A Polyphenol Enriched Variety of Apple Alters Circulating Immune Cell Gene Expression and Faecal Microbiota Composition in Healthy Adults: A Randomized Controlled Trial
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Apples
2.2. Analysis of Apple Polyphenols
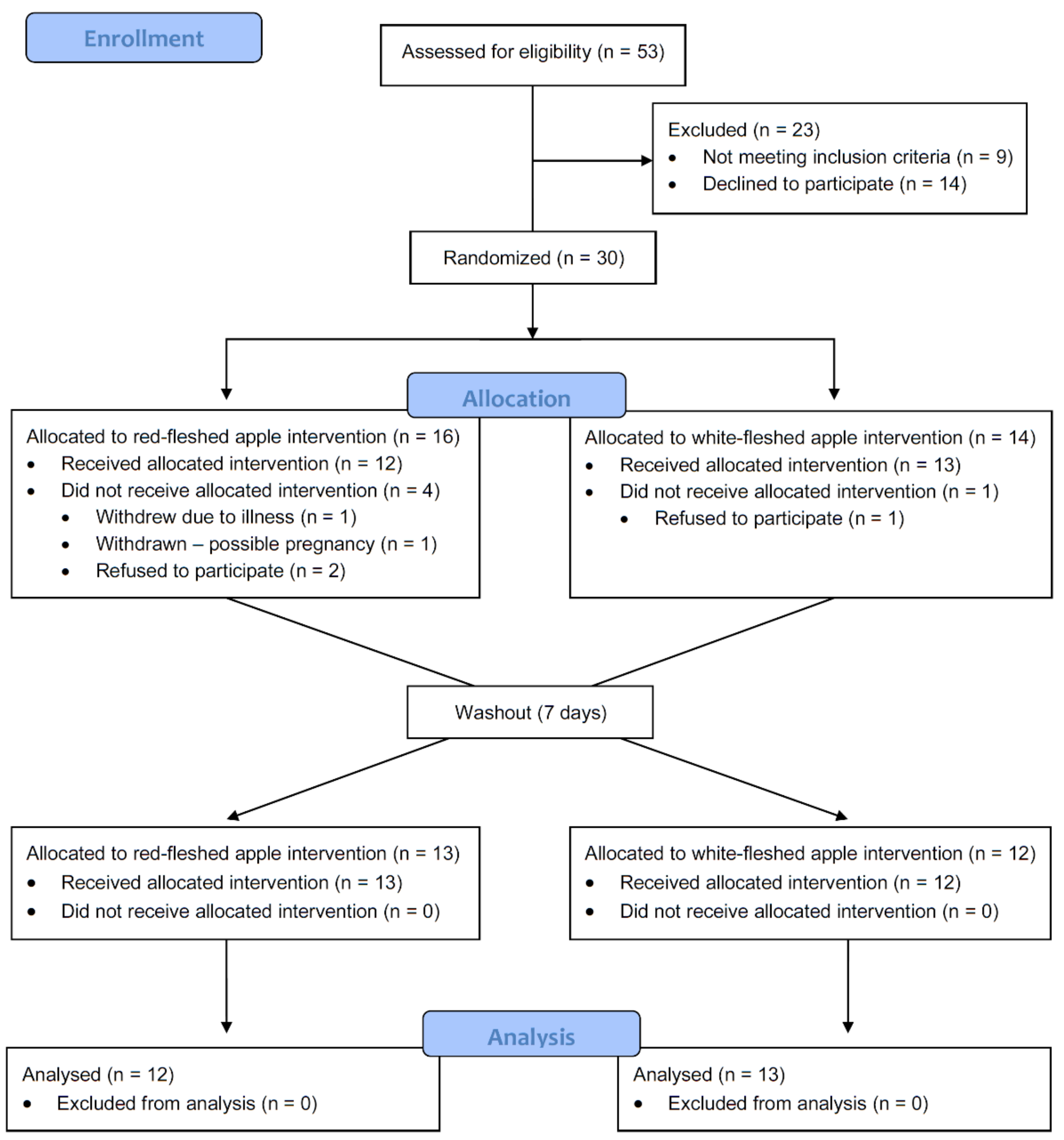
2.3. Clinical Trial Design
2.4. Blood Processing
2.5. PBMC mRNA Sequencing and Analysis
2.6. Analysis of Faecal Microbial Composition
2.7. Statistical Analyses
2.8. Sequence Data Access
3. Results
3.1. Apple Polyphenol Content
3.2. Participant Characteristics
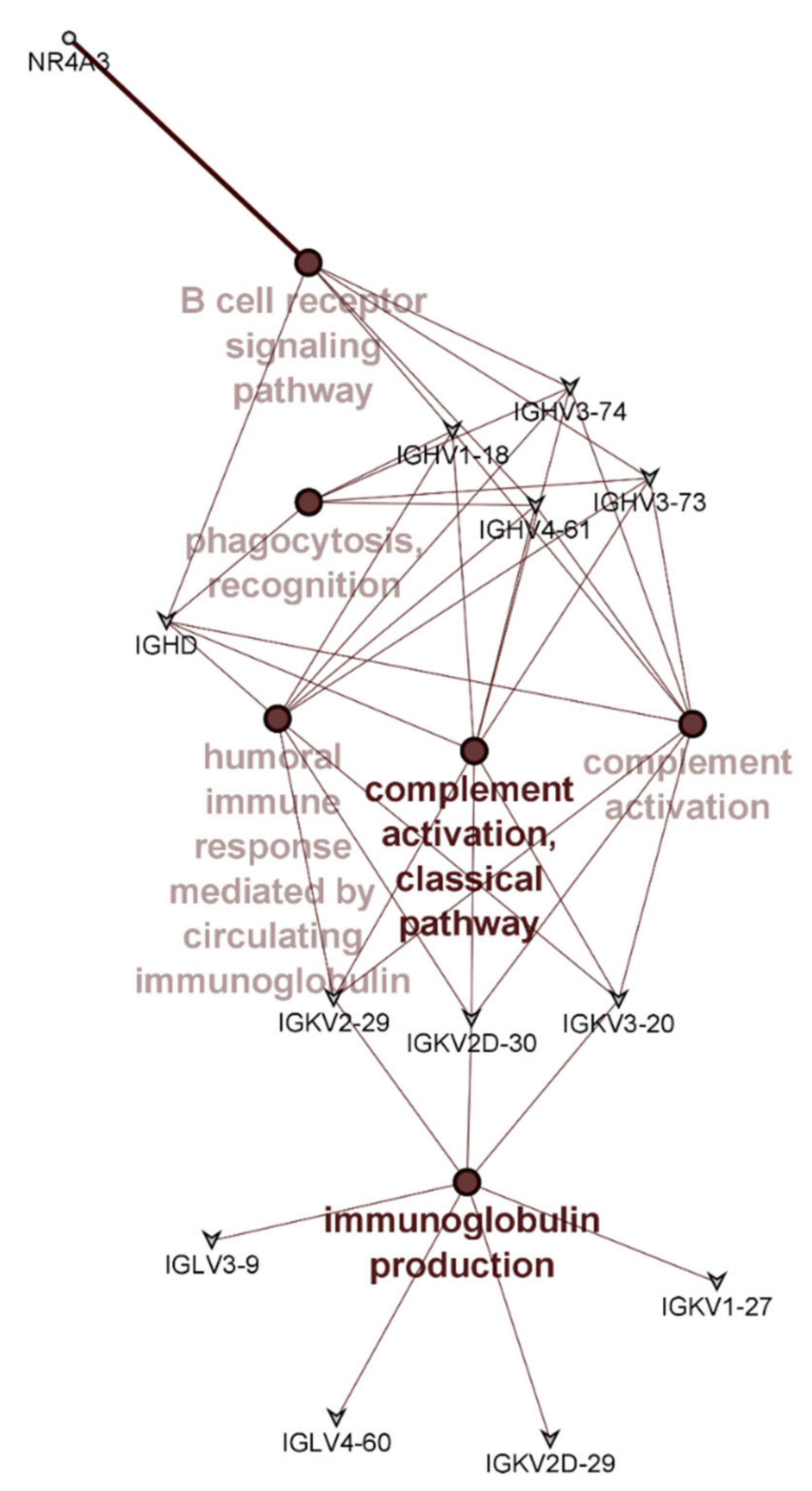
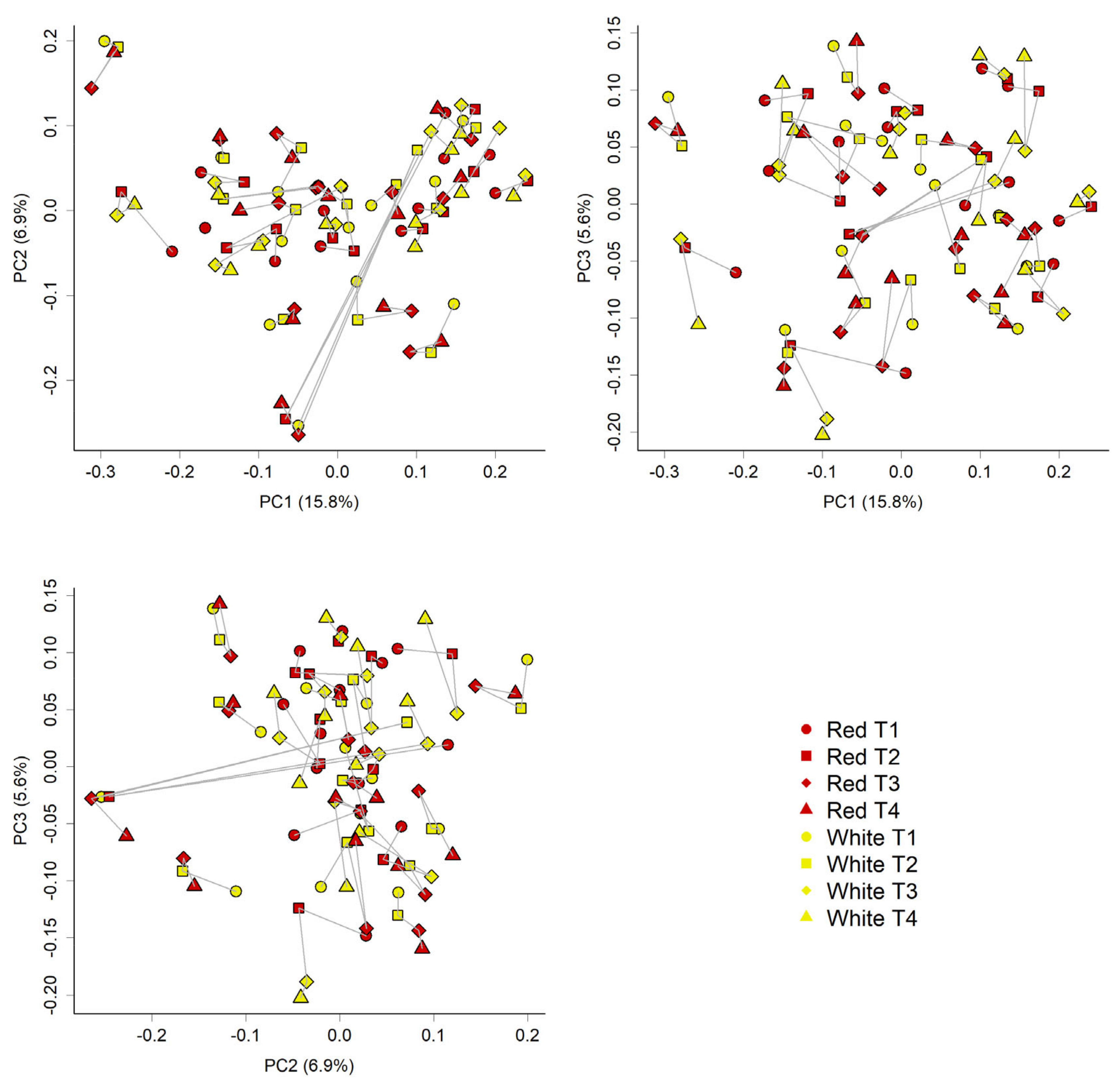
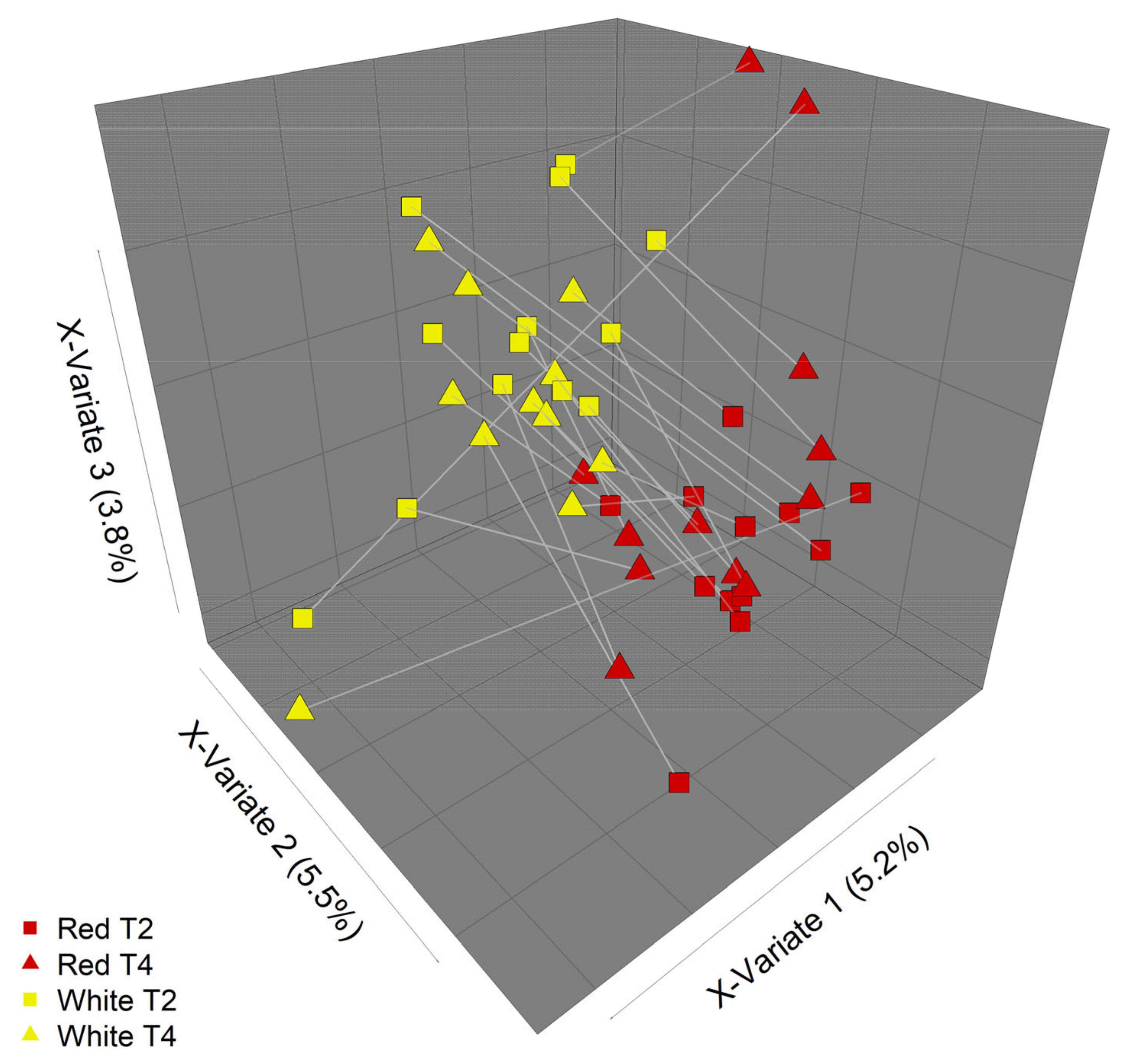
3.3. PMBC Gene Expression
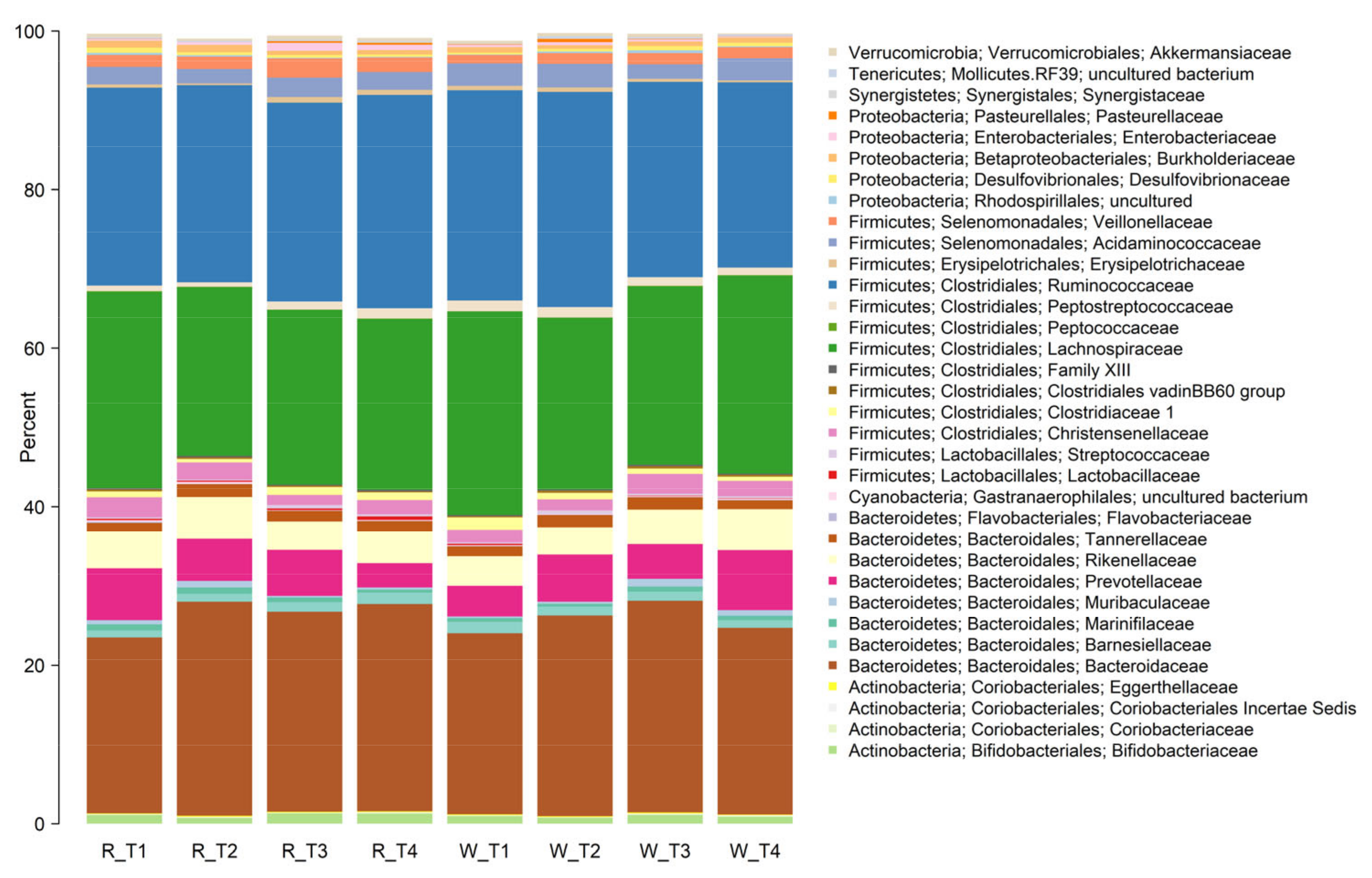
3.4. Faecal Microbiota Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, J.P. A Pembrokeshire Proverb. Notes Queries 1866, 127, 153. [Google Scholar] [CrossRef]
- Davis, M.A.; Bynum, J.P.; Sirovich, B.E. Association between Apple Consumption and Physician Visits: Appealing the Conventional Wisdom That an Apple a Day Keeps the Doctor Away. JAMA Intern. Med. 2015, 175, 777–783. [Google Scholar] [CrossRef]
- Chatzi, L.; Apostolaki, G.; Bibakis, I.; Skypala, I.; Bibaki-Liakou, V.; Tzanakis, N.; Kogevinas, M.; Cullinan, P. Protective Effect of Fruits, Vegetables and the Mediterranean Diet on Asthma and Allergies among Children in Crete. Thorax 2007, 62, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Tosti, V.; Bertozzi, B.; Fontana, L. Health Benefits of the Mediterranean Diet: Metabolic and Molecular Mechanisms. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 318–326. [Google Scholar] [CrossRef]
- Zamora-Ros, R.; Andres-Lacueva, C.; Lamuela-Raventós, R.M.; Berenguer, T.; Jakszyn, P.; Barricarte, A.; Ardanaz, E.; Amiano, P.; Dorronsoro, M.; Larrañaga, N.; et al. Estimation of Dietary Sources and Flavonoid Intake in a Spanish Adult Population (EPIC-Spain). J. Am. Diet. Assoc. 2010, 110, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Martín-Peláez, S.; Fito, M.; Castaner, O. Mediterranean Diet Effects on Type 2 Diabetes Prevention, Disease Progression, and Related Mechanisms. A Review. Nutrients 2020, 12, 2236. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.; Liu, R.H. Apple Phytochemicals and Their Health Benefits. Nutr. J. 2004, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Gerhauser, C. Cancer Chemopreventive Potential of Apples, Apple Juice, and Apple Components. Planta Med. 2008, 74, 1608–1624. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.S.F.; Meijerink, M.; Zeuner, B.; Holck, J.; Louis, P.; Meyer, A.S.; Wells, J.M.; Flint, H.J.; Duncan, S.H. Prebiotic Potential of Pectin and Pectic Oligosaccharides to Promote Anti-Inflammatory Commensal Bacteria in the Human Colon. FEMS Microbiol. Ecol. 2017, 93, fix127. [Google Scholar] [CrossRef] [PubMed]
- Ravn-Haren, G.; Dragsted, L.O.; Buch-Andersen, T.; Jensen, E.N.; Jensen, R.I.; Németh-Balogh, M.; Paulovicsová, B.; Bergström, A.; Wilcks, A.; Licht, T.R.; et al. Intake of Whole Apples or Clear Apple Juice Has Contrasting Effects on Plasma Lipids in Healthy Volunteers. Eur. J. Nutr. 2013, 52, 1875–1889. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Harris, P.J.; Ferguson, L.R. Potential Benefits of Dietary Fibre Intervention in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2016, 17, 919. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R. Role of Plant Polyphenols in Genomic Stability. Mutat. Res. 2001, 475, 89–111. [Google Scholar] [CrossRef]
- Tu, S.H.; Chen, L.C.; Ho, Y.S. An Apple a Day to Prevent Cancer Formation: Reducing Cancer Risk with Flavonoids. J. Food Drug Anal. 2017, 25, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Greenwood, J.M.; Walker, E.G.; Rassam, M.; Sullivan, M.; Evers, D.; Perry, N.B.; Laing, W.A. Anti-Inflammatory Procyanidins and Triterpenes in 109 Apple Varieties. J. Agric. Food Chem. 2012, 60, 10546–10554. [Google Scholar] [CrossRef]
- Ardite, E.; Panés, J.; Miranda, M.; Salas, A.; Elizalde, J.I.; Sans, M.; Arce, Y.; Bordas, J.M.; Fernández-Checa, J.C.; Piqué, J.M. Effects of Steroid Treatment on Activation of Nuclear Factor KappaB in Patients with Inflammatory Bowel Disease. Br. J. Pharmacol. 1998, 124, 431–433. [Google Scholar] [CrossRef]
- Clark, I.A.; Vissel, B. Broader Insights into Understanding Tumor Necrosis Factor and Neurodegenerative Disease Pathogenesis Infer New Therapeutic Approaches. J. Alzheimers Dis. 2021, 79, 931–948. [Google Scholar] [CrossRef]
- Espley, R.V.; Butts, C.A.; Laing, W.A.; Martell, S.; Smith, H.; McGhie, T.K.; Zhang, J.; Paturi, G.; Hedderley, D.; Bovy, A.; et al. Dietary Flavonoids from Modified Apple Reduce Inflammation Markers and Modulate Gut Microbiota in Mice. J. Nutr. 2014, 144, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Lavefve, L.; Howard, L.R.; Carbonero, F. Berry Polyphenols Metabolism and Impact on Human Gut Microbiota and Health. Food Funct. 2020, 11, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Knight, R. Expanding Our Understanding of the Role of the Microbiome in Health and Disease. Arch. Med. Res. 2017, 48, 663–665. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the Microbiome in Human Development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef]
- Stanislawski, M.A.; Dabelea, D.; Wagner, B.D.; Iszatt, N.; Dahl, C.; Sontag, M.K.; Knight, R.; Lozupone, C.A.; Eggesbo, M. Gut Microbiota in the First 2 Years of Life and the Association with Body Mass Index at Age 12 in a Norwegian Birth Cohort. mBio 2018, 9, e01751-18. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.Q.; Mills, R.H.; Peters, N.V.; Holder, M.K.; de Vries, G.J.; Knight, R.; Chassaing, B.; Gonzalez, D.J.; Gewirtz, A.T. Associations of the Fecal Microbial Proteome Composition and Proneness to Diet-Induced Obesity. Mol. Cell. Proteom. 2019, 18, 1864–1879. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A Gut Microbiome Signature for Cirrhosis Due to Nonalcoholic Fatty Liver Disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Rajagopala, S.V.; Vashee, S.; Oldfield, L.M.; Suzuki, Y.; Venter, J.C.; Telenti, A.; Nelson, K.E. The Human Microbiome and Cancer. Cancer Prev. Res. (Phila.) 2017, 10, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Callewaert, C.; Debelius, J.; Hyde, E.; Marotz, C.; Morton, J.T.; Swafford, A.; Vrbanac, A.; Dorrestein, P.C.; Knight, R. Impacts of the Human Gut Microbiome on Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 253–270. [Google Scholar] [CrossRef]
- Volz, R.K.; Oraguzie, N.C.; Whitworth, C.J.; How, N.; Chagné, D.; Carlisle, C.M.; Gardiner, S.E.; Rikkerink, E.H.A.; Lawrence, T. Breeding for Red Flesh Colour in Apple: Progress and Challenges. Acta Hortic. 2009, 814, 337–342. [Google Scholar] [CrossRef]
- Ferguson, L.R.; Barnett, M.P. Why Are Omics Technologies Important to Understanding the Role of Nutrition in Inflammatory Bowel Diseases? Int. J. Mol. Sci. 2016, 17, 1763. [Google Scholar] [CrossRef] [PubMed]
- van Ommen, B.; Stierum, R. Nutrigenomics: Exploiting Systems Biology in the Nutrition and Health Arena. Curr. Opin. Biotechnol. 2002, 13, 517–521. [Google Scholar] [CrossRef]
- Gomes, A.; Oudot, C.; Macià, A.; Foito, A.; Carregosa, D.; Stewart, D.; Van de Wiele, T.; Berry, D.; Motilva, M.J.; Brenner, C.; et al. Berry-Enriched Diet in Salt-Sensitive Hypertensive Rats: Metabolic Fate of (Poly)Phenols and the Role of Gut Microbiota. Nutrients 2019, 11, 2634. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Sánchez-Alcoholado, L.; Pérez-Martínez, P.; Andrés-Lacueva, C.; Cardona, F.; Tinahones, F.; Queipo-Ortuño, M.I. Red Wine Polyphenols Modulate Fecal Microbiota and Reduce Markers of the Metabolic Syndrome in Obese Patients. Food Funct. 2016, 7, 1775–1787. [Google Scholar] [CrossRef] [PubMed]
- Dodt, M.; Roehr, J.T.; Ahmed, R.; Dieterich, C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology 2012, 1, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. Mixomics: An R Package for ‘Omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Trošt, K.; Ulaszewska, M.M.; Stanstrup, J.; Albanese, D.; De Filippo, C.; Tuohy, K.M.; Natella, F.; Scaccini, C.; Mattivi, F. Host: Microbiome Co-Metabolic Processing of Dietary Polyphenols—An Acute, Single Blinded, Cross-over Study with Different Doses of Apple Polyphenols in Healthy Subjects. Food Res. Int. 2018, 112, 108–128. [Google Scholar] [CrossRef]
- Ivey, K.L.; Chan, A.T.; Izard, J.; Cassidy, A.; Rogers, G.B.; Rimm, E.B. Role of Dietary Flavonoid Compounds in Driving Patterns of Microbial Community Assembly. mBio 2019, 10, e01205-19. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Daza, M.C.; Daoust, L.; Boutkrabt, L.; Pilon, G.; Varin, T.; Dudonné, S.; Levy, E.; Marette, A.; Roy, D.; Desjardins, Y. Wild Blueberry Proanthocyanidins Shape Distinct Gut Microbiota Profile and Influence Glucose Homeostasis and Intestinal Phenotypes in High-Fat High-Sucrose Fed Mice. Sci. Rep. 2020, 10, 2217. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Morató, J.; Matthan, N.R.; Liu, J.; de la Torre, R.; Chen, C.O. Cranberries Attenuate Animal-Based Diet-Induced Changes in Microbiota Composition and Functionality: A Randomized Crossover Controlled Feeding Trial. J. Nutr. Biochem. 2018, 62, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Kemperman, R.A.; Gross, G.; Mondot, S.; Possemiers, S.; Marzorati, M.; Van de Wiele, T.; Doré, J.; Vaughan, E.E. Impact of Polyphenols from Black Tea and Red Wine/Grape Juice on a Gut Model Microbiome. Food Res. Int. 2013, 53, 659–669. [Google Scholar] [CrossRef]
- Queipo-Ortuño, M.I.; Boto-Ordóñez, M.; Murri, M.; Gomez-Zumaquero, J.M.; Clemente-Postigo, M.; Estruch, R.; Cardona Diaz, F.; Andrés-Lacueva, C.; Tinahones, F.J. Influence of Red Wine Polyphenols and Ethanol on the Gut Microbiota Ecology and Biochemical Biomarkers. Am. J. Clin. Nutr. 2012, 95, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Massot-Cladera, M.; Abril-Gil, M.; Torres, S.; Franch, À.; Castell, M.; Pérez-Cano, F.J. Impact of Cocoa Polyphenol Extracts on the Immune System and Microbiota in Two Strains of Young Rats. Br. J. Nutr. 2014, 112, 1944–1954. [Google Scholar] [CrossRef] [PubMed]
- Holscher, H.D. Dietary Fiber and Prebiotics and the Gastrointestinal Microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef]
- Yang, J.; Martínez, I.; Walter, J.; Keshavarzian, A.; Rose, D.J. In Vitro Characterization of the Impact of Selected Dietary Fibers on Fecal Microbiota Composition and Short Chain Fatty Acid Production. Anaerobe 2013, 23, 74–81. [Google Scholar] [CrossRef]
- Mano, F.; Ikeda, K.; Joo, E.; Fujita, Y.; Yamane, S.; Harada, N.; Inagaki, N. The Effect of White Rice and White Bread as Staple Foods on Gut Microbiota and Host Metabolism. Nutrients 2018, 10, 1323. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient Metabolism by the Human Gut Microbiome: Major Fermentation by-Products and Their Impact on Host Health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef]
- Jones, R.B.; Alderete, T.L.; Kim, J.S.; Millstein, J.; Gilliland, F.D.; Goran, M.I. High Intake of Dietary Fructose in Overweight/Obese Teenagers Associated with Depletion of Eubacterium and Streptococcus in Gut Microbiome. Gut Microbes 2019, 10, 712–719. [Google Scholar] [CrossRef]
- Hervert-Hernández, D.; Pintado, C.; Rotger, R.; Goñi, I. Stimulatory Role of Grape Pomace Polyphenols on Lactobacillus Acidophilus Growth. Int. J. Food Microbiol. 2009, 136, 119–122. [Google Scholar] [CrossRef]
- Volstatova, T.; Marsik, P.; Rada, V.; Geigerova, M.; Havlik, J. Effect of Apple Extracts and Selective Polyphenols on the Adhesion of Potential Probiotic Strains of Lactobacillus Gasseri R and Lactobacillus Casei Fmp. J. Funct. Foods 2017, 35, 391–397. [Google Scholar] [CrossRef]
- Boesmans, L.; Valles-Colomer, M.; Wang, J.; Eeckhaut, V.; Falony, G.; Ducatelle, R.; Van Immerseel, F.; Raes, J.; Verbeke, K. Butyrate Producers as Potential Next-Generation Probiotics: Safety Assessment of the Administration of Butyricicoccus Pullicaecorum to Healthy Volunteers. mSystems 2018, 3, e00094-18. [Google Scholar] [CrossRef]
- Eeckhaut, V.; Machiels, K.; Perrier, C.; Romero, C.; Maes, S.; Flahou, B.; Steppe, M.; Haesebrouck, F.; Sas, B.; Ducatelle, R.; et al. Butyricicoccus Pullicaecorum in Inflammatory Bowel Disease. Gut 2013, 62, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, C.; Huang, J.; Xie, M.; Li, X.; Fu, L. Butyricicoccus Plays a Key Role in Mediating the Antagonism between Probiotic and Antibiotic on Food Allergy. Food Agric. Immunol. 2019, 30, 446–461. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, Metabolism and Microbial Ecology of Butyrate-Producing Bacteria from the Human Large Intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased Abundance of Sutterella Spp. And Ruminococcus Torques in Feces of Children with Autism Spectrum Disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Rúa, R.; Keijer, J.; Caimari, A.; van Schothorst, E.M.; Palou, A.; Oliver, P. Peripheral Blood Mononuclear Cells as a Source to Detect Markers of Homeostatic Alterations Caused by the Intake of Diets with an Unbalanced Macronutrient Composition. J. Nutr. Biochem. 2015, 26, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Lear, R.; O’Leary, M.; O’Brien Andersen, L.; Holt, C.C.; Stensvold, C.R.; van der Giezen, M.; Bowtell, J.L. Tart Cherry Concentrate Does Not Alter the Gut Microbiome, Glycaemic Control or Systemic Inflammation in a Middle-Aged Population. Nutrients 2019, 11, 1063. [Google Scholar] [CrossRef]
- Kojadinovic, M.; Glibetic, M.; Vucic, V.; Popovic, M.; Vidovic, N.; Debeljak-Martacic, J.; Arsic, A. Short-Term Consumption of Pomegranate Juice Alleviates Some Metabolic Disturbances in Overweight Patients with Dyslipidemia. J. Med. Food 2021. [Google Scholar] [CrossRef] [PubMed]
- Most, J.; Penders, J.; Lucchesi, M.; Goossens, G.H.; Blaak, E.E. Gut Microbiota Composition in Relation to the Metabolic Response to 12-Week Combined Polyphenol Supplementation in Overweight Men and Women. Eur. J. Clin. Nutr. 2017, 71, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Patocka, J.; Bhardwaj, K.; Klimova, B.; Nepovimova, E.; Wu, Q.; Landi, M.; Kuca, K.; Valis, M.; Wu, W. Malus Domestica: A Review on Nutritional Features, Chemical Composition, Traditional and Medicinal Value. Plants 2020, 9, 1408. [Google Scholar] [CrossRef] [PubMed]
- Koutsos, A.; Lima, M.; Conterno, L.; Gasperotti, M.; Bianchi, M.; Fava, F.; Vrhovsek, U.; Lovegrove, J.A.; Tuohy, K.M. Effects of Commercial Apple Varieties on Human Gut Microbiota Composition and Metabolic Output Using an in Vitro Colonic Model. Nutrients 2017, 9, 533. [Google Scholar] [CrossRef]
- Kleiveland, C.R. Peripheral Blood Mononuclear Cells. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 161–167. [Google Scholar]
| Compound | Red-Fleshed | Placebo | p-Value |
|---|---|---|---|
| Chlorogenic acid | 116.7 ± 13.5 | 136.9 ± 9.3 | 0.102 |
| Catechin | 0.3 ± 0.0 | 9.9 ± 1.7 | <0.001 |
| Cyanidin 3-galactoside | 26.3 ± 3.0 | 0.0 ± 0.0 | <0.001 |
| Cyanidin 3-glucoside | 0.3 ± 0.0 | 0.0 ± 0.0 | <0.001 |
| Epicatechin | 3.0 ± 0.4 | 34.7 ± 3.6 | <0.001 |
| 4-p-Coumaryl quinic acid | 0.0 ± 0.0 | 0.0 ± 0.0 | N/A |
| Phloridzin | 11.1 ± 1.1 | 6.0 ± 1.2 | <0.01 |
| Phloridzin xyloside | 119.4 ± 10.7 | 35.2 ± 4.1 | <0.001 |
| Procyanidin B1 | 0.2 ± 0.0 | 13.2 ± 2.1 | <0.001 |
| Procyanidin B2 | 4.3 ± 0.1 | 70.0 ± 10.2 | <0.001 |
| Procyanidin B5 | 0.5 ± 0.1 | 6.6 ± 0.9 | <0.001 |
| Quercetin 3-arabinoside | 2.1 ± 0.1 | 1.2 ± 0.3 | <0.01 |
| Quercetin 3-galactoside | 19.1 ± 0.6 | 6.5 ± 2.7 | <0.001 |
| Quercetin 3-rutinoside | 0.9 ± 0.6 | 0.6 ± 0.3 | 0.5339 |
| Measure | Female (n = 20) | Male (n = 5) | ||
|---|---|---|---|---|
| Sequence 1 | 1 (n = 10) | 2 (n = 10) | 1 (n = 2) | 2 (n = 3) |
| BMI (kg/m) 2 | 23.3 ± 3.0 | 23.4 ± 2.1 | 22.82 | 27.4 ± 4.7 |
| Age (years) | 39.8 ± 15.4 | 38.0 ± 13.0 | 47.3 ± 8.1 | 39.7 ± 14.2 |
| Cholesterol (mmol/L) | 4.87 ± 1.18 | 5.34 ± 1.42 | 6.25 ± 2.62 | 5.20 ± 1.65 |
| HDL cholesterol (mmol/L) | 1.85 ± 0.36 | 1.76 ± 0.20 | 2.25 ± 0.21 | 1.40 ± 0.46 |
| Chol/HDL ratio | 2.66 ± 0.51 | 3.06 ± 0.94 | 2.85 ± 1.48 | 4.03 ± 1.68 |
| LDL cholesterol (mmol/L) | 2.65 ± 0.99 | 3.16 ± 1.36 | 3.40 ± 2.40 | 3.17 ± 1.70 |
| Triglycerides (mmol/L) | 0.81 ± 0.21 | 0.94 ± 0.39 | 1.25 ± 0.92 | 1.40 ± 0.72 |
| C-reactive protein (CRP; mg/L) 3 | 5.70 ± 12.01 | 1.05 ± 0.93 | 1.00 ± 0.00 | 4.67 ± 7.22 |
| Variable | Apple | Estimate (95% CI) | p-Value |
|---|---|---|---|
| CRP | Red | −0.372 (−0.976–0.232) | 0.2208 |
| White | 0 | ||
| Cholesterol | Red | −0.124 (−0.329–0.082) | 0.2315 |
| White | 0 | ||
| Cholesterol/HDL | Red | 0.036 (−0.114–0.186) | 0.6337 |
| White | 0 | ||
| HDL | Red | −0.096 (−0.203–0.011) | 0.0761 |
| White | 0 | ||
| LDL | Red | −0.043 (−0.198–0.111) | 0.5748 |
| White | 0 | ||
| Triglycerides | Red | 0.069 (−0.094–0.231) | 0.3977 |
| White | 0 |
| Variable | Intervention | n | Mean ± SD | Range | p-Value |
|---|---|---|---|---|---|
| CRP (mg L−1) | Before | 48 | 1.354 ± 1.830 | 0.5–9.0 | 0.3648 |
| After | 49 | 1.286 ± 1.373 | 0.5–6.0 | ||
| Cholesterol | Before | 50 | 5.128 ± 1.359 | 3.3–8.1 | 0.0565 |
| After | 49 | 4.986 ± 1.322 | 3.1–8.0 | ||
| Cholesterol/HDL ratio | Before | 50 | 2.956 ± 0.945 | 1.7–6.0 | 0.8714 |
| After | 49 | 2.953 ± 0.845 | 1.7–5.6 | ||
| HDL | Before | 50 | 1.794 ± 0.363 | 1.0–2.6 | 0.1353 |
| After | 49 | 1.737 ± 0.369 | 1.0–2.6 | ||
| LDL | Before | 50 | 2.914 ± 1.211 | 1.2–5.7 | 0.1293 |
| After | 49 | 2.831 ± 1.144 | 1.2–5.5 | ||
| Triglycerides | Before | 50 | 0.918 ± 0.411 | 0.4–2.0 | 0.9594 |
| After | 49 | 0.918 ± 0.356 | 0.3–2.1 |
| Ensembl ID | Gene Name | Description | LogFC | LogCPM | p-Value |
|---|---|---|---|---|---|
| ENSG00000243264 | IGKV2D-29 | Immunoglobulin kappa variable 2D-29 | 1.50 | 2.60 | <0.001 |
| ENSG00000244575 | IGKV1-27 | Immunoglobulin kappa variable 1-27 | −1.38 | 4.00 | 0.002 |
| ENSG00000211976 | IGHV3-73 | Immunoglobulin heavy variable 3-73 | 1.17 | 2.32 | 0.002 |
| ENSG00000253998 | IGKV2-29 | Immunoglobulin kappa variable 2-29 | 1.05 | 0.59 | 0.002 |
| ENSG00000224650 | IGHV3-74 | Immunoglobulin heavy variable 3-74 | 0.77 | 3.73 | 0.004 |
| ENSG00000119508 | NR4A3 | Nuclear receptor subfamily 4 group A member 3 | −1.15 | 1.05 | 0.010 |
| ENSG00000239571 | IGKV2D-30 | Immunoglobulin kappa variable 2D-30 | 0.79 | 0.81 | 0.013 |
| ENSG00000211898 | IGHD | Immunoglobulin heavy constant delta | 0.77 | 7.06 | 0.014 |
| ENSG00000104918 | RETN | Resistin | 0.71 | 2.85 | 0.015 |
| ENSG00000239951 | IGKV3-20 | Immunoglobulin kappa variable 3-20 | −0.90 | 6.79 | 0.019 |
| ENSG00000211970 | IGHV4-61 | Immunoglobulin heavy variable 4-61 | −1.12 | 3.16 | 0.025 |
| ENSG00000211639 | IGLV4-60 | Immunoglobulin lambda variable 4-60 | −1.13 | 1.90 | 0.025 |
| ENSG00000211945 | IGHV1-18 | Immunoglobulin heavy variable 1-18 | −0.81 | 3.23 | 0.033 |
| ENSG00000211670 | IGLV3-9 | Immunoglobulin lambda variable 3-9 | 0.71 | 1.97 | 0.033 |
| ENSG00000239855 | IGKV1-6 | Immunoglobulin kappa variable 1-6 | 0.64 | 2.66 | 0.046 |
| ENSG00000211611 | IGKV6-21 | Immunoglobulin kappa variable 6-21 | 0.65 | 0.88 | 0.047 |
| ENSG00000211668 | IGLV2-11 | Immunoglobulin lambda variable 2-11 | −0.77 | 4.25 | 0.047 |
| ENSG00000211895 | IGHA1 | Immunoglobulin heavy constant alpha 1 | −0.89 | 9.78 | 0.048 |
| Phylum | Genus | Red | White | p-Value |
|---|---|---|---|---|
| Firmicutes | Blautia | 2.53 ± 0.32 | 3.12 ± 0.32 | 0.049 |
| Firmicutes | Roseburia | 2.30 ± 0.26 | 3.07 ± 0.35 | 0.041 |
| Firmicutes | Phascolarctobacterium | 1.75 ± 0.33 | 2.79 ± 0.59 | 0.045 |
| Firmicutes | Ruminococcus 1 | 1.10 ± 0.21 | 1.69 ± 0.28 | 0.024 |
| Proteobacteria | Sutterella | 0.50 ± 0.12 | 0.33 ± 0.07 | 0.026 |
| Firmicutes | Lactobacillus | 0.30 ± 0.19 | 0.04 ± 0.02 | 0.045 |
| Bacteroidetes | Coprobacter | 0.27 ± 0.18 | 0.11 ± 0.04 | 0.001 |
| Firmicutes | Butyricicoccus | 0.23 ± 0.04 | 0.18 ± 0.03 | 0.049 |
| Firmicutes | Streptococcus | 0.18 ± 0.05 | 0.36 ± 0.08 | 0.015 |
| Firmicutes | Intestinibacter | 0.17 ± 0.04 | 0.25 ± 0.05 | 0.050 |
| Proteobacteria | Haemophilus | 0.12 ± 0.12 | 0.24 ± 0.17 | 0.037 |
| Tenericutes | uncl. Mollicutes RF39 | 0.11 ± 0.03 | 0.21 ± 0.06 | 0.033 |
| Firmicutes | Terrisporobacter | 0.09 ± 0.03 | 0.22 ± 0.09 | 0.042 |
| Tenericutes | unclassified Izimaplasmatales | 0.03 ± 0.02 | 0.09 ± 0.06 | 0.041 |
| Firmicutes | Uncl. Ruminococcaceae UCG 011 | 0.02 ± 0.01 | 0.01 ± 0.00 | 0.040 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barnett, M.P.G.; Young, W.; Armstrong, K.; Brewster, D.; Cooney, J.M.; Ellett, S.; Espley, R.V.; Laing, W.; Maclean, P.; McGhie, T.; et al. A Polyphenol Enriched Variety of Apple Alters Circulating Immune Cell Gene Expression and Faecal Microbiota Composition in Healthy Adults: A Randomized Controlled Trial. Nutrients 2021, 13, 1092. https://doi.org/10.3390/nu13041092
Barnett MPG, Young W, Armstrong K, Brewster D, Cooney JM, Ellett S, Espley RV, Laing W, Maclean P, McGhie T, et al. A Polyphenol Enriched Variety of Apple Alters Circulating Immune Cell Gene Expression and Faecal Microbiota Composition in Healthy Adults: A Randomized Controlled Trial. Nutrients. 2021; 13(4):1092. https://doi.org/10.3390/nu13041092
Chicago/Turabian StyleBarnett, Matthew P. G., Wayne Young, Kelly Armstrong, Diane Brewster, Janine M. Cooney, Stephanie Ellett, Richard V. Espley, William Laing, Paul Maclean, Tony McGhie, and et al. 2021. "A Polyphenol Enriched Variety of Apple Alters Circulating Immune Cell Gene Expression and Faecal Microbiota Composition in Healthy Adults: A Randomized Controlled Trial" Nutrients 13, no. 4: 1092. https://doi.org/10.3390/nu13041092
APA StyleBarnett, M. P. G., Young, W., Armstrong, K., Brewster, D., Cooney, J. M., Ellett, S., Espley, R. V., Laing, W., Maclean, P., McGhie, T., Pringle, G., Roy, N. C., & Ferguson, L. R. (2021). A Polyphenol Enriched Variety of Apple Alters Circulating Immune Cell Gene Expression and Faecal Microbiota Composition in Healthy Adults: A Randomized Controlled Trial. Nutrients, 13(4), 1092. https://doi.org/10.3390/nu13041092








