Pathway Preferential Estrogens Prevent Hepatosteatosis Due to Ovariectomy and High-Fat Diets
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Models and Treatments
2.2. Food Intake and Body Composition
2.3. Liver Histology
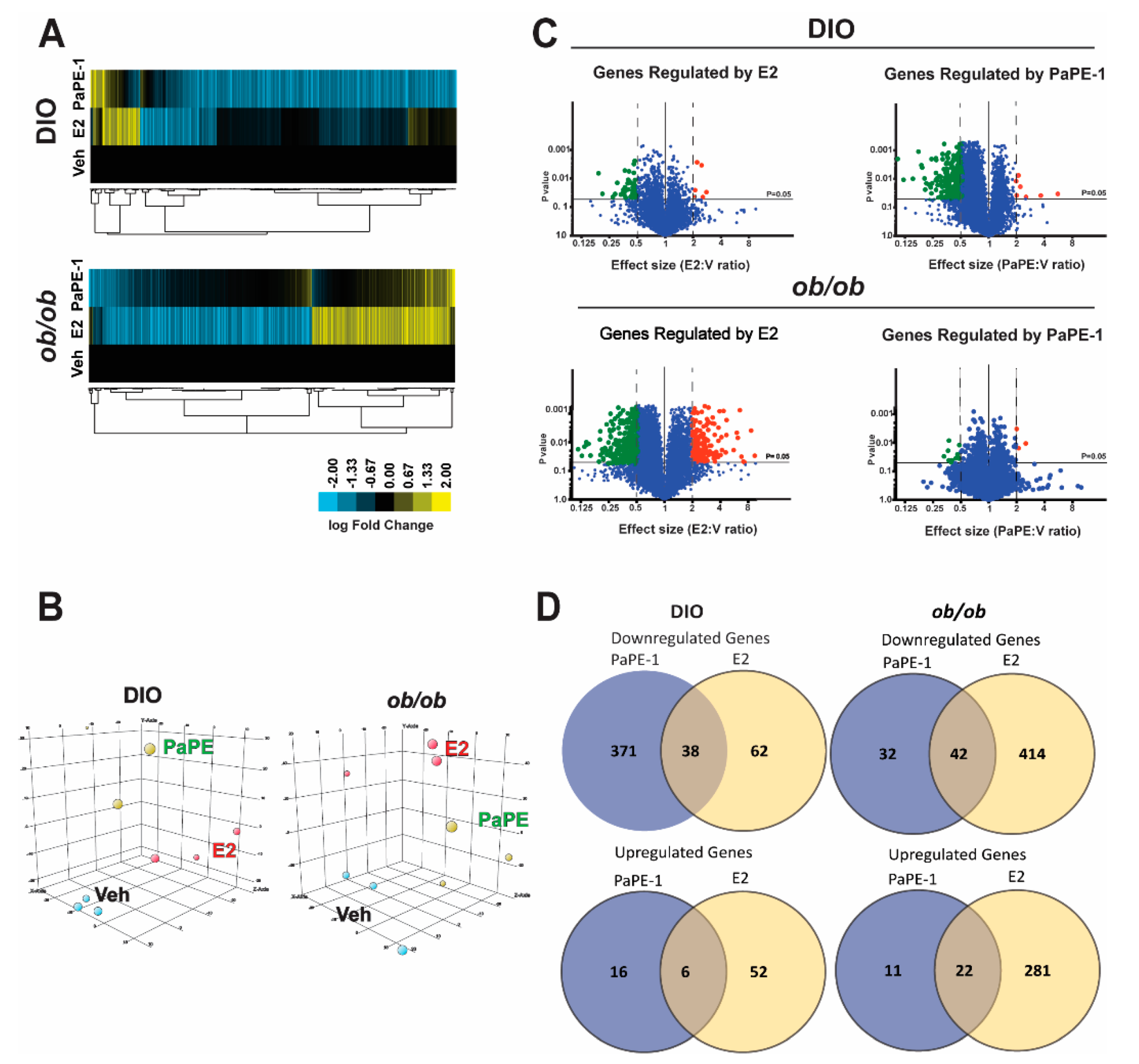
2.4. RNA-Seq and Transcriptional Profiling
2.5. Cell Culture, Ligand Treatments, Reagents
2.6. Real-Time RT-qPCR
2.7. Seahorse Metabolic Profiling Assays
2.8. Neutral Lipid Droplet Staining
2.9. Metabolomics Analysis
2.10. Statistical Analysis
3. Results
3.1. PaPE-1 Was Effective in Decreasing Ovariectomy-Associated Weight Gain and Hepatic Lipid Deposition without Stimulating Reproductive Tissues
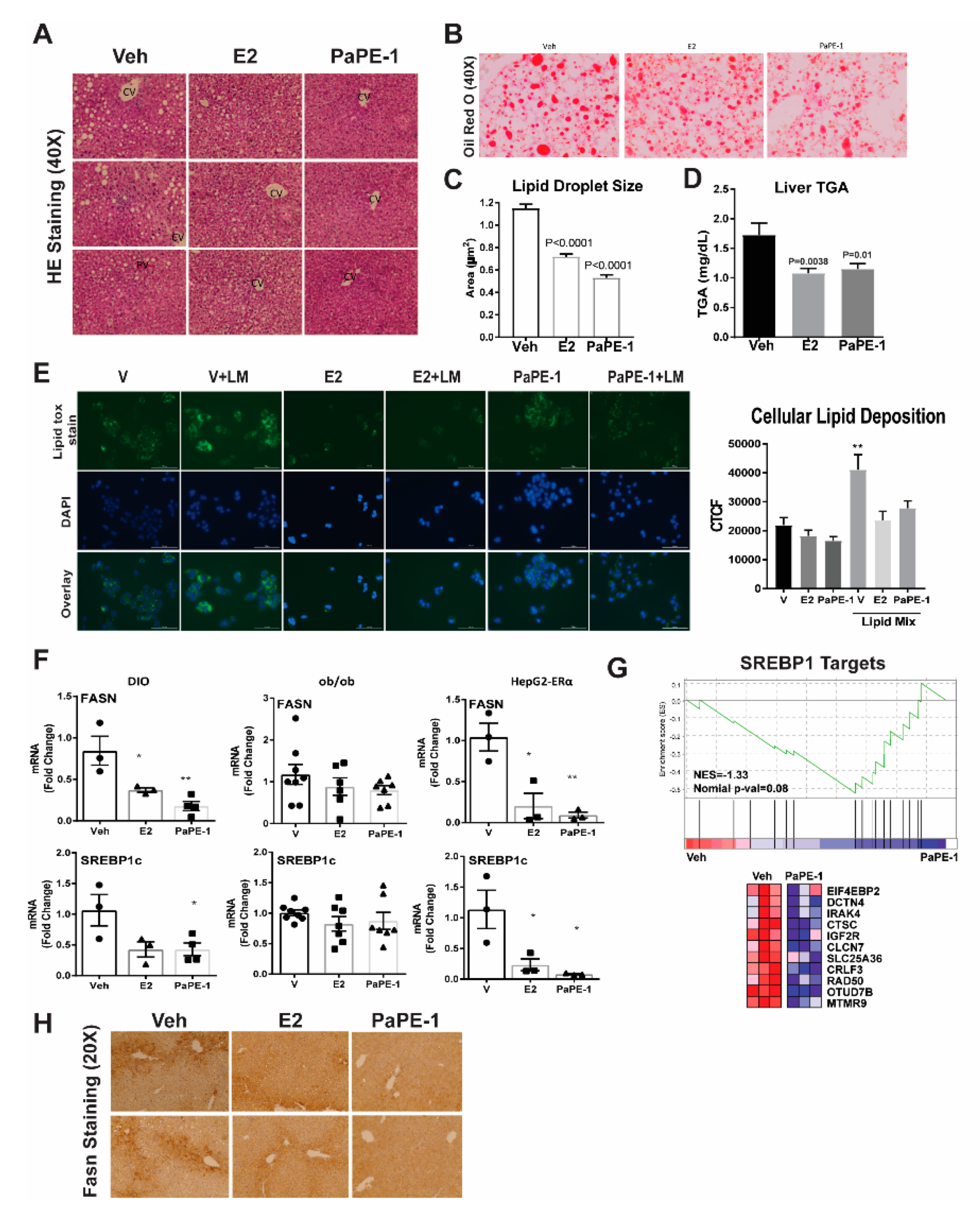
3.2. PaPE-1 Alleviated Hepatic Steatosis in Obese Mice and Decreased Lipid Accumulation in Hepg2-ERα Cells
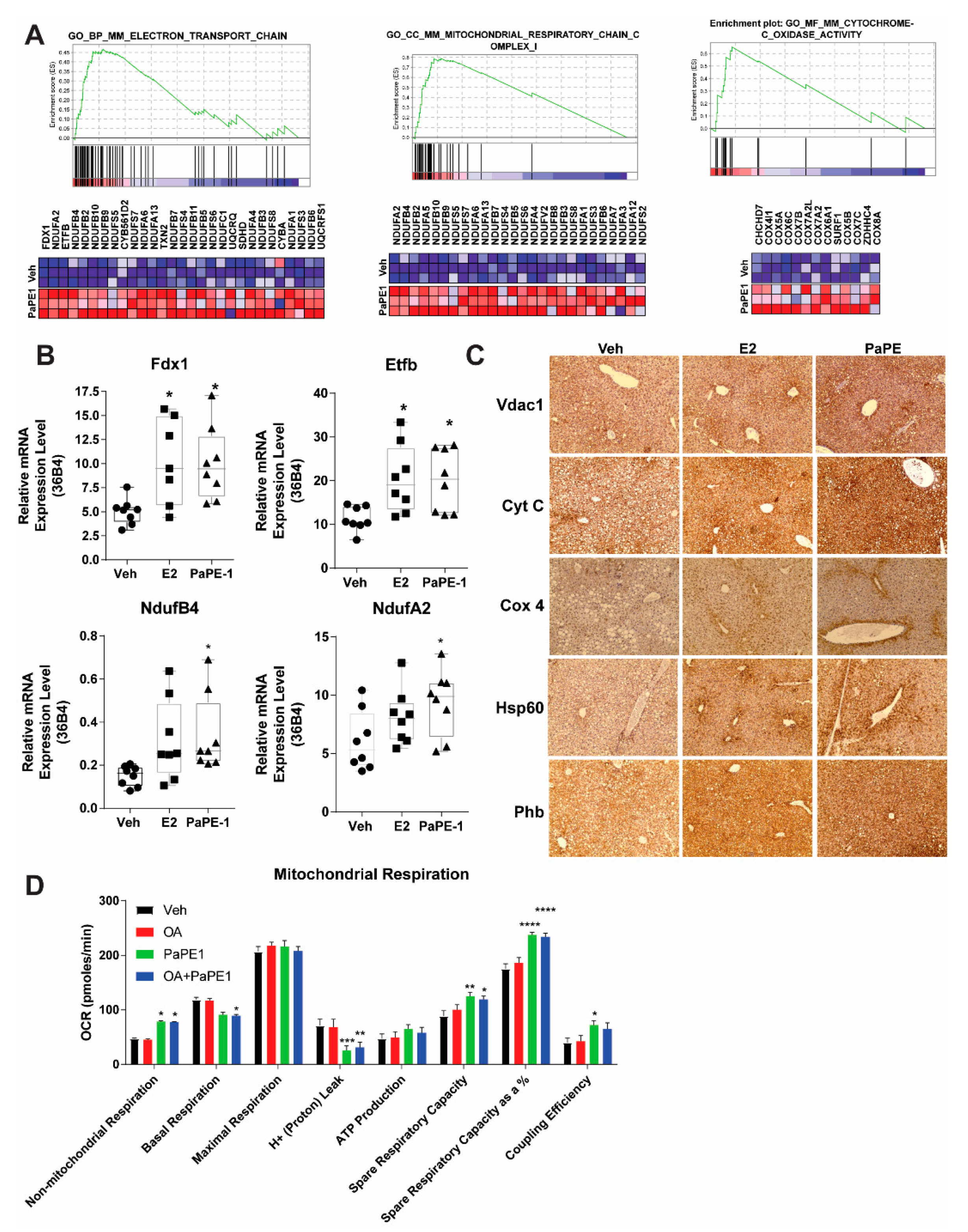
3.3. PaPE-1 Increases Overall Mitochondrial Protein Expression and Activity in Liver
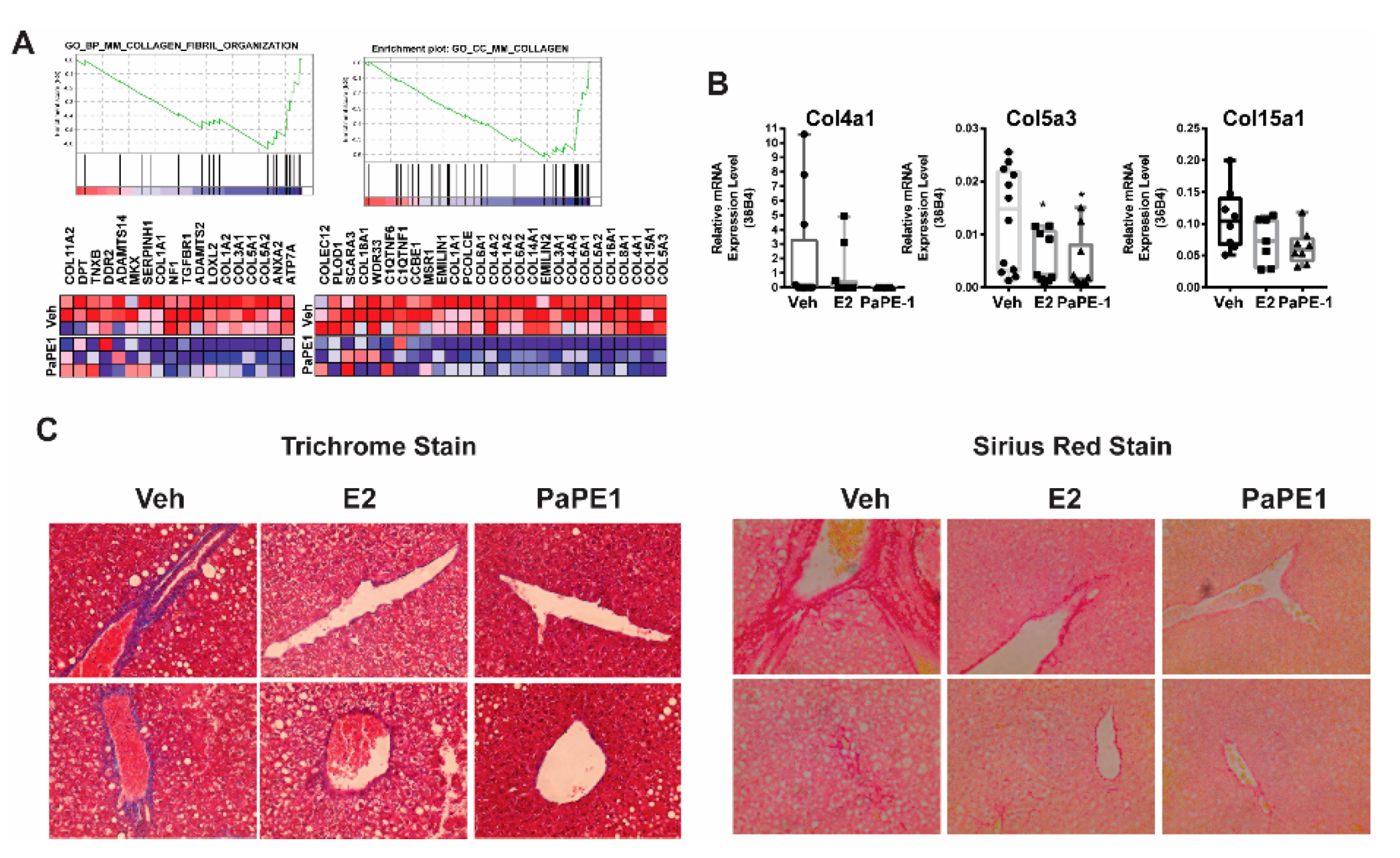
3.4. PaPE-1 Decreased Collagen Deposition in Liver
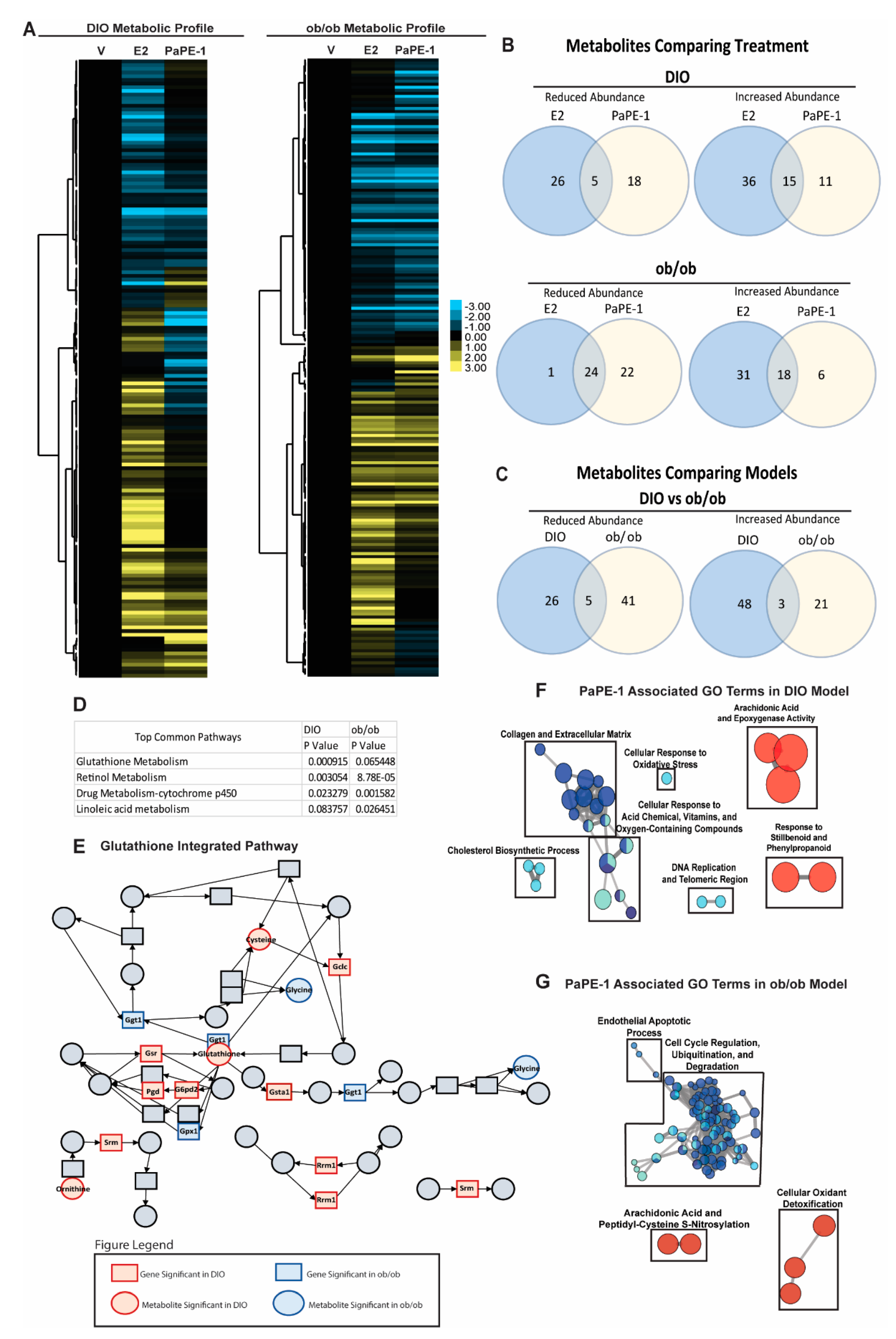
3.5. Metabolomics and Integrated Pathway Analysis Show Pape-1 Reduces Inflammation in the Liver
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henao-Mejia, J.; Elinav, E.; Kin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-mediated dysbiosi regulates progression of NAFLD and obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Scorletti, E.; Calder, P.C.; Byrne, C.D. Non-alcoholic fatty liver disease and cardiovascular risk: Metabolic aspects and novel treatments. Endocrine 2011, 40, 332–343. [Google Scholar] [CrossRef]
- Loguercio, C.; De Simone, T.; D’Auria, M.V.; de Sio, I.; Federico, A.; Tuccillo, C.; Abbatecola, A.M.; Del Vecchio Blnco, C. Non-alcoholic fatty liver disease: A multicentre clinical study by the Italian Association for the Study of the Liver. Dig. Liver Dis. 2004, 36, 398–405. [Google Scholar] [CrossRef]
- Feldstein, A.E.; Werneburg, N.W.; Canbay, A.; Guicciardi, M.E.; Bronk, S.F.; Rydzewski, R.; Burgart, L.J.; Gores, G.J. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via ysosomal pathway. Hepatology 2004, 40, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P. Steatohepatitis: A tale of two ‘hits’? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Yang, L.; McCall, S.; Huang, J.; Yu, X.X.; Pandey, S.K.; Bhanot, S.; Monia, B.P.; Li, Y.X.; Diehl, A.M. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology 2007, 45, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Grobe, Y.; Ponciano-Rodriguez, G.; Ramos, M.H.; Uribe, M.; Mendez-Sanchez, N. Prevalence of non alcoholic fatty liver disease in premenopausal, postmenopausal and polycystic ovary syndrome women. The role of estrogens. Ann. Heptaology 2010, 9, 402–409. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, M.; Hu, Z.; Shrestha, U.K. Prevalence of nonalcoholic fatty liver disease and its metabollic risk factors in women of different ages and body mass index. Menopause 2014, 22, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F.; Clegg, D.J.; Hevener, A.L. The role of estrogens in control of energy balance and glucose homeostasis. Endocr. Rev. 2013, 34, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, S.; Holmang, A.; Bjorntorp, P. The effects of oestrogen and progesterone on insulin sensitivity in female rats. Acta Physiol. Scand 1993, 149, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.D.; Thomas, M.K.; Williams, J.K.; Zhang, L.; Greaves, K.A.; Cefalu, W.T. Insulin sensitivity and cardiovascular risk factors in ovariectomized monkeys with estradiol alone or combined with nomegestrol acetate. J. Clin. Endocrinol. Metab. 1998, 83, 896–901. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Madak-Erdogan, Z.; Kim, S.H.; Gong, P.; Zhao, Y.C.; Chambliss, K.L.; Carlson, K.E.; Mayne, C.G.; Shaul, P.W.; Korach, K.S.; Katzenellenbogen, J.A.; et al. Design of pathway preferential estrogens that provide beneficial metabolic and vascular effects without stimulating reproductive tissues. Sci. Signal 2016, 9, ra53. [Google Scholar] [CrossRef] [PubMed]
- Madak-Erdogan, Z.; Band, S.; Zhao, Y.C.; Smith, B.P.; Kulkoyluoglu-Cotul, E.; Zuo, Q.; Santaliz Casiano, A.; Wrobel, K.; Rossi, G.; Smith, R.L.; et al. Free Fatty Acids Rewire Cancer Metabolism in Obesity-Associated Breast Cancer via Estrogen Receptor and mTOR Signaling. Cancer Res. 2019, 79, 2494–2510. [Google Scholar] [CrossRef] [PubMed]
- Madak-Erdogan, Z.; Gong, P.; Chen Zhao, Y.; Xu, L.; Wrobel, K.U.; Hartman, J.A.; Wang, M.; Cam, A.; Iwaniec, U.T.; Turner, R.T.; et al. Dietary licorice root supplementation reduces diet-induced weight gain, lipid deeposition, and heptaic steatosis in ovariectomized mice without stimulating reproductive tissues and mammary gland. Mol. Nutr. Food Res. 2015, 60, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.; Strissel, K.; Zhao, K.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef]
- Schleicher, M.; Shepherd, B.R.; Suarez, Y.; Fernandez-Hernando, C.; Yu, J.; Pan, Y.; Acevedo, L.M.; Shadel, G.S.; Sessa, W.C. Prohibitin-1 maintains the angiogenic capacity of endothelial cells by regulating mitochondrial function and senescence. J. Cell Biol. 2008, 180, 101–112. [Google Scholar] [CrossRef]
- Flegal, K.M.; Kruszon-Moran, D.; Carroll, M.D.; Fryar, C.D.; Ogden, C.L. Trends in obesity among adults in the United States, 2005–2014. JAMA 2016, 315, 2284–2291. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Y.; Wang, L.; Li, Z.; Zhang, H.; Wu, J.; Rahman, N.; Guo, Y.; Li, D.; Li, N.; et al. Differential effects of estrogen/androgen on the prevention of nonalcoholic fatty liver disease in the male rat. J. Lipid Res. 2013, 54, 345–357. [Google Scholar] [CrossRef]
- Yang, J.D.; Abdelmalek, M.F.; Pang, H.; Guy, C.D.; Smith, A.D.; Diehl, A.M.; Suzuki, A. Gender and menopause impact severity of fibrosis among patients with nonalcoholic steatohepatitis. Hepatology 2014, 59, 1406–1414. [Google Scholar] [CrossRef]
- Clark, J.M.; Brancati, F.L.; Diehl, A.M. Nonalcoholic Fatty Liver Disease. Gastroenterology 2002, 122, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, R.N.; Fisher, C.D.; Canet, M.J.; Lake, A.D.; Cherrington, N.J. Diversity in antioxidant response enzymes in progressive stages of human nonalcoholic fatty liver disease. Drug Metab. Dispos. 2010, 38, 2293–2301. [Google Scholar] [CrossRef]
- Eve, A.A.; Liu, X.; Wang, Y.; Miller, M.J.; Jeffery, E.H.; Madak-Erdogan, Z. Biomarkers of Broccoli Consumption: Implications for Glutathione Metabolism and Liver Health. Nutrients 2020, 12, 2514. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, I.A.; Farrell, G.C.; Schriemer, R.; Robertson, G.R. Leptin is essential for the hepatic fibrogenic response to chronic liver injury. J. Hepatol. 2002, 37, 206–213. [Google Scholar] [CrossRef]
- Ikejima, K.; Honda, H.; Yoshikawa, M.; Hirose, M.; Kitamura, T.; Takei, S.; Sato, N. Leptin augments inflammatory and profibrogenic responses in the murine liver induced by hepatoxic chemicals. Hepatology 2001, 34, 288–297. [Google Scholar] [CrossRef]
- Ito, M.; Suzuki, J.; Tsujioka, S.; Sasaki, M.; Gomori, A.; Shirakura, T.; Hirose, H.; Ito, M.; Ishihara, A.; Iwaasa HKanatani, A. Longitudinal analysis of murine steatohepatitis model induced by chronic exposure to high-fat diet. Hepatol. Res. 2007, 37, 50–57. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, Q.; Chen, K.L.; Arredondo Eve, A.; Liu, Y.-J.; Kim, S.H.; Katzenellenbogen, B.S.; Katzenellenbogen, J.A.; Madak-Erdogan, Z. Pathway Preferential Estrogens Prevent Hepatosteatosis Due to Ovariectomy and High-Fat Diets. Nutrients 2021, 13, 3334. https://doi.org/10.3390/nu13103334
Zuo Q, Chen KL, Arredondo Eve A, Liu Y-J, Kim SH, Katzenellenbogen BS, Katzenellenbogen JA, Madak-Erdogan Z. Pathway Preferential Estrogens Prevent Hepatosteatosis Due to Ovariectomy and High-Fat Diets. Nutrients. 2021; 13(10):3334. https://doi.org/10.3390/nu13103334
Chicago/Turabian StyleZuo, Qianying, Karen L. Chen, Alicia Arredondo Eve, Yu-Jeh Liu, Sung Hoon Kim, Benita S. Katzenellenbogen, John A. Katzenellenbogen, and Zeynep Madak-Erdogan. 2021. "Pathway Preferential Estrogens Prevent Hepatosteatosis Due to Ovariectomy and High-Fat Diets" Nutrients 13, no. 10: 3334. https://doi.org/10.3390/nu13103334
APA StyleZuo, Q., Chen, K. L., Arredondo Eve, A., Liu, Y.-J., Kim, S. H., Katzenellenbogen, B. S., Katzenellenbogen, J. A., & Madak-Erdogan, Z. (2021). Pathway Preferential Estrogens Prevent Hepatosteatosis Due to Ovariectomy and High-Fat Diets. Nutrients, 13(10), 3334. https://doi.org/10.3390/nu13103334