl, B.; Thurl, S.; Henker, J.; Siegel, M.; Finke, B.; Sawatzki, G. Detection of Four Human Milk Groups with Respect to Lewis-Blood-Group-Dependent Oligosaccharides by Serologic and Chromatographic Analysis. Adv. Exp. Med. Biol. 2001, 501, 299–306. [Google Scholar] [CrossRef]
- Kulinich, A.; Liu, L. Human Milk Oligosaccharides: The Role in the Fine-Tuning of Innate Immune Responses. Carbohydr. Res. 2016, 432, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Garrido, D.; Ruiz-Moyano, S.; Lemay, D.G.; Sela, D.A.; German, J.B.; Mills, D.A. Comparative Transcriptomics Reveals Key Differences in the Response to Milk Oligosaccharides of Infant Gut-Associated Bifidobacteria. Sci. Rep. 2015, 5, 13517. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, A.; Katoh, T.; Sakanaka, M.; Ling, Y.; Yamada, C.; Asakuma, S.; Urashima, T.; Tomabechi, Y.; Katayama-Ikegami, A.; Kurihara, S.; et al. Sharing of Human Milk Oligosaccharides Degradants within Bifidobacterial Communities in Faecal Cultures Supplemented with Bifidobacterium Bifidum. Sci. Rep. 2018, 8, 13958. [Google Scholar] [CrossRef]
- Sakanaka, M.; Gotoh, A.; Yoshida, K.; Odamaki, T.; Koguchi, H.; Xiao, J.-Z.; Kitaoka, M.; Katayama, T. Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation. Nutrients 2019, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Thongaram, T.; Hoeflinger, J.L.; Chow, J.; Miller, M.J. Human Milk Oligosaccharide Consumption by Probiotic and Human-Associated Bifidobacteria and Lactobacilli. J. Dairy Sci. 2017, 100, 7825–7833. [Google Scholar] [CrossRef]
- Schwab, C.; Ruscheweyh, H.-J.; Bunesova, V.; Pham, V.T.; Beerenwinkel, N.; Lacroix, C. Trophic Interactions of Infant Bifidobacteria and Eubacterium Hallii during L-Fucose and Fucosyllactose Degradation. Front. Microbiol. 2017, 8, 95. [Google Scholar] [CrossRef]
- Yu, Z.-T.; Nanthakumar, N.N.; Newburg, D.S. The Human Milk Oligosaccharide 2′-Fucosyllactose Quenches Campylobacter Jejuni-Induced Inflammation in Human Epithelial Cells HEp-2 and HT-29 and in Mouse Intestinal Mucosa. J. Nutr. 2016, 146, 1980–1990. [Google Scholar] [CrossRef]
- Weichert, S.; Jennewein, S.; Hüfner, E.; Weiss, C.; Borkowski, J.; Putze, J.; Schroten, H. Bioengineered 2′-Fucosyllactose and 3-Fucosyllactose Inhibit the Adhesion of Pseudomonas Aeruginosa and Enteric Pathogens to Human Intestinal and Respiratory Cell Lines. Nutr. Res. 2013, 33, 831–838. [Google Scholar] [CrossRef]
- Jantscher-Krenn, E.; Lauwaet, T.; Bliss, L.A.; Reed, S.L.; Gillin, F.D.; Bode, L. Human Milk Oligosaccharides Reduce Entamoeba Histolytica Attachment and Cytotoxicity in Vitro. Br. J. Nutr. 2012, 108, 1839–1846. [Google Scholar] [CrossRef]
- Iskarpatyoti, J.A.; Morse, E.A.; McClung, R.P.; Ikizler, M.; Wetzel, J.D.; Contractor, N.; Dermody, T.S. Serotype-Specific Differences in Inhibition of Reovirus Infectivity by Human-Milk Glycans Are Determined by Viral Attachment Protein Σ1. Virology 2012, 433, 489–497. [Google Scholar] [CrossRef]
- Lin, A.E.; Autran, C.A.; Szyszka, A.; Escajadillo, T.; Huang, M.; Godula, K.; Prudden, A.R.; Boons, G.-J.; Lewis, A.L.; Doran, K.S.; et al. Human Milk Oligosaccharides Inhibit Growth of Group B Streptococcus. J. Biol. Chem. 2017, 292, 11243–11249. [Google Scholar] [CrossRef]
- Ackerman, D.L.; Craft, K.M.; Doster, R.S.; Weitkamp, J.-H.; Aronoff, D.M.; Gaddy, J.A.; Townsend, S.D. Antimicrobial and Antibiofilm Activity of Human Milk Oligosaccharides against Streptococcus Agalactiae, Staphylococcus Aureus, and Acinetobacter Baumannii. ACS Infect. Dis. 2018, 4, 315–324. [Google Scholar] [CrossRef]
- Gonia, S.; Tuepker, M.; Heisel, T.; Autran, C.; Bode, L.; Gale, C.A. Human Milk Oligosaccharides Inhibit Candida Albicans Invasion of Human Premature Intestinal Epithelial Cells. J. Nutr. 2015, 145, 1992–1998. [Google Scholar] [CrossRef]
- Donovan, S.M.; Comstock, S.S. Human Milk Oligosaccharides Influence Neonatal Mucosal and Systemic Immunity. Ann. Nutr. Metab. 2016, 69 (Suppl. 2), 42–51. [Google Scholar] [CrossRef]
- Triantis, V.; Bode, L.; van Neerven, R.J.J. Immunological Effects of Human Milk Oligosaccharides. Front. Pediatr. 2018, 6, 190. [Google Scholar] [CrossRef]
- Lane, J.A.; O’Callaghan, J.; Carrington, S.D.; Hickey, R.M. Transcriptional Response of HT-29 Intestinal Epithelial Cells to Human and Bovine Milk Oligosaccharides. Br. J. Nutr. 2013, 110, 2127–2137. [Google Scholar] [CrossRef]
- Newburg, D.S.; Ko, J.S.; Leone, S.; Nanthakumar, N.N. Human Milk Oligosaccharides and Synthetic Galactosyloligosaccharides Contain 3′-, 4-, and 6′-Galactosyllactose and Attenuate Inflammation in Human T84, NCM-460, and H4 Cells and Intestinal Tissue Ex Vivo. J. Nutr. 2016, 146, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; van De Worp, W.R.; Stassen, R.; van Maastrigt, C.; Kettelarij, N.; Stahl, B.; Blijenberg, B.; Overbeek, S.A.; Folkerts, G.; Garssen, J.; et al. Human Milk Oligosaccharides Promote Immune Tolerance via Direct Interactions with Human Dendritic Cells. Eur. J. Immunol. 2019, 49, 1001–1014. [Google Scholar] [CrossRef] [PubMed]
- Ayechu-Muruzabal, V.; Overbeek, S.A.; Kostadinova, A.I.; Stahl, B.; Garssen, J.; Van’t Land, B.; Willemsen, L.E.M. Exposure of Intestinal Epithelial Cells to 2′-Fucosyllactose and CpG Enhances Galectin Release and Instructs Dendritic Cells to Drive Th1 and Regulatory-Type Immune Development. Biomolecules 2020, 10, 784. [Google Scholar] [CrossRef]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a Glance. J. Cell. Sci. 2018, 131. [Google Scholar] [CrossRef]
- El-Hawiet, A.; Chen, Y.; Shams-Ud-Doha, K.; Kitova, E.N.; St-Pierre, Y.; Klassen, J.S. High-Throughput Label- and Immobilization-Free Screening of Human Milk Oligosaccharides Against Lectins. Anal. Chem. 2017, 89, 8713–8722. [Google Scholar] [CrossRef]
- Šuligoj, T.; Vigsnæs, L.K.; Abbeele, P.V.d.; Apostolou, A.; Karalis, K.; Savva, G.M.; McConnell, B.; Juge, N. Effects of Human Milk Oligosaccharides on the Adult Gut Microbiota and Barrier Function. Nutrients 2020, 12, 2808. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.D.; Richard, C.; Larsen, B.M.; Field, C.J. The Importance of Human Milk for Immunity in Preterm Infants. Clin. Perinatol. 2017, 44, 23–47. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Elderman, M.; Cheng, L.; de Haan, B.J.; Nauta, A.; de Vos, P. Modulation of Intestinal Epithelial Glycocalyx Development by Human Milk Oligosaccharides and Non-Digestible Carbohydrates. Mol. Nutr. Food Res. 2019, 63, e1900303. [Google Scholar] [CrossRef]
- Wu, R.Y.; Li, B.; Koike, Y.; Määttänen, P.; Miyake, H.; Cadete, M.; Johnson-Henry, K.C.; Botts, S.R.; Lee, C.; Abrahamsson, T.R.; et al. Human Milk Oligosaccharides Increase Mucin Expression in Experimental Necrotizing Enterocolitis. Mol. Nutr. Food Res. 2019, 63, e1800658. [Google Scholar] [CrossRef]
- Gibson, G.R.; Roberfroid, M.B. Dietary Modulation of the Human Colonic Microbiota: Introducing the Concept of Prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef]
- Miqdady, M.; Al Mistarihi, J.; Azaz, A.; Rawat, D. Prebiotics in the Infant Microbiome: The Past, Present, and Future. Pediatric Gastroenterol. Hepatol. Nutr. 2020, 23, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Reverri, E.J.; Devitt, A.A.; Kajzer, J.A.; Baggs, G.E.; Borschel, M.W. Review of the Clinical Experiences of Feeding Infants Formula Containing the Human Milk Oligosaccharide 2′-Fucosyllactose. Nutrients 2018, 10, 1346. [Google Scholar] [CrossRef]
- Akkerman, R.; Faas, M.M.; de Vos, P. Non-Digestible Carbohydrates in Infant Formula as Substitution for Human Milk Oligosaccharide Functions: Effects on Microbiota and Gut Maturation. Crit. Rev. Food Sci. Nutr. 2019, 59, 1486–1497. [Google Scholar] [CrossRef]
- Baumgärtner, F.; Jurzitza, L.; Conrad, J.; Beifuss, U.; Sprenger, G.A.; Albermann, C. Synthesis of Fucosylated Lacto-N-Tetraose Using Whole-Cell Biotransformation. Bioorg. Med. Chem. 2015, 23, 6799–6806. [Google Scholar] [CrossRef]
- Petschacher, B.; Nidetzky, B. Biotechnological Production of Fucosylated Human Milk Oligosaccharides: Prokaryotic Fucosyltransferases and Their Use in Biocatalytic Cascades or Whole Cell Conversion Systems. J. Biotechnol. 2016, 235, 61–83. [Google Scholar] [CrossRef]
- Wejryd, E.; Martí, M.; Marchini, G.; Werme, A.; Jonsson, B.; Landberg, E.; Abrahamsson, T.R. Low Diversity of Human Milk Oligosaccharides Is Associated with Necrotising Enterocolitis in Extremely Low Birth Weight Infants. Nutrients 2018, 10, 1556. [Google Scholar] [CrossRef]
- Gómez-Gallego, C.; Morales, J.M.; Monleón, D.; du Toit, E.; Kumar, H.; Linderborg, K.M.; Zhang, Y.; Yang, B.; Isolauri, E.; Salminen, S.; et al. Human Breast Milk NMR Metabolomic Profile across Specific Geographical Locations and Its Association with the Milk Microbiota. Nutrients 2018, 10, 1355. [Google Scholar] [CrossRef]
- Williams, J.E.; Price, W.J.; Shafii, B.; Yahvah, K.M.; Bode, L.; McGuire, M.A.; McGuire, M.K. Relationships Among Microbial Communities, Maternal Cells, Oligosaccharides, and Macronutrients in Human Milk. J. Hum. Lact. 2017, 33, 540–551. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Kunz, C.; Rudloff, S.; García-Mantrana, I.; Crehuá-Gaudiza, E.; Martínez-Costa, C.; Collado, M.C. Association of Maternal Secretor Status and Human Milk Oligosaccharides With Milk Microbiota: An Observational Pilot Study. J. Pediatric Gastroenterol. Nutr. 2019, 68, 256–263. [Google Scholar] [CrossRef]
- Aakko, J.; Kumar, H.; Rautava, S.; Wise, A.; Autran, C.; Bode, L.; Isolauri, E.; Salminen, S. Human Milk Oligosaccharide Categories Define the Microbiota Composition in Human Colostrum. Benef. Microbes 2017, 8, 563–567. [Google Scholar] [CrossRef]
- Borewicz, K.; Gu, F.; Saccenti, E.; Arts, I.C.W.; Penders, J.; Thijs, C.; van Leeuwen, S.S.; Lindner, C.; Nauta, A.; van Leusen, E.; et al. Correlating Infant Faecal Microbiota Composition and Human Milk Oligosaccharide Consumption by Microbiota of One-Month Old Breastfed Infants. Mol. Nutr. Food Res. 2019, e1801214. [Google Scholar] [CrossRef]
- Sela, D.A.; Mills, D.A. Nursing Our Microbiota: Molecular Linkages between Bifidobacteria and Milk Oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef]
- James, K.; Motherway, M.O.; Bottacini, F.; van Sinderen, D. Bifidobacterium Breve UCC2003 Metabolises the Human Milk Oligosaccharides Lacto-N-Tetraose and Lacto-N-Neo-Tetraose through Overlapping, yet Distinct Pathways. Sci. Rep. 2016, 6, 38560. [Google Scholar] [CrossRef]
- James, K.; O’Connell Motherway, M.; Penno, C.; O’Brien, R.L.; van Sinderen, D. Bifidobacterium Breve UCC2003 Employs Multiple Transcriptional Regulators To Control Metabolism of Particular Human Milk Oligosaccharides. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.R.; Ling, P.-R.; Blackburn, G.L. Review of Infant Feeding: Key Features of Breast Milk and Infant Formula. Nutrients 2016, 8, 279. [Google Scholar] [CrossRef]
- Collados-Gómez, L.; Ferrera-Camacho, P.; Fernandez-Serrano, E.; Camacho-Vicente, V.; Flores-Herrero, C.; García-Pozo, A.M.; Jiménez-García, R. Randomised Crossover Trial Showed That Using Breast Milk or Sucrose Provided the Same Analgesic Effect in Preterm Infants of at Least 28 Weeks. Acta Paediatr. 2018, 107, 436–441. [Google Scholar] [CrossRef]
- Baudesson de Chanville, A.; Brevaut-Malaty, V.; Garbi, A.; Tosello, B.; Baumstarck, K.; Gire, C. Analgesic Effect of Maternal Human Milk Odor on Premature Neonates: A Randomized Controlled Trial. J. Hum. Lact. 2017, 33, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Su, F.; Li, J.; Chen, W. The Analgesic Effects of Maternal Milk Odor on Newborns: A Meta-Analysis. Breastfeed Med. 2018, 13, 327–334. [Google Scholar] [CrossRef]
- Berger, B.; Porta, N.; Foata, F.; Grathwohl, D.; Delley, M.; Moine, D.; Charpagne, A.; Siegwald, L.; Descombes, P.; Alliet, P.; et al. Linking Human Milk Oligosaccharides, Infant Fecal Community Types, and Later Risk To Require Antibiotics. mBio 2020, 11. [Google Scholar] [CrossRef]
- Yu, Z.-T.; Chen, C.; Newburg, D.S. Utilization of Major Fucosylated and Sialylated Human Milk Oligosaccharides by Isolated Human Gut Microbes. Glycobiology 2013, 23, 1281–1292. [Google Scholar] [CrossRef]
- Zeuner, B.; Meyer, A.S. Enzymatic Transfucosylation for Synthesis of Human Milk Oligosaccharides. Carbohydr. Res. 2020, 493, 108029. [Google Scholar] [CrossRef]
- Zeuner, B.; Teze, D.; Muschiol, J.; Meyer, A.S. Synthesis of Human Milk Oligosaccharides: Protein Engineering Strategies for Improved Enzymatic Transglycosylation. Molecules 2019, 24, 33. [Google Scholar] [CrossRef]
- Zeuner, B.; Vuillemin, M.; Holck, J.; Muschiol, J.; Meyer, A.S. Loop Engineering of an α-1,3/4-l-Fucosidase for Improved Synthesis of Human Milk Oligosaccharides. Enzyme Microb. Technol. 2018, 115, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, B.; Muschiol, J.; Holck, J.; Lezyk, M.; Gedde, M.R.; Jers, C.; Mikkelsen, J.D.; Meyer, A.S. Substrate Specificity and Transfucosylation Activity of GH29 α-l-Fucosidases for Enzymatic Production of Human Milk Oligosaccharides. New Biotechnol. 2018, 41, 34–45. [Google Scholar] [CrossRef]
- Guzmán-Rodríguez, F.; Alatorre-Santamaría, S.; Gómez-Ruiz, L.; Rodríguez-Serrano, G.; García-Garibay, M.; Cruz-Guerrero, A. Employment of Fucosidases for the Synthesis of Fucosylated Oligosaccharides with Biological Potential. Biotechnol. Appl. Biochem. 2019, 66, 172–191. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.R.; Newburg, D.S. Clinical Applications of Bioactive Milk Components. Nutr. Rev. 2015, 73, 463–476. [Google Scholar] [CrossRef] [PubMed]
| Macronutrients | |
| Protein | 0.9 g/dL |
| Fat | 3.5 g/dL |
| Carbohydrates (mainly glucose) | 6.7 g/dL |
| Minerals | |
| Calcium | 200–250 mg/L |
| Magnesium | 30–35 mg/L |
| Phosphorus | 120–140 mg/L |
| Sodium | 120–250 mg/L |
| Potassium | 400–550 mg/L |
| Iron | 0.3–0.9 mg/L |
| Fluoride | 4–15 µg/L |
| Fat-Soluble Vitamins | |
| Vitamin A | 0.3–0.6 mg/L |
| Vitamin D | 15 IU/day (in exclusively breastfed infants) or 0.33 µg/L |
| Vitamin E | 3–8 mg/L |
| Vitamin K | 2–3 µg/L |
| Water-Soluble Vitamins | |
| Ascorbic acid | 100 mg/L |
| Thiamine (vitamin B1) | 200 µg/L |
| Riboflavin (vitamin B2) | 0.35–0.39 mg/L |
| Niacin | 1.8–6 mg/L |
| Vitamin B6 | 0.09–0.31 mg/L |
| Folate | 80–140 µg/L |
| Vitamin B12 | 0.5–1 µg/L |
| Neutral Fucosylated HMOs | Neutral Nonfucosylated HMOs | Acidic HMOs |
|---|---|---|
| 2′-Fucosyllactose (2′-FL) | Lacto-N-biose (LNB) | Disialyllacto-N-tetraose (DSLNT) |
| 3-Fucosyllactose (3-FL) | Lacto-N-tetraose (LNT) | Sialyllacto-N-neo-tetraose b (LST b) |
| Lacto-N-fucopentaose I (LNFP I) | Lacto-N-neotetraose(LNnT) | Sialyllacto-N-neo-tetraose c (LST c) |
| Lacto-N-fucopentaose II (LNFP II) | Lacto-N-hexaose (LNH) | Sialyl lacto-N-tetraose a (LST a) |
| Lacto-N-fucopentaose III (LNFP III) | Lacto-N-neohexaose (LNnH) | 3′-Sialyllactose (3′-SL) |
| Difucosyllactose (DFL) | 6′-Sialyllactose (6′-SL) |
| Milk Group | Prevalence | Fucosylated HMO Linkages | Examples of Fucosylated HMOs |
|---|---|---|---|
| Se+Le+ | 70% | α1-2 α1-4 α1-3 | 2′-Fucosyllactose (2′-FL) Lacto-N-difuco-hexaose I (LNDFH I) Lacto-N-fucopentaose II (LNFP II) 3′-Fucosyllactose (3′-FL) Difucosyllactose (DFL) |
| Se-Le+ | 20% | α1-4 α1-3 | Lacto-N-difuco-hexaose II (LNDFH II) LNFP II 3′-FL |
| Se+Le- | 10% | α1-2 α1-3 | Lacto-N-fucopentaose I (LNFP I) 2′-FL 3′-FL |
| Se-Le- | 1% | α1-3 only | Difucosyl-para-lacto-N-neohexaose (DFpLNnH) 3′-FL |
| HMO | Structure | Approval in EU (European Food Safety Authority) | Approval in USA (US Food and Drug Administration) |
|---|---|---|---|
| 2′-Fucosyllactose (2′-FL) |  | 2015 | 2016 |
| Lacto-N-neotetraose (LNnT) | 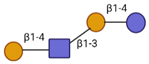 | 2015 | 2016 |
| 3′-Sialyllactose (3′-SL) | 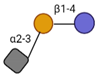 | - | 2019 |
 .
.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moubareck, C.A. Human Milk Microbiota and Oligosaccharides: A Glimpse into Benefits, Diversity, and Correlations. Nutrients 2021, 13, 1123. https://doi.org/10.3390/nu13041123
Moubareck CA. Human Milk Microbiota and Oligosaccharides: A Glimpse into Benefits, Diversity, and Correlations. Nutrients. 2021; 13(4):1123. https://doi.org/10.3390/nu13041123
Chicago/Turabian StyleMoubareck, Carole Ayoub. 2021. "Human Milk Microbiota and Oligosaccharides: A Glimpse into Benefits, Diversity, and Correlations" Nutrients 13, no. 4: 1123. https://doi.org/10.3390/nu13041123
APA StyleMoubareck, C. A. (2021). Human Milk Microbiota and Oligosaccharides: A Glimpse into Benefits, Diversity, and Correlations. Nutrients, 13(4), 1123. https://doi.org/10.3390/nu13041123






