Artificial Intelligence in Nutrients Science Research: A Review
Abstract
1. Introduction
1.1. Artificial Neural Networks (ANNs)
1.2. Machine Learning (ML)
1.3. Internet of Things (IoT)
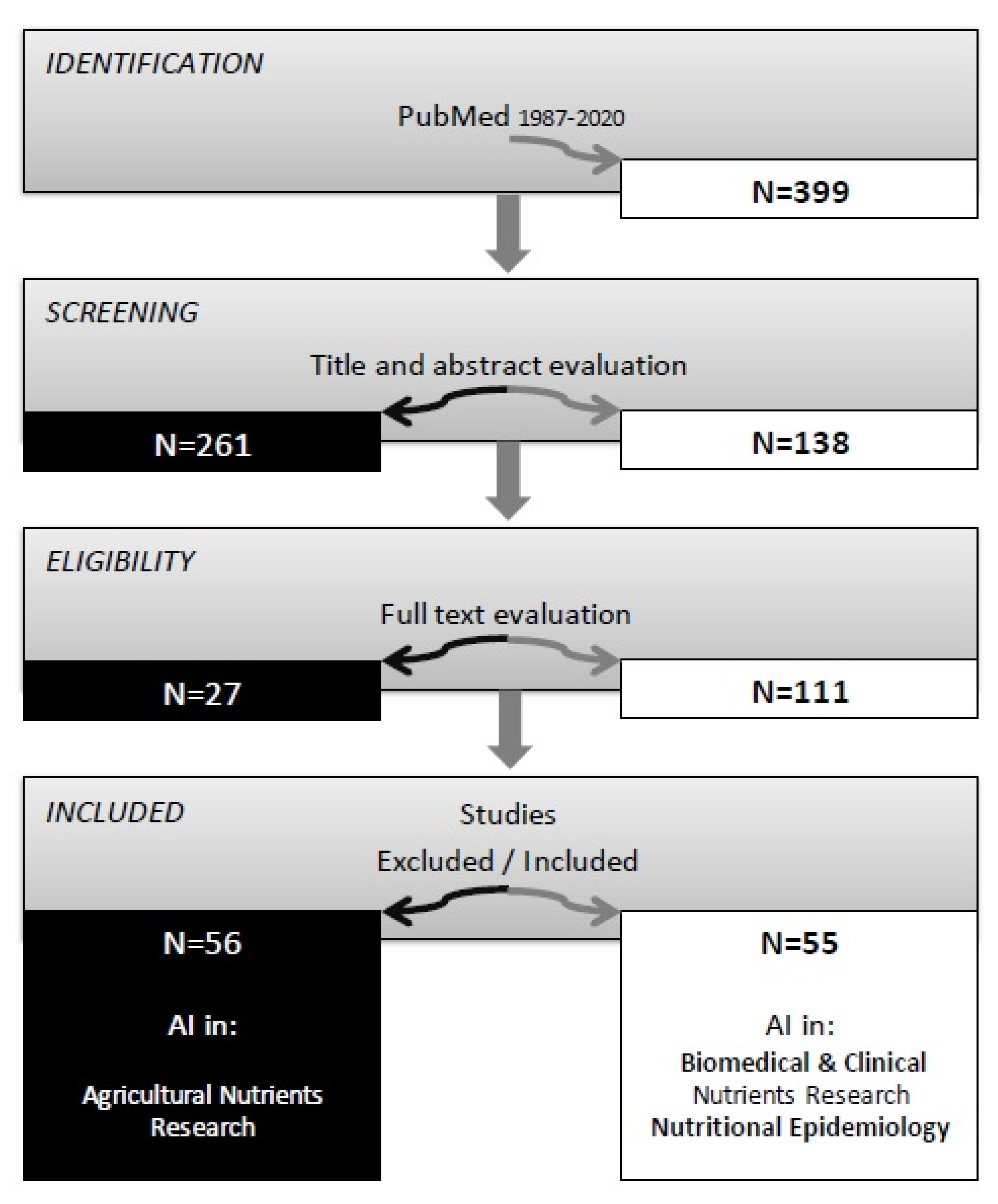
2. Materials and Methods
3. Results
3.1. AI in Biomedical Nutrients Research
3.1.1. AI in Food Composition Study
3.1.2. AI in Research on Production of Nutrients
3.1.3. AI in Research on the Influence of Nutrients on Physiological and Pathophysiological Functions
3.1.4. AI in Research on Gut Microbiota
3.2. AI in Clinical Nutrients Research
3.2.1. AI in Clinical Nutrients Intake
3.2.2. AI in Evaluating Diseases Risks in Relations to Food and Nutrients Patterns
3.2.3. AI in Studying the Relationships between Disease and Trace Elements Levels
3.2.4. AI in Studying on Supplementations
3.3. AI in Nutritional Epidemiology
3.3.1. AI in Dietary Assessment
3.3.2. AI in Physical Monitoring Systems
3.3.3. AI in Environmental Trace Elements Monitoring Systems
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McCarthy, J.; Minsky, M.; Rochester, N.; Shannon, C.E. A Proposal for the Dartmouth Summer Research Project on Artificial Intelligence. 1955. Available online: http://raysolomonoff.com/dartmouth/boxa/dart564props.pdf (accessed on 6 November 2020).
- Nilsson, N.J. The Quest for Artificial Intelligence; Cambridge University Press: Cambrige, UK; New York, NY, USA, 2010. [Google Scholar]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2018, 103, 167–175. [Google Scholar] [CrossRef]
- Yasaka, K.; Abe, O. Deep learning and artificial intelligence in radiology: Current applications and future directions. PLoS Med. 2018, 15, e1002707. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.W.; Torres Soto, J.; Glicksberg, B.S.; Shameer, K.; Miotto, R.; Ali, M.; Ashley, E.; Dudley, J.T. Artificial intelligence in cardiology. J. Am. Coll. Cardiol. 2018, 71, 2668–2679. [Google Scholar] [CrossRef] [PubMed]
- Hessler, G.; Baringhaus, K.-H. Artificial intelligence in drug design. Molecules 2018, 23, 2520. [Google Scholar] [CrossRef] [PubMed]
- Heydarian, H.; Adam, M.T.P.; Burrows, T.; Collins, C.E.; Rollo, M.E. Assessing eating behaviour using upper limb mounted motion sensors: A systematic review. Nutrients 2019, 11, 1168. [Google Scholar] [CrossRef] [PubMed]
- Demirci, F.; Akan, P.; Kume, T.; Sisman, A.R.; Erbayraktar, Z.; Sevinc, S. Artificial neural network approach in laboratory test reporting: Learning algorithms. Am. J. Clin. Pathol. 2016, 146, 227–237. [Google Scholar] [CrossRef]
- Valletta, E.; Kučera, L.; Prokeš, L.; Amato, F.; Pivetta, T.; Hampl, A.; Havel, J.; Vaňhara, P. Multivariate calibration approach for quantitative determination of cell-line cross contamination by intact cell mass spectrometry and artificial neural networks. PLoS ONE 2016, 11, e0147414. [Google Scholar] [CrossRef]
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Gallucci, M.; Pallucca, C.; Di Battista, M.E.; Fougère, B.; Grossi, E.; Fougèreand, B. Artificial neural networks help to better understand the interplay between cognition, mediterranean diet, and physical performance: Clues from TRELONG study. J. Alzheimer’s Dis. 2019, 71, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.R.; Abbod, M.F.; Liu, Q.; Shieh, J.-S.; Chao, T.Y.; Hsieh, C.Y.; Yang, Y.C. Ensembled artificial neural networks to predict the fitness score for body composition analysis. J. Nutr. Heal. Aging 2010, 15, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Szymkuć, S.; Gajewska, E.P.; Klucznik, T.; Molga, K.; Dittwald, P.; Startek, M.; Bajczyk, M.; Grzybowski, B.A. Computer-assisted synthetic planning: The end of the beginning. Angew. Chem. Int. Ed. 2016, 55, 5904–5937. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C. Machine learning in medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Lee, M.J.; Asadi, H. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Woldaregay, A.Z.; Årsand, E.; Walderhaug, S.; Albers, D.; Mamykina, L.; Botsis, T.; Hartvigsen, G. Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes. Artif. Intell. Med. 2019, 98, 109–134. [Google Scholar] [CrossRef]
- Danneskiold-Samsøe, N.B.; Dias de Freitas Queiroz Barros, H.; Santos, R.; Bicas, J.L.; Cazarin, C.B.B.; Madsen, L.; Kristiansen, K.; Pastore, G.M.; Brix, S.; Júnior, M.R.M. Interplay between food and gut microbiota in health and disease. Food Res. Int. 2019, 115, 23–31. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Ni, Y.; Cheung, C.K.; Lam, K.S.; Wang, Y.; Xia, Z.; Ye, D.; Guo, J.; Tse, M.A.; et al. Gut microbiome fermentation determines the efficacy of exercise for diabetes prevention. Cell Metab. 2020, 31, 77–91.e5. [Google Scholar] [CrossRef]
- Li, J.-P.O.; Liu, H.; Ting, D.S.; Jeon, S.; Chan, R.V.P.; Kim, J.E.; Sim, D.A.; Thomas, P.B.; Lin, H.; Chen, Y.; et al. Digital technology, tele-medicine and artificial intelligence in ophthalmology: A global perspective. Prog. Retin. Eye Res. 2020, 100900. [Google Scholar] [CrossRef]
- Sadoughi, F.; Behmanesh, A.; Sayfouri, N. Internet of things in medicine: A systematic mapping study. J. Biomed. Inform. 2020, 103, 103383. [Google Scholar] [CrossRef]
- Jæger, B.; Mishra, A. IoT platform for seafood farmers and consumers. Sensors 2020, 20, 4230. [Google Scholar] [CrossRef]
- Dettmar, H.; Barbour, G.; Blackwell, K.T.; Vogl, T.; Alkon, D.; Fry, F.S., Jr.; Totah, J.; Chambers, T. Orange juice classification with a biologically based neural network. Comput. Chem. 1996, 20, 261–266. [Google Scholar] [CrossRef]
- Yang, M.; Cao, X.; Wu, R.; Liu, B.; Ye, W.; Yue, X.; Wu, J. Comparative proteomic exploration of whey proteins in human and bovine colostrum and mature milk using iTRAQ-coupled LC-MS/MS. Int. J. Food Sci. Nutr. 2017, 68, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.S.; Chagas, B.C.; Pacheco, C.S.V.; Santos, H.M.; de Menezes, L.H.S.; Nascimento, M.M.; Batista, M.A.S.; de Jesus, R.M.; Amorim, F.A.C.; Santos, L.N.; et al. Development of procedure for sample preparation of cashew nuts using mixture design and evaluation of nutrient profiles by Kohonen neural network. Food Chem. 2019, 273, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Li, W.; Zhang, X.; Kong, W.; Liu, F.; Wang, W.; Peng, J. High-sensitivity determination of nutrient elements in panax notoginseng by laser-induced breakdown spectroscopy and chemometric methods. Molecules 2019, 24, 1525. [Google Scholar] [CrossRef]
- Rasouli, Z.; Hassanzadeh, Z.; Ghavami, R. Application of a new version of GA-RBF neural network for simultaneous spectrophotometric determination of Zn(II), Fe(II), Co(II) and Cu(II) in real samples: An exploratory study of their complexation abilities toward MTB. Talanta 2016, 160, 86–98. [Google Scholar] [CrossRef]
- Soltani, S.; Haghaei, H.; Shayanfar, A.; Vallipour, J.; Asadpour Zeynali, K.; Jouyban, A. QSBR study of bitter taste of peptides: Application of GA-PLS in combination with MLR, SVM, and ANN approaches. Biomed. Res. Int. 2013, 2013, 501310. [Google Scholar] [CrossRef]
- Huang, S.-M.; Li, H.-J.; Liu, Y.-C.; Kuo, C.-H.; Shieh, C.J. An efficient approach for lipase-catalyzed synthesis of retinyl laurate nutraceutical by combining ultrasound assistance and artificial neural network optimization. Molecules 2017, 22, 1972. [Google Scholar] [CrossRef]
- Zheng, Z.-Y.; Guo, X.-N.; Zhu, K.-X.; Peng, W.; Zhou, H.-M. Artificial neural network—Genetic algorithm to optimize wheat germ fermentation condition: Application to the production of two anti-tumor benzoquinones. Food Chem. 2017, 227, 264–270. [Google Scholar] [CrossRef]
- Kumar Saini, D.; Yadav, D.; Pabbi, S.; Chhabra, D.; Shukla, P. Phycobiliproteins from Anabaena variabilis CCC421 and its production enhancement strategies using combinatory evolutionary algorithm approach. Bioresour. Technol. 2020, 309, 123347. [Google Scholar] [CrossRef]
- Pavani, A.; Naushad, S.M.; Lakshmitha, G.; Nivetha, S.; Stanley, B.A.; Malempati, A.R.; Kutala, V.K. Development of neuro-fuzzy model to explore gene–nutrient interactions modulating warfarin dose requirement. Pharmacogenomics 2016, 17, 1315–1325. [Google Scholar] [CrossRef]
- Yu, P.; Song, H.; Gao, J.; Li, B.; Liu, Y.; Wang, Y. Vitamin D (1,25-(OH)2D3) regulates the gene expression through competing endogenous RNAs networks in high glucose-treated endothelial progenitor cells. J. Steroid Biochem. Mol. Biol. 2019, 193, 105425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Gu, M.; Xu, Y.; Wu, Z. A comprehensive analysis on the effects of 1,25(OH)2D3 on primary chondrocytes cultured from patients with osteoarthritis. Gene 2020, 730, 144322. [Google Scholar] [CrossRef] [PubMed]
- Kolhe, R.; Mondal, A.K.; Pundkar, C.; Periyasamy-Thandavan, S.; Mendhe, B.; Hunter, M.; Isales, C.M.; Hill, W.D.; Hamrick, M.W.; Fulzele, S. Modulation of miRNAs by vitamin C in human bone marrow stromal cells. Nutrients 2018, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Y.; Wang, L.J.; Wang, J.J.; Feng, W.J.; Yang, Z.Q.; Ni, S.H.; Huang, Y.S.; Li, H.; Yang, Y.; Wang, M.Q.; et al. Hispaglabridin B, a constituent of liquorice identified by a bioinformatics and machine learning approach, relieves protein-energy wasting by inhibiting forkhead box O1. Br. J. Pharmacol. 2019, 176, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, X.; Yu, F.; Xu, L.; Miao, J.-H.; Xiao, P. In Silico Investigation of the pharmacological mechanisms of beneficial effects of ginkgo biloba l. on Alzheimer’s disease. Nutrients 2018, 10, 589. [Google Scholar] [CrossRef]
- Panwar, B.; Gupta, S.; Raghava, G.P. Prediction of vitamin interacting residues in a vitamin binding protein using evolutionary information. BMC Bioinform. 2013, 14, 44. [Google Scholar] [CrossRef]
- Yu, D.-J.; Hu, J.; Yan, H.; Yang, X.; Yang, J.-Y.; Shen, H.-B. Enhancing protein-vitamin binding residues prediction by multiple heterogeneous subspace SVMs ensemble. BMC Bioinform. 2014, 15, 297. [Google Scholar] [CrossRef]
- Devika, N.T.; Raman, K. Deciphering the metabolic capabilities of Bifidobacteria using genome-scale metabolic models. Sci. Rep. 2019, 9, 18222. [Google Scholar] [CrossRef]
- Shima, H.; Masuda, S.; Date, Y.; Shino, A.; Tsuboi, Y.; Kajikawa, M.; Inoue, Y.; Kanamoto, T.; Kikuchi, J. Exploring the impact of food on the gut ecosystem based on the combination of machine learning and network visualization. Nutrients 2017, 9, 1307. [Google Scholar] [CrossRef]
- Mohammed, A.; Guda, C. Application of a hierarchical enzyme classification method reveals the role of gut microbiome in human metabolism. BMC Genom. 2015, 16, S16. [Google Scholar] [CrossRef]
- Lu, Y.; Stathopoulou, T.; Vasiloglou, M.F.; Christodoulidis, S.; Blum, B.; Walser, T.; Meier, V.; Stanga, Z.; Mougiakakou, S.G. An artificial intelligence-based system for nutrient intake assessment of hospitalised patients. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 2019, 5696–5699. [Google Scholar] [CrossRef] [PubMed]
- Oka, R.; Nomura, A.; Yasugi, A.; Kometani, M.; Gondoh, Y.; Yoshimura, K.; Yoneda, T. Study protocol for the effects of Artificial Intelligence (AI)-supported automated nutritional intervention on glycemic control in patients with type 2 diabetes mellitus. Diabetes Ther. 2019, 10, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Vasiloglou, M.F.; Mougiakakou, S.; Aubry, E.; Bokelmann, A.; Fricker, R.; Gomes, F.; Guntermann, C.; Meyer, A.L.; Studerus, D.; Stanga, Z. A Comparative study on carbohydrate estimation: GoCARB vs. dietitians. Nutrients 2018, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Chin, E.L.; Simmons, G.; Bouzid, Y.Y.; Kan, A.; Burnett, D.J.; Tagkopoulos, I.; Lemay, D.G. Nutrient estimation from 24-hour food recalls using machine learning and database mapping: A case study with lactose. Nutrients 2019, 11, 3045. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, D.; Dimitropoulos, K.; Langlet, B.; Daras, P.; Ioakimidis, I. Validation of a deep learning system for the full automation of bite and meal duration analysis of experimental meal videos. Nutrients 2020, 12, 209. [Google Scholar] [CrossRef]
- Chi, Y.-L.; Chen, T.-Y.; Tsai, W.-T. A chronic disease dietary consultation system using OWL-based ontologies and semantic rules. J. Biomed. Informatics 2015, 53, 208–219. [Google Scholar] [CrossRef]
- Posada-Quintero, H.F.; Reljin, N.; Moutran, A.; Georgopalis, D.; Lee, E.C.; Giersch, G.E.; Casa, D.J.; Chon, K.H. Mild dehydration identification using machine learning to assess autonomic responses to cognitive stress. Nutrients 2019, 12, 42. [Google Scholar] [CrossRef]
- Khan, A.S.; Hoffmann, A. Building a case-based diet recommendation system without a knowledge engineer. Artif. Intell. Med. 2003, 27, 155–179. [Google Scholar] [CrossRef][Green Version]
- Buisson, J.-C. Nutri-Educ, a nutrition software application for balancing meals, using fuzzy arithmetic and heuristic search algorithms. Artif. Intell. Med. 2008, 42, 213–227. [Google Scholar] [CrossRef]
- Baek, J.-W.; Kim, J.-C.; Chun, J.; Chung, K. Hybrid clustering based health decision-making for improving dietary habits. Technol. Health Care 2019, 27, 459–472. [Google Scholar] [CrossRef]
- Mezgec, S.; Koroušić Seljak, B. NutriNet: A deep learning food and drink image recognition system for dietary assessment. Nutrients 2017, 9, 657. [Google Scholar] [CrossRef] [PubMed]
- Panaretos, D.; Koloverou, E.; Dimopoulos, A.C.; Kouli, G.-M.; Vamvakari, M.; Tzavelas, G.; Pitsavos, C.; Panagiotakos, D. A comparison of statistical and machine-learning techniques in evaluating the association between dietary patterns and 10-year cardiometabolic risk (2002–2012): The ATTICA study. Br. J. Nutr. 2018, 120, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.E.; Valdes, A.M.; Drew, D.A.; Asnicar, F.; Mazidi, M.; Wolf, J.; Capdevila, J.; Hadjigeorgiou, G.; Davies, R.; Al Khatib, H.; et al. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 2020, 26, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Naushad, S.M.; Janaki Ramaiah, M.; Pavithrakumari, M.; Jayapriya, J.; Hussain, T.; Alrokayan, S.A.; Gottumukkala, S.R.; Digumarti, R.; Kutala, V.K. Artificial neural network-based exploration of gene-nutrient interactions in folate and xeno-biotic metabolic pathways that modulate susceptibility to breast cancer. Gene 2016, 580, 159–168. [Google Scholar] [CrossRef]
- Shiao, S.P.K.; Grayson, J.; Lie, A.; Yu, C.H. Predictors of the healthy eating index and glycemic index in multi-ethnic colorectal cancer families. Nutrients 2018, 10, 674. [Google Scholar] [CrossRef]
- Tan, C.; Chen, H.; Xia, C. The prediction of cardiovascular disease based on trace element contents in hair and a classifier of boosting decision stumps. Biol. Trace Element Res. 2008, 129, 9–19. [Google Scholar] [CrossRef]
- Chen, H.; Tan, C. Prediction of Type-2 diabetes based on several element levels in blood and chemometrics. Biol. Trace Element Res. 2011, 147, 67–74. [Google Scholar] [CrossRef]
- Chen, H.; Tan, C.; Lin, Z.; Wu, T. The diagnostics of diabetes mellitus based on ensemble modeling and hair/urine element level analysis. Comput. Biol. Med. 2014, 50, 70–75. [Google Scholar] [CrossRef]
- Lin, T.; Liu, T.; Lin, Y.; Yan, L.; Chen, Z.; Wang, J. Comparative study on serum levels of macro and trace elements in schizophrenia based on supervised learning methods. J. Trace Elements Med. Biol. 2017, 43, 202–208. [Google Scholar] [CrossRef]
- Li, R.; Wu, K.; Li, Y.; Liang, X.; Tse, W.K.F.; Yang, L.; Lai, K.P. Revealing the targets and mechanisms of vitamin A in the treatment of COVID-19. Aging 2020, 12, 15784–15796. [Google Scholar] [CrossRef]
- Chen, L.; Hu, C.; Hood, M.; Zhang, X.; Zhang, L.; Kan, J.; Du, J. A Novel Combination of vitamin C, curcumin and glycyrrhizic acid potentially regulates immune and inflammatory response associated with coronavirus infections: A perspective from system biology analysis. Nutrients 2020, 12, 1193. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Qi, X.; Sweet, R.A.; Wang, L. Network systems pharmacology-based mechanism study on the beneficial effects of vitamin d against psychosis in Alzheimer’s disease. Sci. Rep. 2020, 10, 6136. [Google Scholar] [CrossRef]
- Sun, M.; Liu, Q.; Schmidt, K.; Yang, J.; Yao, N.; Fernstrom, J.D.; Fernstrom, M.H.; Delany, J.P.; Sclabassi, R.J. Determination of food portion size by image processing. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2008, 2008, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Stathopoulou, T.; Vasiloglou, M.F.; Pinault, L.F.; Kiley, C.; Spanakis, E.K.; Mougiakakou, S. goFOOD(TM): An artificial intelligence system for dietary assessment. Sensors (Basel) 2020, 20, 4283. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ambayo, H.; De Baets, B.; Kolsteren, P.; Thanintorn, N.; Hawwash, D.; Bouwman, J.; Bronselaer, A.; Pattyn, F.; Lachat, C. An ontology to standardize research output of nutritional epidemiology: From paper-based standards to linked content. Nutrients 2019, 11, 1300. [Google Scholar] [CrossRef]
- Lo, F.P.-W.; Sun, Y.; Qiu, J.; Lo, B. Food volume estimation based on deep learning view synthesis from a single depth map. Nutrients 2018, 10, 2005. [Google Scholar] [CrossRef]
- Fang, S.; Shao, Z.; Kerr, D.A.; Boushey, C.J.; Zhu, F. An end-to-end image-based automatic food energy estimation technique based on learned energy distribution images: Protocol and methodology. Nutrients 2019, 11, 877. [Google Scholar] [CrossRef]
- Ji, Y.; Plourde, H.; Bouzo, V.; Kilgour, R.D.; Cohen, T.R. Validity and usability of a smartphone image-based dietary assessment app compared to 3-day food diaries in assessing dietary intake among canadian adults: Randomized controlled trial. JMIR Mhealth Uhealth 2020, 8, e16953. [Google Scholar] [CrossRef]
- Hsu, M.-H.; Huang, L.-C.; Chen, T.M.; Chen, L.-F.; Chao, J.C.-J. A web-based decision support system for dietary analysis and recommendations. Telemed. J. E. Health 2011, 17, 68–75. [Google Scholar] [CrossRef]
- Manogaran, G.; Shakeel, P.M.; Fouad, H.; Nam, Y.; Baskar, S.; Chilamkurti, N.; Sundarasekar, R. Wearable IoT Smart-Log Patch: An edge computing-based bayesian deep learning network system for multi access physical monitoring system. Sensors (Basel) 2019, 19, 3030. [Google Scholar] [CrossRef]
- Tragomalou, A.; Moschonis, G.; Manios, Y.; Kassari, P.; Ioakimidis, I.; Diou, C.; Stefanopoulos, L.; Lekka, E.; Maglaveras, N.; Delopoulos, A.; et al. Novel e-health applications for the management of cardiometabolic risk factors in children and adolescents in Greece. Nutrients 2020, 12, 1380. [Google Scholar] [CrossRef] [PubMed]
- Ramyaa, R.; Hosseini, O.; Krishnan, G.P.; Krishnan, S. Phenotyping women based on dietary macronutrients, physical activity, and body weight using machine learning tools. Nutrients 2019, 11, 1681. [Google Scholar] [CrossRef] [PubMed]
- Novič, M.; Grošelj, N. Bottle-neck type of neural network as a mapping device towards food specifications. Anal. Chim. Acta 2009, 649, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Drivelos, S.A.; Danezis, G.P.; Halagarda, M.; Popek, S.; Georgiou, C.A. Geographical origin and botanical type honey authentication through elemental metabolomics via chemometrics. Food Chem. 2021, 338, 127936. [Google Scholar] [CrossRef] [PubMed]
- Tunakova, Y.; Novikova, S.; Ragimov, A.; Faizullin, R.; Valiev, V. A Method for assessing the retention of trace elements in human body using neural network technology. J. Heal. Eng. 2017, 2017, 3471616. [Google Scholar] [CrossRef] [PubMed]
- Gedrich, K.; Hensel, A.; Binder, I.; Karg, G. How optimal are computer-calculated optimal diets? Eur. J. Clin. Nutr. 1999, 53, 309–318. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Verma, M.; Hontecillas, R.; Tubau-Juni, N.; Abedi, V.; Bassaganya-Riera, J. Challenges in personalized nutrition and health. Front. Nutr. 2018, 5, 117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, M.; Ma, H.; Li, X.; Heianza, Y.; Qi, L. Dietary fiber, genetic variations of gut microbiota-derived short-chain fatty acids, and bone health in UK biobank. J. Clin. Endocrinol. Metab. 2020, 106, 201–210. [Google Scholar] [CrossRef]
- Akyazi, T.; Goti, A.; Oyarbide-Zubillaga, A.; Alberdi, E.; Bayon, F. A Guide for the food industry to meet the future skills requirements emerging with industry 4. Foods 2020, 9, 492. [Google Scholar] [CrossRef]
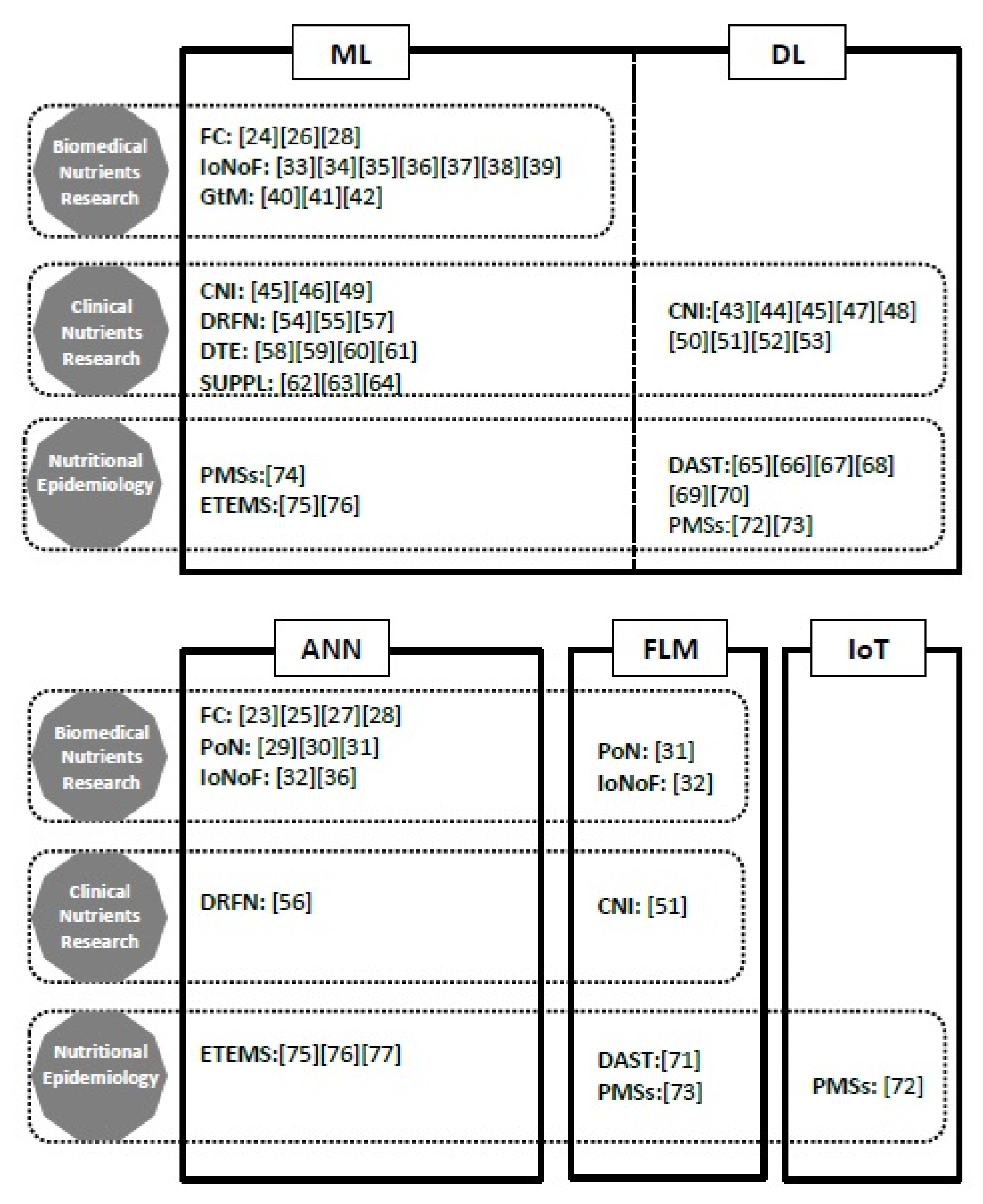
| Biomedical Nutrients Research | Topic | Number of Studies [Ref] | Nutrients | Domains | Algorithms | Years |
| Food composition | 6 [23,24,25,26,27,28] | Proteins, Minerals (K, Ca, Mg), Trace elements | ANN, ML | SVM, LS-SVM, SVR, GA-RBFN, PLS, GA-PLS, KohNN, LASSO, CLAs | 1996, 2013, 2016, 2017, 2019 | |
| Production of nutrients | 3 [29,30,31] | Retinol, Benzoquinones, Phycobiliproteins | ANN, FLM | LM, GA, ANN-GAR, FFD, GA-Fuzzy | 2017, 2020 | |
| Influence of nutrients on phys./path. functions | 8 [32,33,34,35,36,37,38,39] | Proteins, Vitamins (A,B,C,D,K) | ANN, FLM, ML | SVM, BN, NB, RF, CLAs | 2013, 2014, 2016, 2018, 2019 | |
| Gut microbiota | 3 [40,41,42] | Nutrients from food | ML, NV | SVM, kNN, RF, CLAs | 2015, 2017, 2019 | |
| Total | 20 | 1996–2020 |
| Clinical Nutrients Research | Topic | Number of Studies [Ref] | Nutrients | Domains | Algorithms | Years |
| Clinical nutrients intake | 11 [43,44,45,46,47,48,49,50,51,52,53] | Carbohydrate, Lactose, Protein, Minerals | ML, DL FLM | LASSO, FFNN, SVM, kNN, | 2003, 2008, 2015, 2017–2019 | |
| Diseases risks to food and nutrients patterns | 4 [54,55,56,57] | Carbohydrate, Triglyceride, Micronutrients (folate, B12) | ANN, ML | kNN, DTA LR, RF | 2016, 2018, 2020 | |
| Disease and trace elements levels | 4 [58,59,60,61] | Trace elements (lithium, zinc, chromium, copper, iron, manganese) | ML | SVM, DTA, RF, NB | 2009, 2012, 2014, 2017 | |
| Supplementations | 3 [62,63,64] | Vitamins (A, C, D) Curcumin, Glycyrrhizic acid | ML | CLAs | 2020 | |
| Total | 22 | 2003–2020 |
| Nutritional Epidemiology | Topic | Number of Studies [Ref] | Nutrients | Domains | Algorithms | Years |
| Dietary assessment | 7 [65,66,67,68,69,70,71] | Macronutrients | ML, DL FLM | ICP, CLAs | 2008, 2011, 2018–2020 | |
| Physical monitoring systems | 3 [72,73,74] | Macronutrients | IoT, ML, DL FLM | kNN, SVM, BDLN | 2019–2020 | |
| Environmental trace elements monitoring systems | 3 [75,76,77] | Trace elements | ANN, ML | PNN, KohNN, PLS | 2009, 2017, 2020 | |
| Total | 13 | 2008–2020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sak, J.; Suchodolska, M. Artificial Intelligence in Nutrients Science Research: A Review. Nutrients 2021, 13, 322. https://doi.org/10.3390/nu13020322
Sak J, Suchodolska M. Artificial Intelligence in Nutrients Science Research: A Review. Nutrients. 2021; 13(2):322. https://doi.org/10.3390/nu13020322
Chicago/Turabian StyleSak, Jarosław, and Magdalena Suchodolska. 2021. "Artificial Intelligence in Nutrients Science Research: A Review" Nutrients 13, no. 2: 322. https://doi.org/10.3390/nu13020322
APA StyleSak, J., & Suchodolska, M. (2021). Artificial Intelligence in Nutrients Science Research: A Review. Nutrients, 13(2), 322. https://doi.org/10.3390/nu13020322






