Vitamin D Receptor (VDR) Gene Polymorphism in Patients Diagnosed with Colorectal Cancer
Abstract
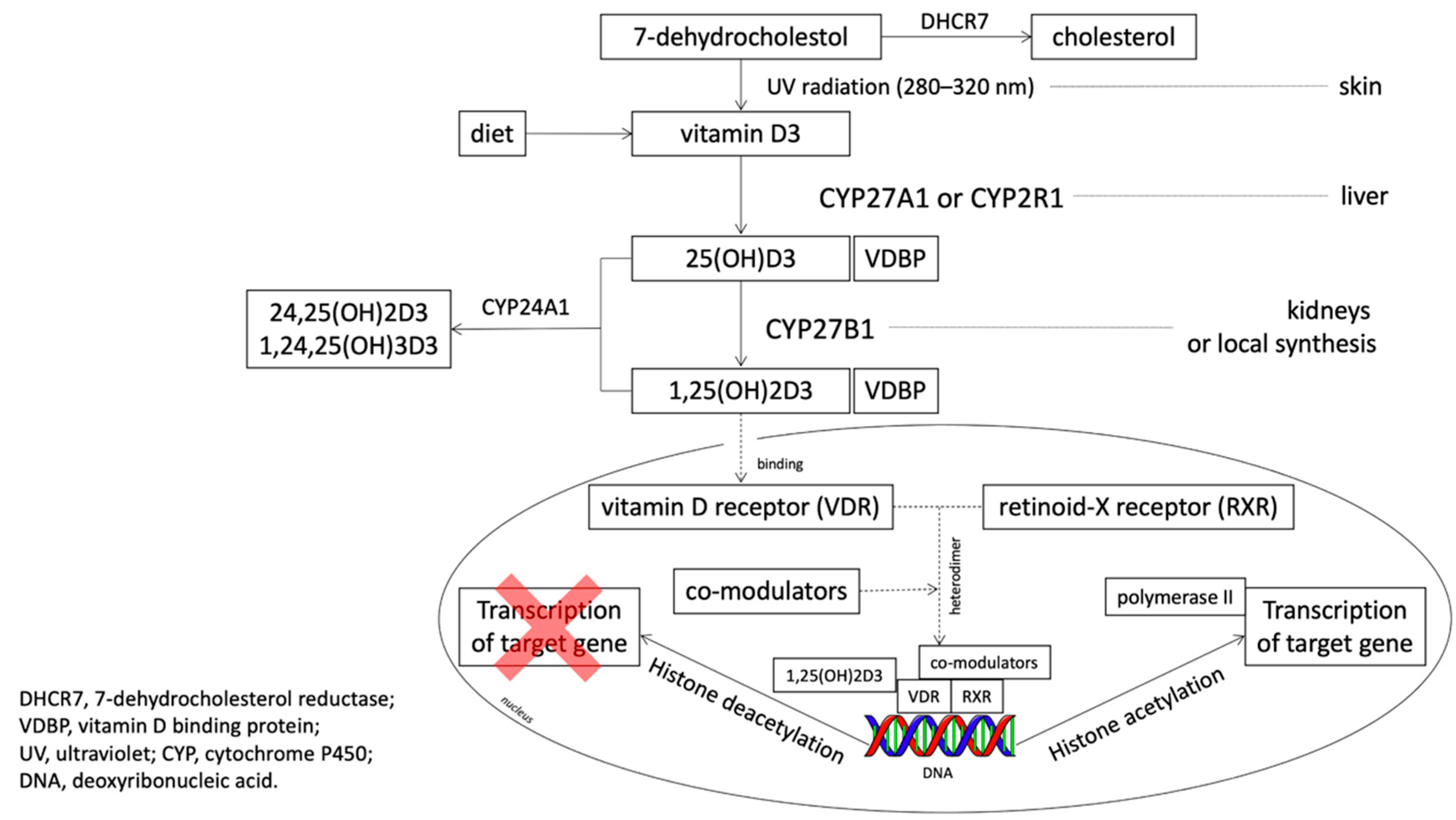
1. Introduction
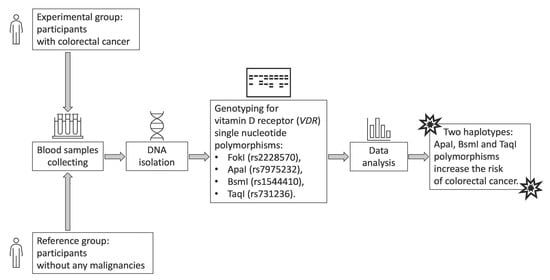
2. Materials and Methods
2.1. Control and Patient Characteristics
2.2. Material Analysis
2.3. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO: Cancer Tomorrow. Available online: http://gco.iarc.fr/tomorrow/home (accessed on 8 November 2020).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Colorectal Cancer Prevention (PDQ®)–Health Professional Version—National Cancer Institute. Available online: https://www.cancer.gov/types/colorectal/hp/colorectal-prevention-pdq (accessed on 8 November 2020).
- Feldman, D.; Krishnan, A.V.; Swami, S.; Giovannucci, E.; Feldman, B.J. The role of vitamin D in reducing cancer risk and progression. Nat. Rev. Cancer 2014, 14, 342–357. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Mayorga, G.; Gómez-López, G.; Barbáchano, A.; Fernández-Barral, A.; Peña, C.; Pisano, D.G.; Cantero, R.; Rojo, F.; Muñoz, A.; Larriba, M.J. Vitamin D receptor expression and associated gene signature in tumour stromal fibroblasts predict clinical outcome in colorectal cancer. Gut 2017, 1449–1462. [Google Scholar] [CrossRef] [PubMed]
- Munemitsu, S.; Albert, I.; Souza, B.; Rubinfeld, B.; Polakis, P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc. Natl. Acad. Sci. USA 1995, 92, 3046–3050. [Google Scholar] [CrossRef]
- Pereira, F.; Larriba, M.J.; Muñoz, A. Vitamin D and colon cancer. Endocr. Relat. Cancer 2012, 19, R51–R71. [Google Scholar] [CrossRef]
- Larriba, M.J.; Muñoz, A. SNAIL vs. vitamin D receptor expression in colon cancer: Therapeutics implications. Br. J. Cancer 2005, 92, 985–989. [Google Scholar] [CrossRef]
- Veldman, C.M.; Cantorna, M.T.; DeLuca, H.F. Expression of 1,25-dihydroxyvitamin D(3) receptor in the immune system. Arch. Biochem. Biophys. 2000, 374, 334–338. [Google Scholar] [CrossRef]
- Zinser, G.M.; Sundberg, J.P.; Welsh, J. Vitamin D(3) receptor ablation sensitizes skin to chemically induced tumorigenesis. Carcinogenesis 2002, 23, 2103–2109. [Google Scholar] [CrossRef]
- Bises, G.; Kállay, E.; Weiland, T.; Wrba, F.; Wenzl, E.; Bonner, E.; Kriwanek, S.; Obrist, P.; Cross, H.S. 25-Hydroxyvitamin D3-1α-hydroxylase Expression in Normal and Malignant Human Colon. J. Histochem. Cytochem. 2004, 52, 985–989. [Google Scholar] [CrossRef]
- VDR vitamin D Receptor [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=7421 (accessed on 8 November 2020).
- Jenkinson, C. The vitamin D metabolome: An update on analysis and function. Cell Biochem. Funct. 2019, 37, 408–423. [Google Scholar] [CrossRef]
- Dou, R.; Ng, K.; Giovannucci, E.L.; Manson, J.E.; Qian, Z.R.; Ogino, S. Vitamin D and colorectal cancer: Molecular, epidemiological and clinical evidence. Br. J. Nutr. 2016, 115, 1643–1660. [Google Scholar] [CrossRef] [PubMed]
- Gandini, S.; Gnagnarella, P.; Serrano, D.; Pasquali, E.; Raimondi, S. Vitamin D Receptor Polymorphisms and Cancer. In Sunlight, Vitamin D and Skin Cancer; Reichrath, J., Ed.; Springer: New York, NY, USA, 2014; Chapter 5; pp. 69–105. [Google Scholar]
- Maalmi, H.; Sassi, F.H.; Berraies, A.; Ammar, J.; Hamzaoui, K.; Hamzaoui, A. Association of vitamin D receptor gene polymorphisms with susceptibility to asthma in Tunisian children: A case control study. Hum. Immunol. 2013, 74, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Mansour, L.; Sedky, M.; AbdelKhader, M.; Sabry, R.; Kamal, M.; El-Sawah, H. The role of vitamin D receptor genes (FOKI and BSMI) polymorphism in osteoporosis. Middle East Fertil. Soc. J. 2010, 15, 79–83. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Shi, Y.Y.; He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15, 97–98. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; He, Z.; Tang, W.; Li, T.; Zeng, Z.; He, L.; Shi, Y. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009, 19, 519–523. [Google Scholar] [CrossRef]
- Smouse, P.E.; Whitehead, M.R.; Peakall, R. An informational diversity framework, illustrated with sexually deceptive orchids in early stages of speciation. Mol. Ecol. Resour. 2015, 15, 1375–1384. [Google Scholar] [CrossRef]
- Colorectal Cancer—Stages. Available online: https://www.cancer.net/cancer-types/colorectal-cancer/stages (accessed on 8 November 2020).
- LDlink. Available online: https://ldlink.nci.nih.gov/?tab=home (accessed on 8 November 2020).
- Lichtenstein, P.; Holm, N.V.; Verkasalo, P.K.; Iliadou, A.; Kaprio, J.; Koskenvuo, M.; Pukkala, E.; Skytthe, A.; Hemminki, K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000, 343, 78–85. [Google Scholar] [CrossRef]
- Shea, M.K.; Benjamin, E.J.; Dupuis, J.; Massaro, J.M.; Jacques, P.F.; D’Agostino, R.B., Sr.; Ordovas, J.M.; O’Donnell, C.J.; Dawson-Hughes, B.; Vasan, R.S.; et al. Genetic and non-genetic correlates of vitamins K and D. Eur. J. Clin. Nutr. 2009, 63, 458–464. [Google Scholar] [CrossRef]
- Uitterlinden, A.G.; Fang, Y.; Van Meurs, J.B.; Pols, H.A.; Van Leeuwen, J.P. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004, 338, 143–156. [Google Scholar] [CrossRef]
- Laczmanska, I.; Laczmanski, L.; Bebenek, M.; Karpinski, P.; Czemarmazowicz, H.; Ramsey, D.; Milewicz, A.; Sasiadek, M.M. Vitamin D receptor gene polymorphisms in relation to the risk of colorectal cancer in the Polish population. Tumour Biol. 2004, 35, 12397–12401. [Google Scholar] [CrossRef] [PubMed]
- Touvier, M.; Chan, D.S.; Lau, R.; Aune, D.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Riboli, E.; Hercberg, S.; Norat, T. Meta-analyses of vitamin D intake, 25-hydroxyvitamin D status, vitamin D receptor polymorphisms, and colorectal cancer risk. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Serrano, D.; Gnagnarella, P.; Raimondi, S.; Gandini, S. Meta-analysis on vitamin D receptor and cancer risk: Focus on the role of TaqI, ApaI, and Cdx2 polymorphisms. Eur. J. Cancer Prev. 2016, 25, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Chen, M.; Hu, X.; Wang, H.; Yang, J.; Zhang, C.; Pan, F.; Sun, G. Associations between VDR gene polymorphisms and colorectal cancer susceptibility: An updated meta-analysis based on 39 case-control studies. Oncotarget 2018, 9, 13068–13076. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Messaritakis, I.; Koulouridi, A.; Sfakianaki, M.; Vogiatzoglou, K.; Gouvas, N.; Athanasakis, E.; Tsiaoussis, J.; Xynos, E.; Mavroudis, D.; Tzardi, M.; et al. The Role of Vitamin D Receptor Gene Polymorphisms in Colorectal Cancer Risk. Cancers 2020, 12, 1379. [Google Scholar] [CrossRef] [PubMed]
- Suksawatamnuay, S.; Sriphoosanaphan, S.; Aumpansub, P.; Aniwan, S.; Thanapirom, K.; Tanasanvimon, S.; Thaimai, P.; Wiangngoen, S.; Ponauthai, Y.; Sumdin, S.; et al. Association between Vitamin D Receptor Single-Nucleotide Polymorphisms and Colorectal Cancer in the Thai Population: A Case-Control Study. BioMed Res. Int. 2020, 2020, 7562958. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, P.P.; Zhai, G.; Bapat, B.; Savas, S.; Woodrow, J.R.; Sharma, I.; Li, Y.; Zhou, X.; Yang, N.; et al. Vitamin D receptor and calcium-sensing receptor polymorphisms and colorectal cancer survival in the Newfoundland population. Br. J. Cancer 2017, 117, 898–906. [Google Scholar] [CrossRef][Green Version]
- Ashmore, J.H.; Gallagher, C.J.; Lesko, S.M.; Muscat, J.E.; Hartman, T.J.; Lazarus, P. No Association Between Vitamin D Intake, VDR Polymorphisms, and Colorectal Cancer in a Population-Based Case-Control Study. Cancer Epidemiol Biomark. Prev. 2015, 24, 1635–1637. [Google Scholar] [CrossRef]
- Al-Ghafari, A.B.; Balamash, K.S.; Al Doghaither, H.A. Relationship between Serum Vitamin D and Calcium Levels and Vitamin D Receptor Gene Polymorphisms in Colorectal Cancer. BioMed Res. Int. 2019, 2019, 8571541. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, Y.J.; Feng, X.L.; Abulimiti, A.; Huang, C.Y.; Luo, H.; Zhang, C.X. Interactions Between Vitamin D and Calcium Intake, Vitamin D Receptor Genetic Polymorphisms, and Colorectal Cancer Risk. Dig. Dis. Sci. 2020. (online ahead of print). [Google Scholar]

| CRC Group (n = 103) | Control Group (n = 109) | p-Value | |
|---|---|---|---|
| Age: years (mean ± SEM) | 71.4 ± 0.8 | 58.6 ± 0.6 | <0.01 |
| Gender, n (%) | |||
| Female | 42 (41) | 59 (54) | 0.052 |
| Male | 61 (59) | 50 (46) | |
| Tumor stage, n (%) | |||
| I | 23 (22.3) | ||
| II | 30 (29.1) | ||
| III | 27 (26.3) | ||
| IV | 23 (22.3) | ||
| Pathological tumor stage (pT), n (%) | |||
| T1 | 0 (0) | ||
| T2 | 21 (20.4) | ||
| T3 | 63 (61.2) | ||
| T4 | 19 (18.4) | ||
| Pathological nodal status (pN), n (%) | |||
| no lymph node metastasis | 49 (47.6) | ||
| lymph node metastasis | 54 (52.4) | ||
| Pathological metastasis status (pM), n (%) | |||
| no distant metastasis | 64 (62.1) | ||
| metastasis to distant organs | 39 (37.9) |
| SNP | Primer Sequence | Restriction Enzyme | Source |
|---|---|---|---|
| FokI | Fok1R: 5-ATGGAAACACCTTGCTTCTTCTCCCTC-3 Fok11F: 5-AGCTGGCCCTGGCACTGACTCtGGCTCT-3 | FastDigest Fok-I | Maalmi et al., 2013, Mansour et al., 2010 [16,17] with own modifications |
| ApaI | ATaq1F: 5-GGATCCTAAATGCACGGAGA-3 ATaq1R: 5-AGGAAAGGGGTTAGGTTGGA-3 | FastDigest Apa-I | Designed with Primer 3 application [18] |
| BsmI | ABsm1F: 5-CGGGGAGTATGAAGGACAAA-3 ABsm1R: 5-CCATCTCTCAGGCTCCAAAG-3 | FastDigest Hha-I | Designed with Primer 3 application [18] |
| TaqI | ATaq1F: 5-GGATCCTAAATGCACGGAGA-3 ATaq1R: 5-AGGAAAGGGGTTAGGTTGGA-3 | FastDigest Taq-I | Designed with Primer 3 application [18] |
| SNP | Pattern for Genotyping |
|---|---|
| FokI | FF homozygote: 267 bp Ff heterozygote: 267, 198, 69 bp ff homozygote: 198, 69 bp PCR product: 267 bp |
| ApaI | AA homozygote: 630 bp Aa heterozygote: 630, 484, 146 bp aa homozygote: 484, 146 bp PCR product: 630 bp |
| BsmI | BB homozygote: 348 bp Bb heterozygote: 348, 243, 105 bp bb homozygote: 243, 105 bp PCR product: 348 bp |
| TaqI | TT homozygote: 425, 205 bp Tt heterozygote: 425, 225, 205, 200 bp tt homozygote: 225, 205, 200 bp PCR product: 630 bp |
| SNP | x2 | p-Value |
|---|---|---|
| FokI | 0.563 | 0.755 |
| ApaI | 0.120 | 0.942 |
| BsmI | 3.218 | 0.200 |
| TaqI | 0.580 | 0.748 |
| Polymorphism | Genotype/ Allele | CRC n (%) | Control n (%) | Crude OR (95% CI) | p-Value | Adjusted OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|
| TT | 30 (29) | 56 (51) | - | - | - | - | |
| TaqI | Tt | 57 (55) | 42 (39) | 0.39 (0.22–0.72) | 0.002 | 0.36 (0.15–0.83) | 0.016 |
| rs731236 | tt | 16 (16) | 11 (10) | 0.37 (0.15–0.89) | 0.027 | 0.31 (0.09–1.09) | 0.068 |
| T | 117 (57) | 154 (71) | - | - | - | - | |
| t | 89 (43) | 64 (29) | 0.55 (0.37–0.82) | 0.003 | 0.50 (0.29–0.88) | 0.017 | |
| Tt + tt | 89 (60) | 64 (36) | - | - | - | - | |
| vs. TT | 60 (40) | 112 (64) | 2.60 (1.66–4.07) | <0.0001 | - | - | |
| AA | 23 (22) | 19 (17) | - | - | - | - | |
| ApaI | Aa | 49 (48) | 55 (50) | 1.36 (0.63–2.79) | 0.40 | 0.81 (0.29–2.25) | 0.68 |
| rs7975232 | aa | 31 (30) | 35 (32) | 1.37 (0.63–2.97) | 0.43 | 0.81 (0.40–3.72) | 0.72 |
| A | 95 (46) | 93 (43) | - | - | - | - | |
| a | 111 (54) | 125 (57) | 1.13 (0.78–1.67) | 0.51 | 1.15 (0.67–1.98) | 0.60 | |
| Aa + aa | 111 (71) | 125 (77) | - | - | - | - | |
| vs. AA | 46 (29) | 38 (23) | 1.36 (0.83–2.25) | 0.22 | - | - | |
| FF | 32 (31) | 43 (39) | - | - | - | - | |
| FokI | Ff | 50 (49) | 48 (44) | 0.71 (0.39–1.31) | 0.28 | 0.70 (0.29–1.65) | 0.41 |
| rs2228570 | ff | 21 (20) | 18 (17) | 0.64 (0.29–1.39) | 0.26 | 0.58 (0.19–1.72) | 0.32 |
| F | 114 (55) | 134 (61) | - | - | - | - | |
| f | 92 (45) | 84 (39) | 0.78 (0.53–1.14) | 0.20 | 0.74 (0.43–1.28) | 0.28 | |
| Ff + ff | 92 (59) | 84 (48) | - | - | - | - | |
| vs. FF | 64 (41) | 86 (52) | 1.47 (0.95–2.28) | 0.08 | - | - | |
| bb | 50 (49) | 38 (35) | - | - | - | - | |
| BsmI | Bb | 40 (39) | 60 (55) | 1.97 (1.10–3.53) | 0.02 | 1.05 (0.30–3.71) | 0.19 |
| rs1544410 | BB | 13 (13) | 11 (10) | 1.11 (0.45–2.76) | 0.82 | 1.73 (0.77–3.91) | 0.93 |
| b | 140 (68) | 136 (62) | - | - | - | - | |
| B | 66 (32) | 82 (38) | 0.78 (0.52–1.17) | 0.23 | 1.20 (0.69–2.11) | 0.52 | |
| bb vs. | 66 (40) | 82 (52) | - | - | - | - | |
| BB + Bb | 100 (60) | 76 (48) | 1.64 (1.05–2.54) | 0.03 | - | - | |
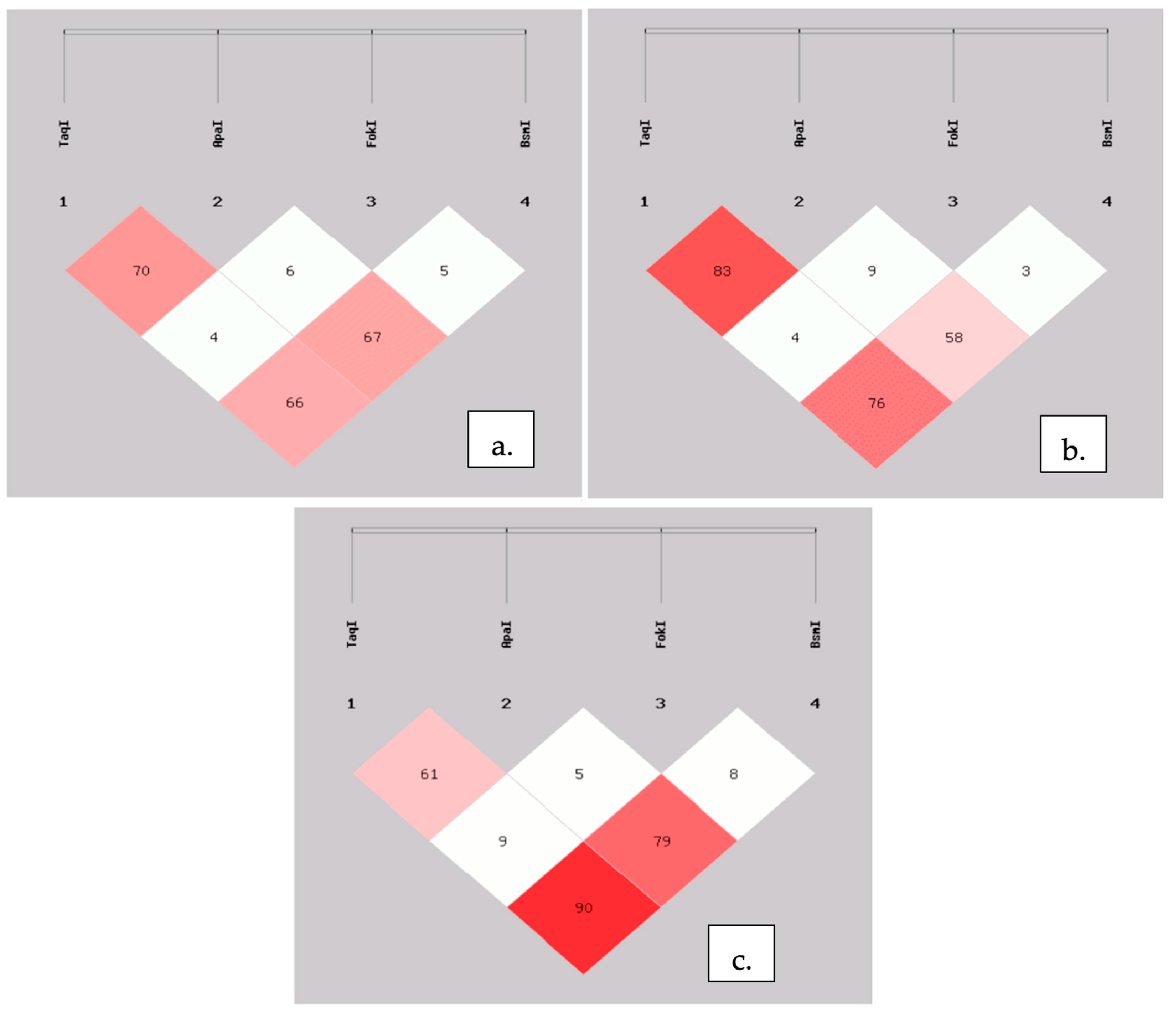
| Haplotype | CRC Frequency (%) | Control Frequency (%) | OR (95% CI) | p-Value |
|---|---|---|---|---|
| TAB | 1.7 | 3.8 | 0.43 (0.12–1.53) | 0.18 |
| TAb | 10.2 | 12.1 | 2.84 (0.46–1.53) | 0.56 |
| TaB | 0.0 | 8.9 | - | - |
| Tab | 44.9 | 45.9 | 0.96 (0.65–1.40) | 0.82 |
| tAB | 27.0 | 24.8 | 1.12 (0.72–1.73) | 0.61 |
| tAb | 7.2 | 2.0 | 3.84 (1.29–11.38) | 0.01 |
| taB | 3.4 | 0.1 | 30.22 (2.81–325.31) | 0.01 |
| tab | 5.6 | 2.4 | 2.39 (0.84–6.82) | 0.09 |
| Genotype | CRC n (%) | Control n (%) | p-Value |
|---|---|---|---|
| TtAaBb | 26 (25.24) | 33 (30.28) | - |
| TtAabb | 11 (10.68) | 1 (0.92) | 0.01 |
| TTAaBb | 1 (0.97) | 8 (7.34) | 0.92 |
| TTAabb | 8 (7.77) | 11 (10.09) | 0.88 |
| ttAABB | 10 (9.71) | 8 (7.34) | 0.39 |
| TTaaBb | 0 | 12 (11.01) | 0.04 |
| TTaabb | 19 (18.45) | 19 (17.43) | 0.56 |
| TtAABb | 6 (5.83) | 3 (2.75) | 0.22 |
| Ttaabb | 8 (7.77) | 1 (0.92) | 0.03 |
| TtAAbb | 2 (1.94) | 1 (0.92) | - |
| TTAABb | 1 (0.97) | 2 (1.83) | - |
| TTAAbb | 1 (0.97) | 3 (2.75) | - |
| TTaaBB | 0 | 1 (0.92) | - |
| TtAaBB | 0 | 1 (0.92) | - |
| TtaaBb | 3 (2.91) | 1 (0.92) | - |
| TtAABB | 1 (0.97) | 1 (0.92) | - |
| ttAAbb | 1 (0.97) | 1 (0.92) | - |
| ttAABb | 1 (0.97) | 0 | - |
| ttAaBb | 2 (1.94) | 1 (0.92) | - |
| ttAaBB | 1 (0.97) | 0 | - |
| ttaaBB | 1 (0.97) | 0 | - |
| ttaabb | 0 | 1 (0.92) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latacz, M.; Rozmus, D.; Fiedorowicz, E.; Snarska, J.; Jarmołowska, B.; Kordulewska, N.; Savelkoul, H.; Cieślińska, A. Vitamin D Receptor (VDR) Gene Polymorphism in Patients Diagnosed with Colorectal Cancer. Nutrients 2021, 13, 200. https://doi.org/10.3390/nu13010200
Latacz M, Rozmus D, Fiedorowicz E, Snarska J, Jarmołowska B, Kordulewska N, Savelkoul H, Cieślińska A. Vitamin D Receptor (VDR) Gene Polymorphism in Patients Diagnosed with Colorectal Cancer. Nutrients. 2021; 13(1):200. https://doi.org/10.3390/nu13010200
Chicago/Turabian StyleLatacz, Maria, Dominika Rozmus, Ewa Fiedorowicz, Jadwiga Snarska, Beata Jarmołowska, Natalia Kordulewska, Huub Savelkoul, and Anna Cieślińska. 2021. "Vitamin D Receptor (VDR) Gene Polymorphism in Patients Diagnosed with Colorectal Cancer" Nutrients 13, no. 1: 200. https://doi.org/10.3390/nu13010200
APA StyleLatacz, M., Rozmus, D., Fiedorowicz, E., Snarska, J., Jarmołowska, B., Kordulewska, N., Savelkoul, H., & Cieślińska, A. (2021). Vitamin D Receptor (VDR) Gene Polymorphism in Patients Diagnosed with Colorectal Cancer. Nutrients, 13(1), 200. https://doi.org/10.3390/nu13010200