Armillaria luteo-virens Sacc Ameliorates Dextran Sulfate Sodium Induced Colitis through Modulation of Gut Microbiota and Microbiota-Related Bile Acids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of ALS Powder
2.2. Mice and Dietary Supplementation
2.3. Induction of Colitis
2.4. Quantitative Real-Time PCR
2.5. Western Blotting
2.6. Immunofluorescent Staining
2.7. Hematoxylin and Eosin (H&E Staining)
2.8. Alcian Blue Staining
2.9. Transmission Electron Microscopy
2.10. Analysis of Inflammatory Cytokine and Myeloperoxidase (MPO)
2.11. 16S rRNA Gene Sequence Analysis
2.12. Bile Acid Detection
2.13. Statistical Analysis
3. Results
3.1. ALS Alleviates Colitis Symptoms
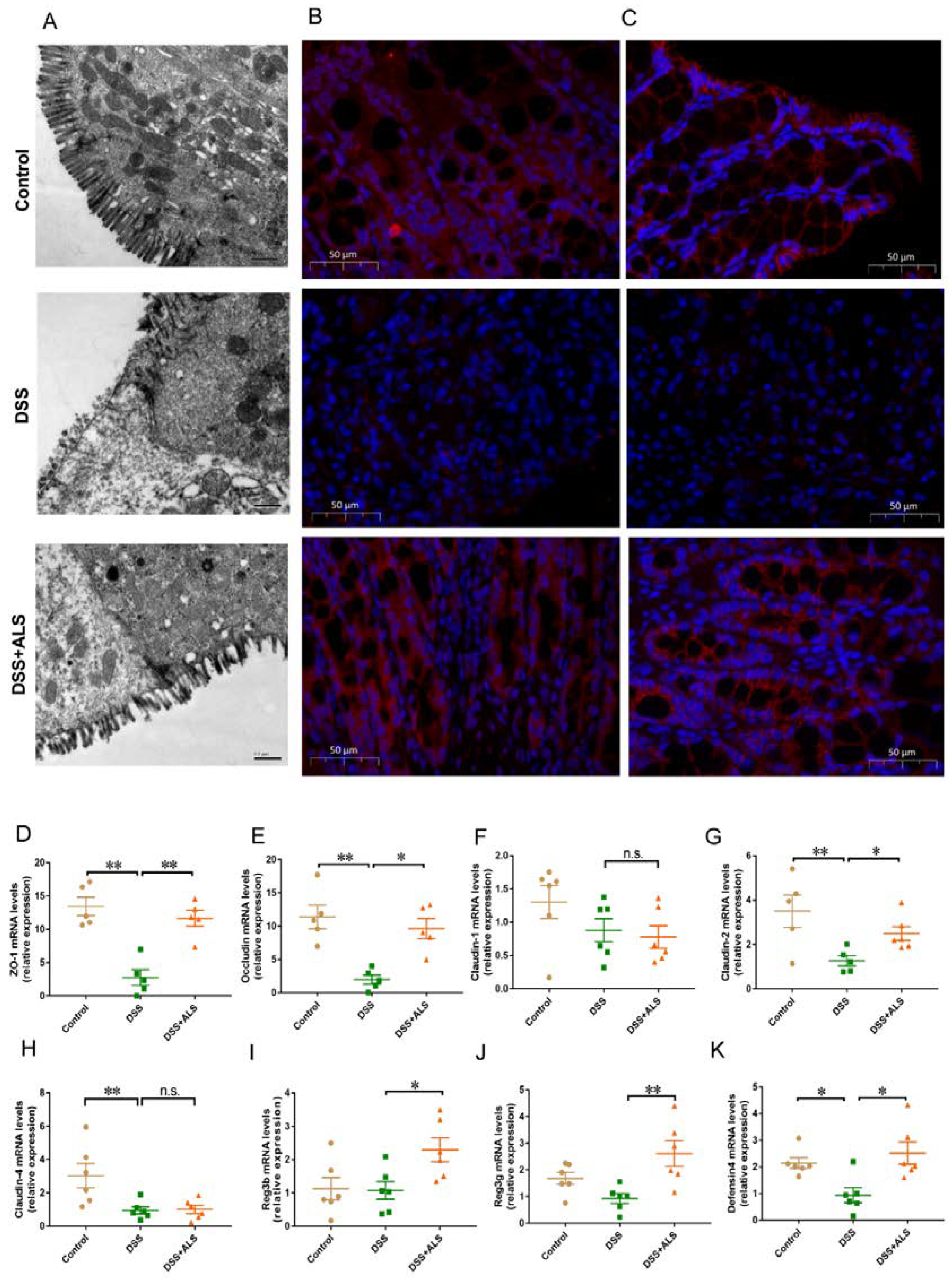
3.2. ALS Improves Intestinal Barrier Function
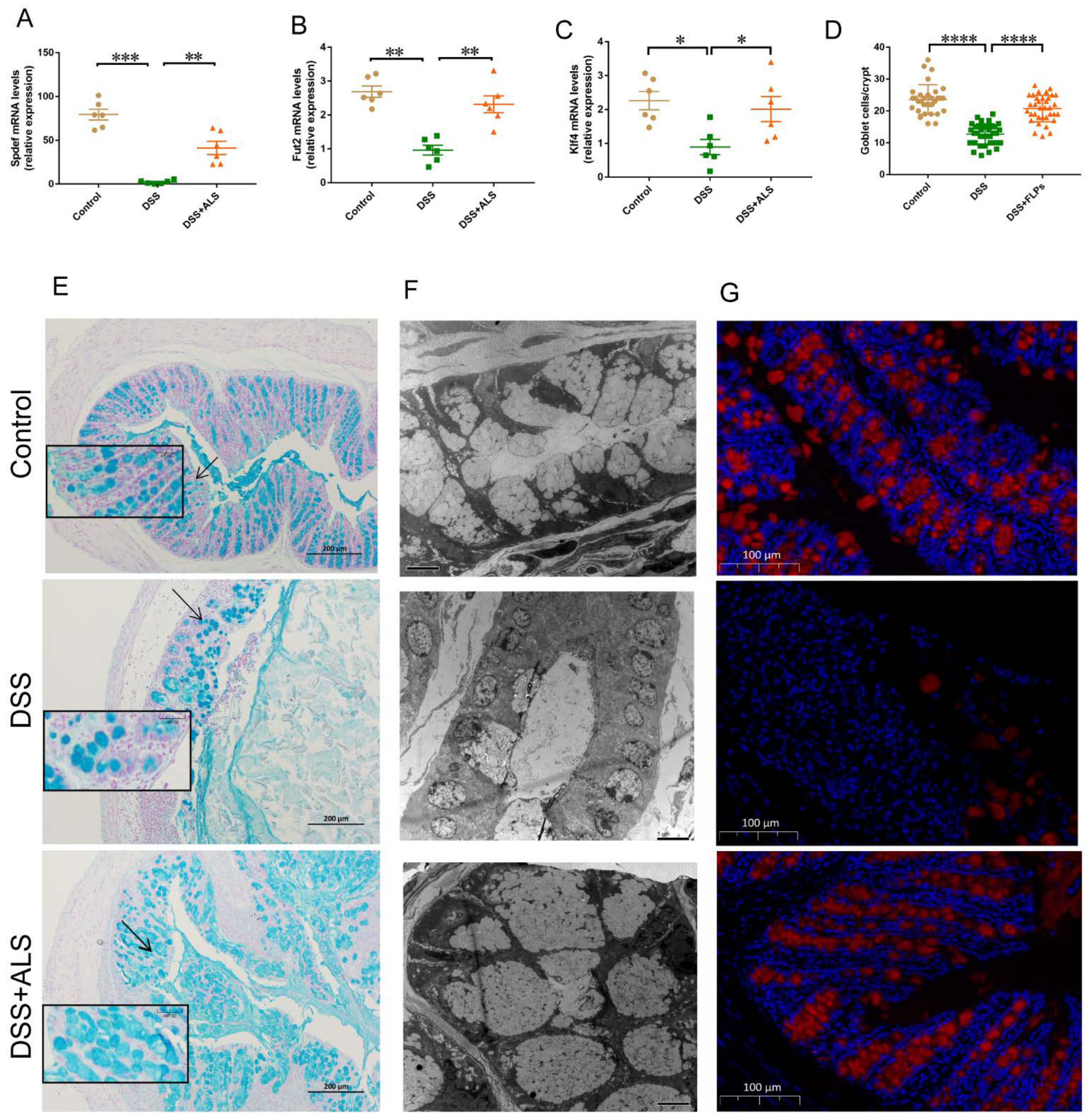
3.3. ALS Recovers the Intestinal Goblet Loss
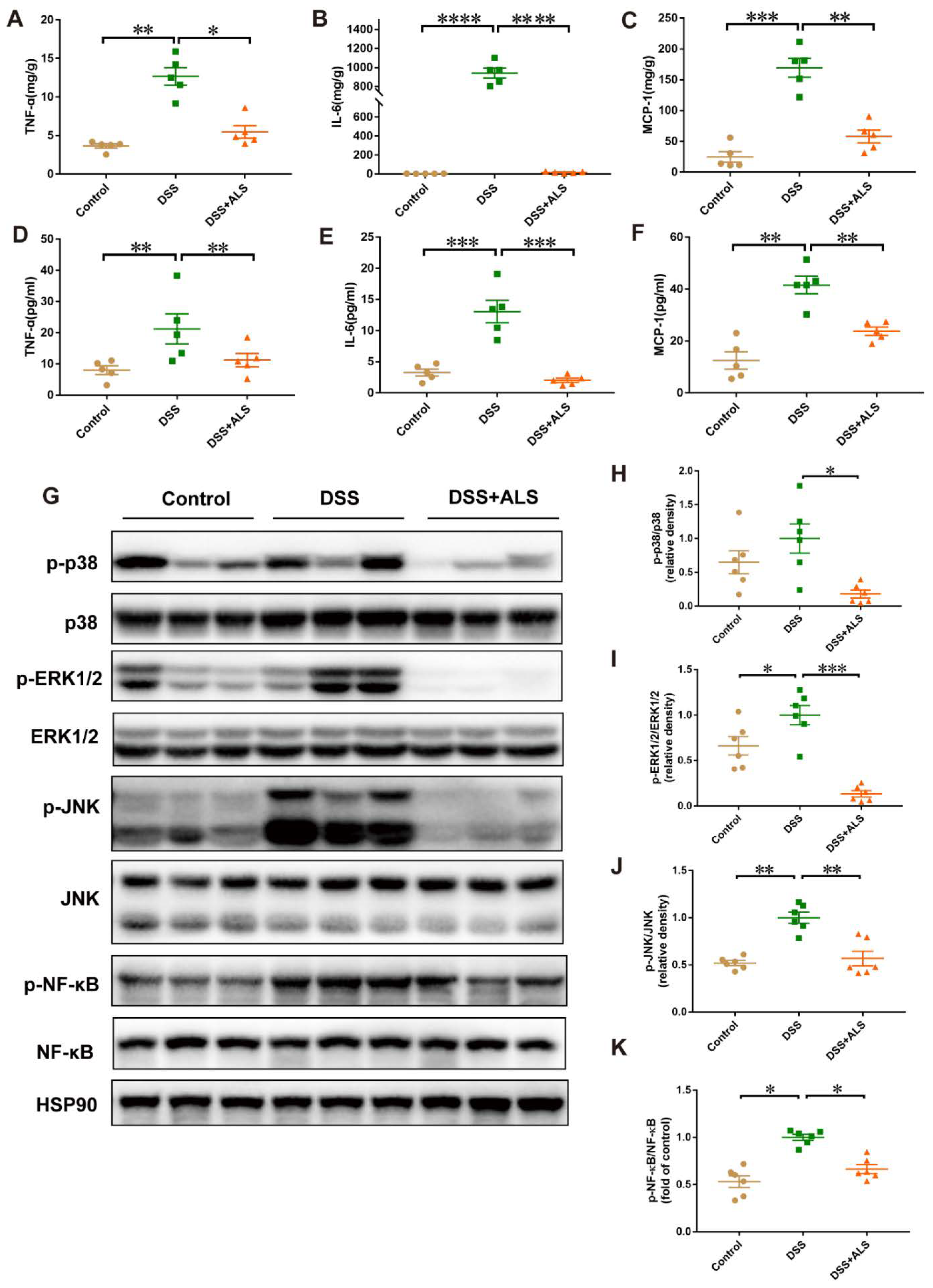
3.4. ALS Mitigates Inflammation in DSS-Induced Colitis
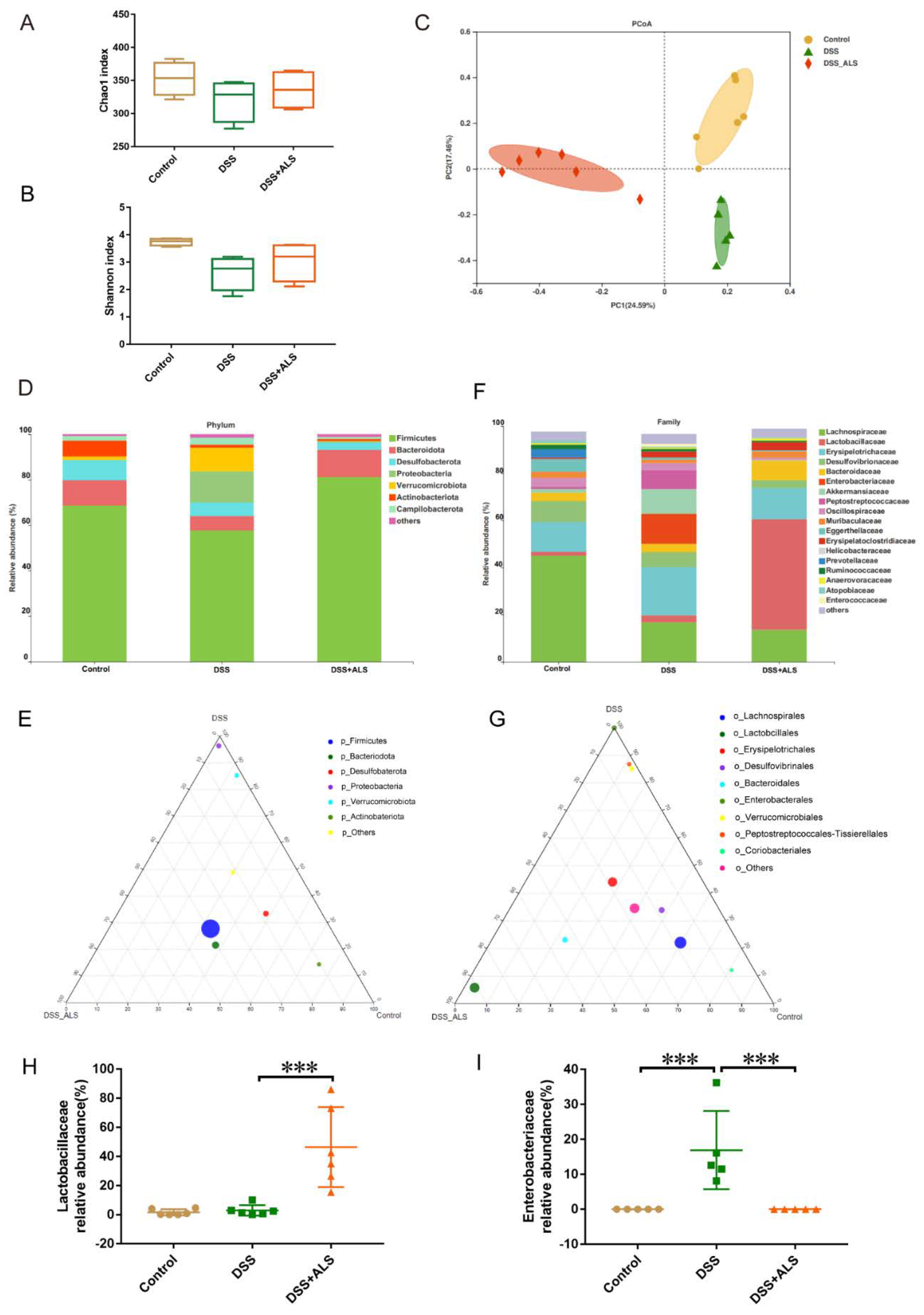
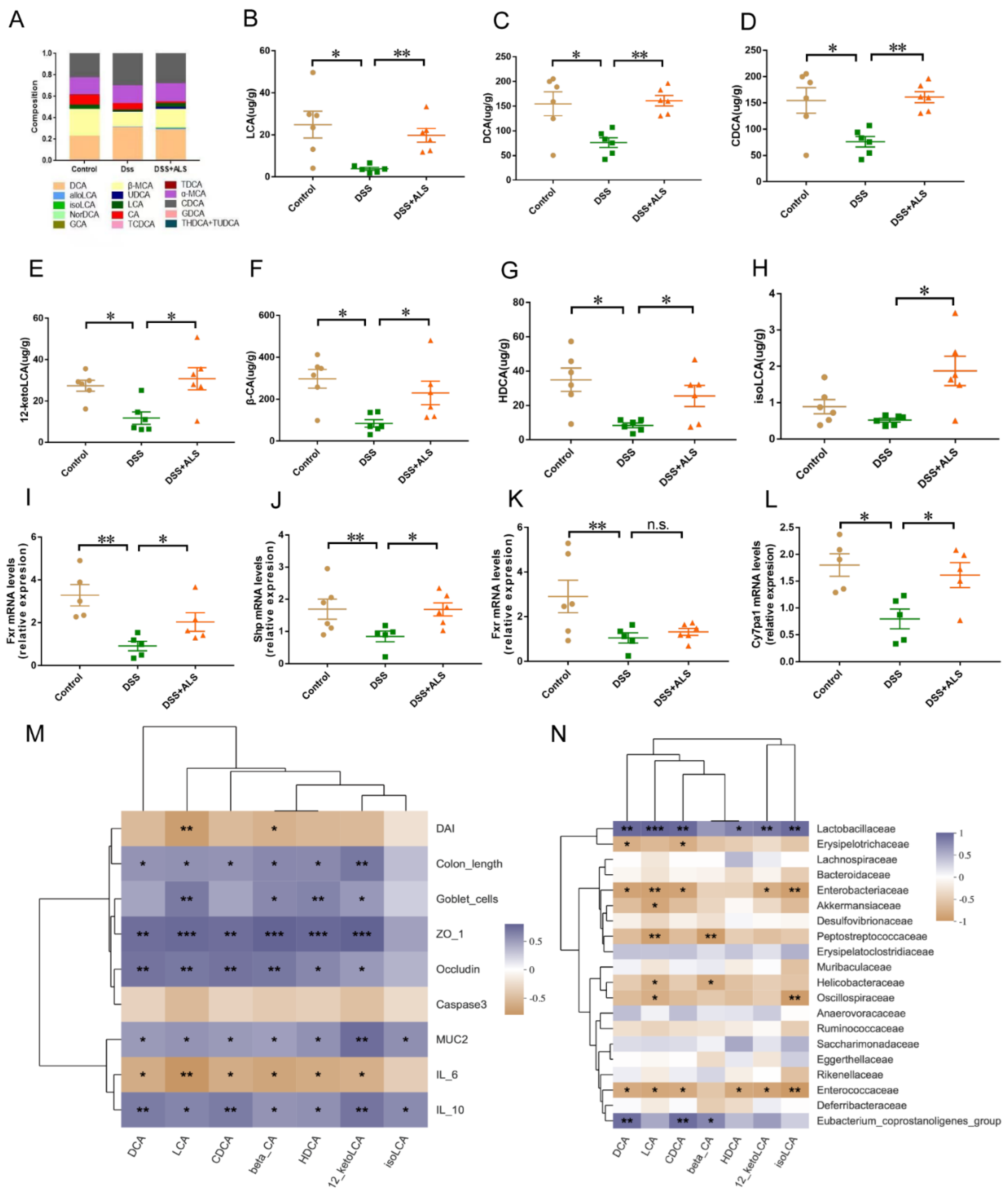
3.5. ALS Ameliorates Gut Microbiota Dysbiosis
3.6. ALS Increases the Production of Microbial Bile Acids (BAs)
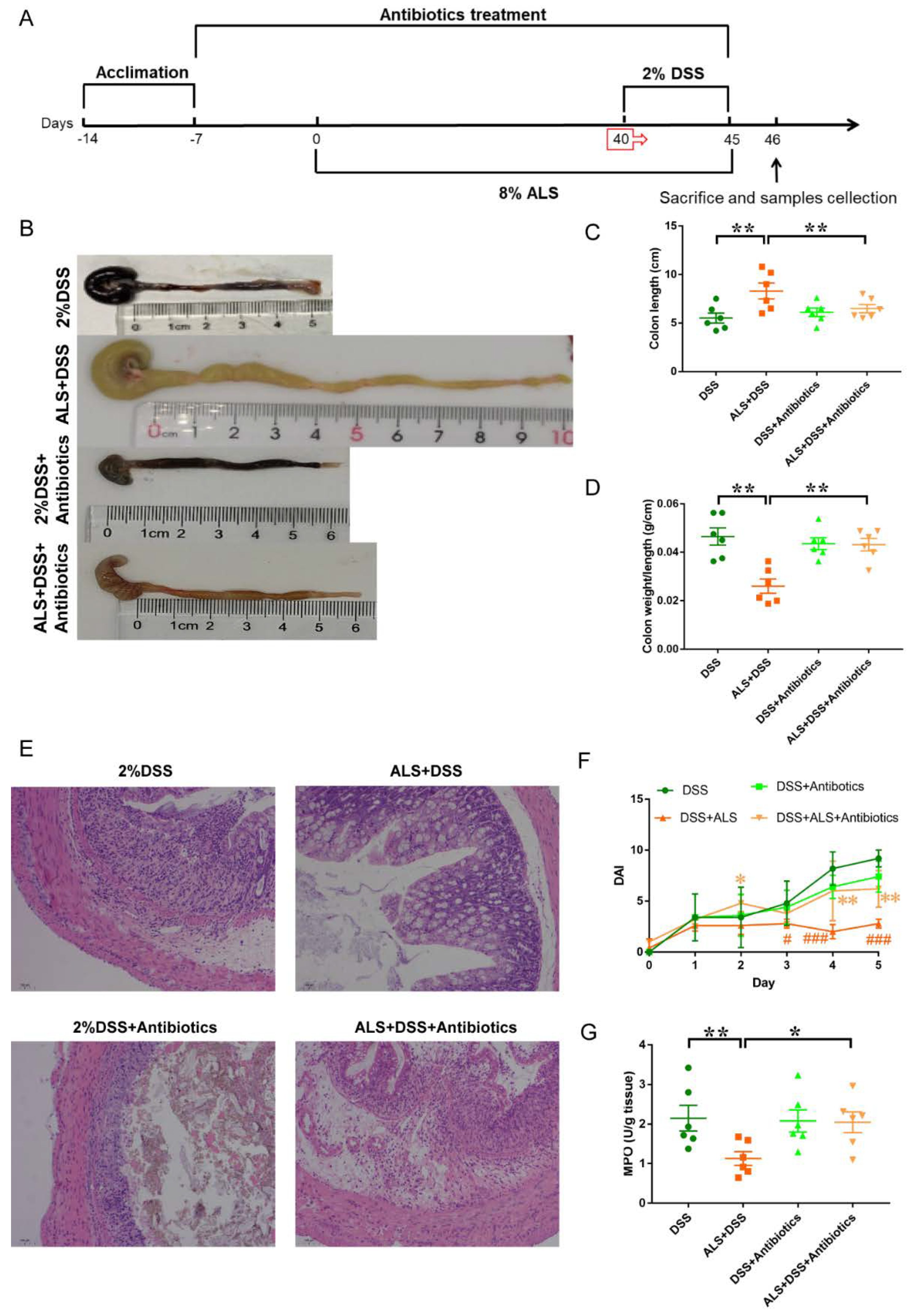
3.7. Microbiota Ablation with Antibiotics Reduces the Efficacy of ALS Supplementation in Colitis Impairment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xavier, R.J.; Podolsky, D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007, 448, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Varum, F.; Bravo, R.; Furrer, E.; Bojic, D.; Basit, A.W. Inflammatory bowel disease: Exploring gut pathophysiology for novel therapeutic targets. Transl. Res. 2016, 176, 38–68. [Google Scholar] [CrossRef]
- Kaplan, G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Sartor, R.B. Microbial Influences in Inflammatory Bowel Diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef]
- Ungaro, R.; Mehandru, S.; Allen, P.B.; Peyrin-Biroulet, L.; Colombel, J.-F. Ulcerative colitis. Lancet 2016, 389, 1756–1770. [Google Scholar] [CrossRef]
- Danese, S. New therapies for inflammatory bowel disease: From the bench to the bedside. Gut 2011, 61, 918–932. [Google Scholar] [CrossRef]
- Caprilli, R.; Latella, G.; Frieri, G. Treatment of inflammatory bowel diseases: To heal the wound or to heal the sick? J. Crohns Coliti 2012, 6, 621–625. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2013, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Franzosa, E.A.; Sirota-Madi, A.; Avila, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2018, 4, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Jie, Z.; Cui, B.; Wang, H.; Feng, Q.; Zou, Y.; Zhang, X.; Yang, H.; Wang, J.; Zhang, F.; et al. Fecal microbiota transplantation results in bacterial strain displacement in patients with inflammatory bowel diseases. FEBS Open Bio 2019, 10, 41–55. [Google Scholar] [CrossRef]
- Jang, Y.J.; Kim, W.-K.; Han, D.H.; Lee, K.; Ko, G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes. 2019, 10, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jiao, Y.; Lu, H.; Shu, X.; Chen, Q. Chemical characterization, antioxidant properties and anticancer activity of exopolysaccharides from Floccularia luteovirens. Carbohydr. Polym. 2020, 229, 115432. [Google Scholar] [CrossRef]
- Chen, C.; Shao, Y.; Tao, Y.; Wen, X. Optimization of dynamic microwave-assisted extraction of Armillaria poly-saccharides using RSM, and their biological activity. LWT-Food Sci. Technol. 2015, 64, 7. [Google Scholar] [CrossRef]
- Feng, K.; Liu, Q.; Ng, T.; Liu, H.; Li, J.; Chen, G.; Sheng, H.; Xie, Z.; Wang, H. Isolation and characterization of a novel lectin from the mushroom Armillaria luteo-virens. Biochem. Biophys. Res. Commun. 2006, 345, 1573–1578. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, J.; Hou, L.; Gao, Y.; Sun, J.; Zhang, N.; Fan, B.; Wang, F. The Small Molecule Fractions of Floccularia luteovirens Induce Apoptosis of NSCLC Cells through Activating Caspase-3 Activity. Int. J. Mol. Sci. 2021, 22, 10609. [Google Scholar] [CrossRef]
- Gao, X.; Cao, Q.; Cheng, Y.; Zhao, D.; Wang, Z.; Yang, H.; Wu, Q.; You, L.; Wang, Y.; Lin, Y.; et al. Chronic stress promotes colitis by disturbing the gut microbiota and triggering immune system response. Proc. Natl. Acad. Sci. USA 2018, 115, E2960–E2969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murthy, S.N.S.; Cooper, H.S.; Shim, H.; Shah, R.S.; Ibrahim, S.A.; Sedergran, D.J. Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin. Dig. Dis. Sci. 1993, 38, 1722–1734. [Google Scholar] [CrossRef]
- Rachmilewitz, D.; Karmeli, F.; Takabayashi, K.; Hayashi, T.; Leider-Trejo, L.; Lee, J.; Leoni, L.M.; Raz, E. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology 2002, 122, 1428–1441. [Google Scholar] [CrossRef]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Belle, N.M.; Ji, Y.; Herbine, K.; Wei, Y.; Park, J.; Zullo, K.; Hung, L.-Y.; Srivatsa, S.; Young, T.; Oniskey, T.; et al. TFF3 interacts with LINGO2 to regulate EGFR activation for protection against colitis and gastrointestinal helminths. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zobeiri, M.; Momtaz, S.; Parvizi, F.; Tewari, D.; Farzaei, M.H.; Nabavi, S.M. Targeting Mitogen-Activated Protein Kinases by Natural Products: A Novel Therapeutic Approach for Inflammatory Bowel Diseases. Curr. Pharm. Biotechnol. 2020, 21, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef]
- Ding, L.; Yang, L.; Wang, Z.; Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 2015, 5, 135–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadaleta, R.M.; van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.L.; Renooij, W.; Murzilli, S.; Klomp, L.W.J.; Siersema, P.D.; Schipper, M.; Danese, S.; et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef]
- Kong, B.; Wang, L.; Chiang, J.Y.; Zhang, Y.; Klaassen, C.D.; Guo, G.L. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 2012, 56, 1034–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef]
- Nickerson, K.P.; Chanin, R.; McDonald, C. Deregulation of intestinal anti-microbial defense by the dietary additive, maltodextrin. Gut Microbes 2015, 6, 78–83. [Google Scholar] [CrossRef] [Green Version]
- Witkowski, M.; Gagliani, N.; Huber, S. Recipe for IBD: Can we use food to control inflammatory bowel disease? Semin. Immunopathol. 2017, 40, 145–156. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.X.; Lee, J.S.; Campbell, E.; Colgan, S.P. Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc. Natl. Acad. Sci. USA 2020, 117, 11648–11657. [Google Scholar] [CrossRef]
- Lee, M.; Chang, E.B. Inflammatory Bowel Diseases (IBD) and the Microbiome—Searching the Crime Scene for Clues. Gastroenterology 2021, 160, 524–537. [Google Scholar] [CrossRef]
- Van Baarlen, P.; Wells, J.M.; Kleerebezem, M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 2013, 34, 208–215. [Google Scholar] [CrossRef]
- Ahl, D.; Liu, H.; Schreiber, O.; Roos, S.; Phillipson, M.; Holm, L. Lactobacillus reuteriincreases mucus thickness and ameliorates dextran sulphate sodium-induced colitis in mice. Acta Physiol. 2016, 217, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xie, S.; Miao, J.; Li, Y.; Wang, Z.; Wang, M.; Yu, Q. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes 2020, 11, 997–1014. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Paramsothy, S.; Nielsen, S.; Kamm, M.A.; Deshpande, N.P.; Faith, J.J.; Clemente, J.C.; Paramsothy, R.; Walsh, A.J.; Bogaerde, J.V.D.; Samuel, D.; et al. Specific Bacteria and Metabolites Associated With Response to Fecal Microbiota Transplantation in Patients With Ulcerative Colitis. Gastroenterology 2019, 156, 1440–14540.e2. [Google Scholar] [CrossRef] [Green Version]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 2020, 27, 659–670.e5. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, G.; Perino, A.; Yildiz, E.; El Alam, G.; Sleiman, M.B.; Gioiello, A.; Pellicciari, R.; Schoonjans, K. Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 2020, 159, 956–968.e8. [Google Scholar] [CrossRef]
- Baars, A.; Oosting, A.; Knol, J.; Garssen, J.; Van Bergenhenegouwen, J. The Gut Microbiota as a Therapeutic Target in IBD and Metabolic Disease: A Role for the Bile Acid Receptors FXR and TGR5. Microorganisms 2015, 3, 641–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Feng, J.; Li, J.; Yu, Q.; Ji, J.; Wu, J.; Dai, W.; Guo, C. The gut microbiome-bile acid axis in hepatocarcinogenesis. Biomed. Pharmacother. 2020, 133, 111036. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2012, 62, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Citi, S. Intestinal barriers protect against disease. Science 2018, 359, 1097–1098. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, K.M.; Büller, H.A.; Opdam, F.J.; Kim, Y.S.; Einerhand, A.W.; Dekker, J. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterology 1994, 107, 1352–1363. [Google Scholar] [CrossRef]
- Hansson, G.C. Mucus and mucins in diseases of the intestinal and respiratory tracts. J. Intern. Med. 2019, 285, 479–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Liu, J.; Guo, X.; Li, S.; Wang, F.; Wang, M. Armillaria luteo-virens Sacc Ameliorates Dextran Sulfate Sodium Induced Colitis through Modulation of Gut Microbiota and Microbiota-Related Bile Acids. Nutrients 2021, 13, 3926. https://doi.org/10.3390/nu13113926
Zhang N, Liu J, Guo X, Li S, Wang F, Wang M. Armillaria luteo-virens Sacc Ameliorates Dextran Sulfate Sodium Induced Colitis through Modulation of Gut Microbiota and Microbiota-Related Bile Acids. Nutrients. 2021; 13(11):3926. https://doi.org/10.3390/nu13113926
Chicago/Turabian StyleZhang, Nana, Jianlin Liu, Xinxin Guo, Shuying Li, Fengzhong Wang, and Minjie Wang. 2021. "Armillaria luteo-virens Sacc Ameliorates Dextran Sulfate Sodium Induced Colitis through Modulation of Gut Microbiota and Microbiota-Related Bile Acids" Nutrients 13, no. 11: 3926. https://doi.org/10.3390/nu13113926
APA StyleZhang, N., Liu, J., Guo, X., Li, S., Wang, F., & Wang, M. (2021). Armillaria luteo-virens Sacc Ameliorates Dextran Sulfate Sodium Induced Colitis through Modulation of Gut Microbiota and Microbiota-Related Bile Acids. Nutrients, 13(11), 3926. https://doi.org/10.3390/nu13113926





