Links between Nutrition, Infectious Diseases, and Microbiota: Emerging Technologies and Opportunities for Human-Focused Research
Abstract
1. Introduction
2. Limitations of Traditional In Vivo and In Vitro Models
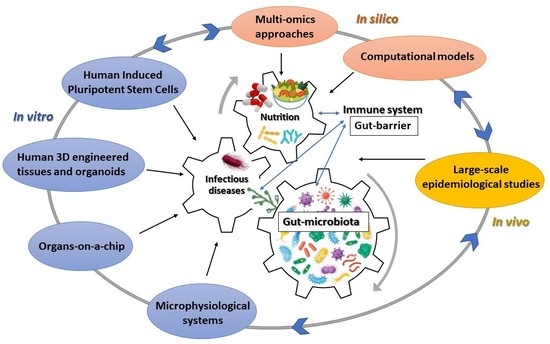
3. Emerging Technologies and Opportunities for a Human-Based Research Approach
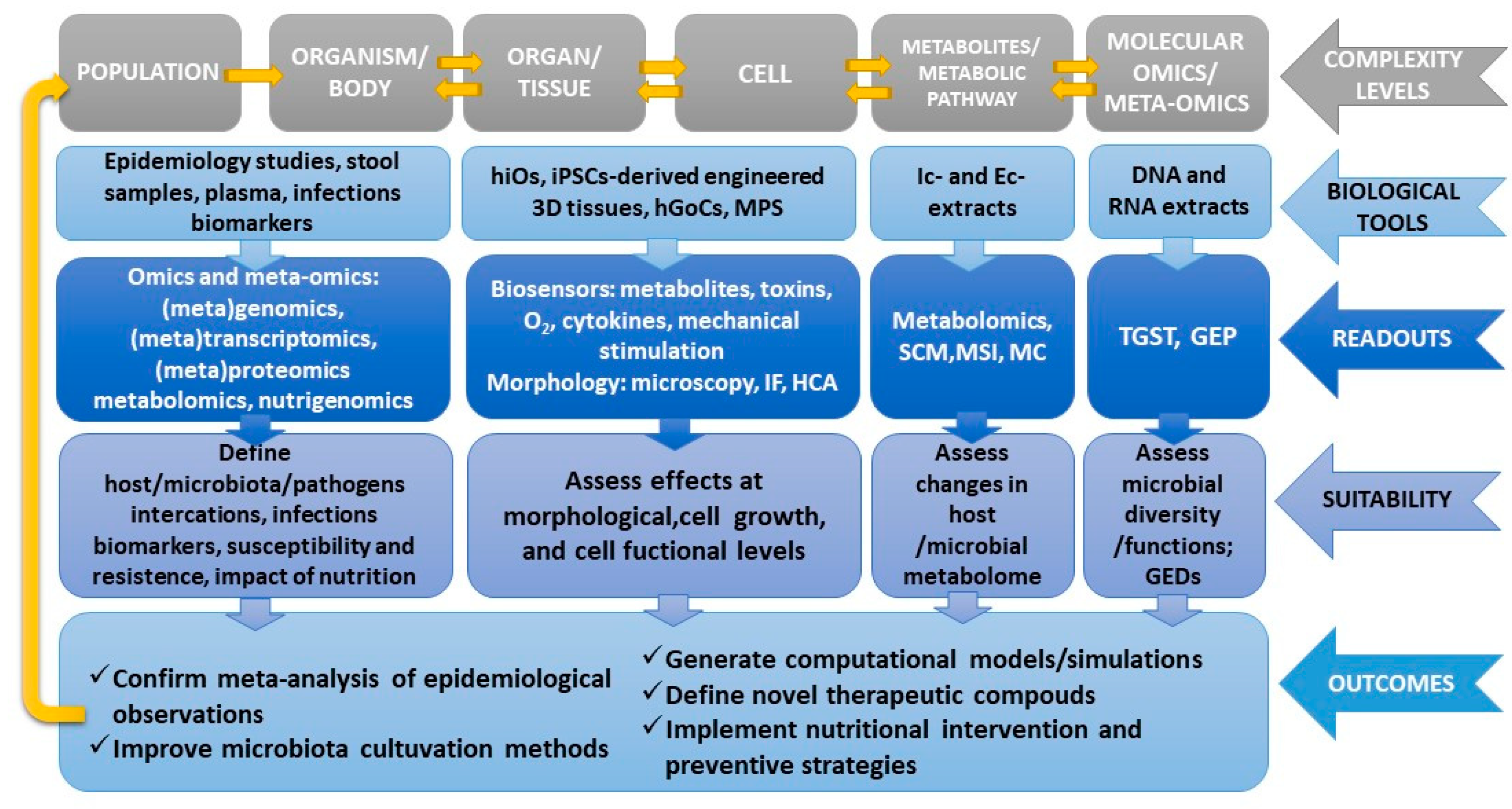
3.1. Multi-Omics Approaches and Computational Models
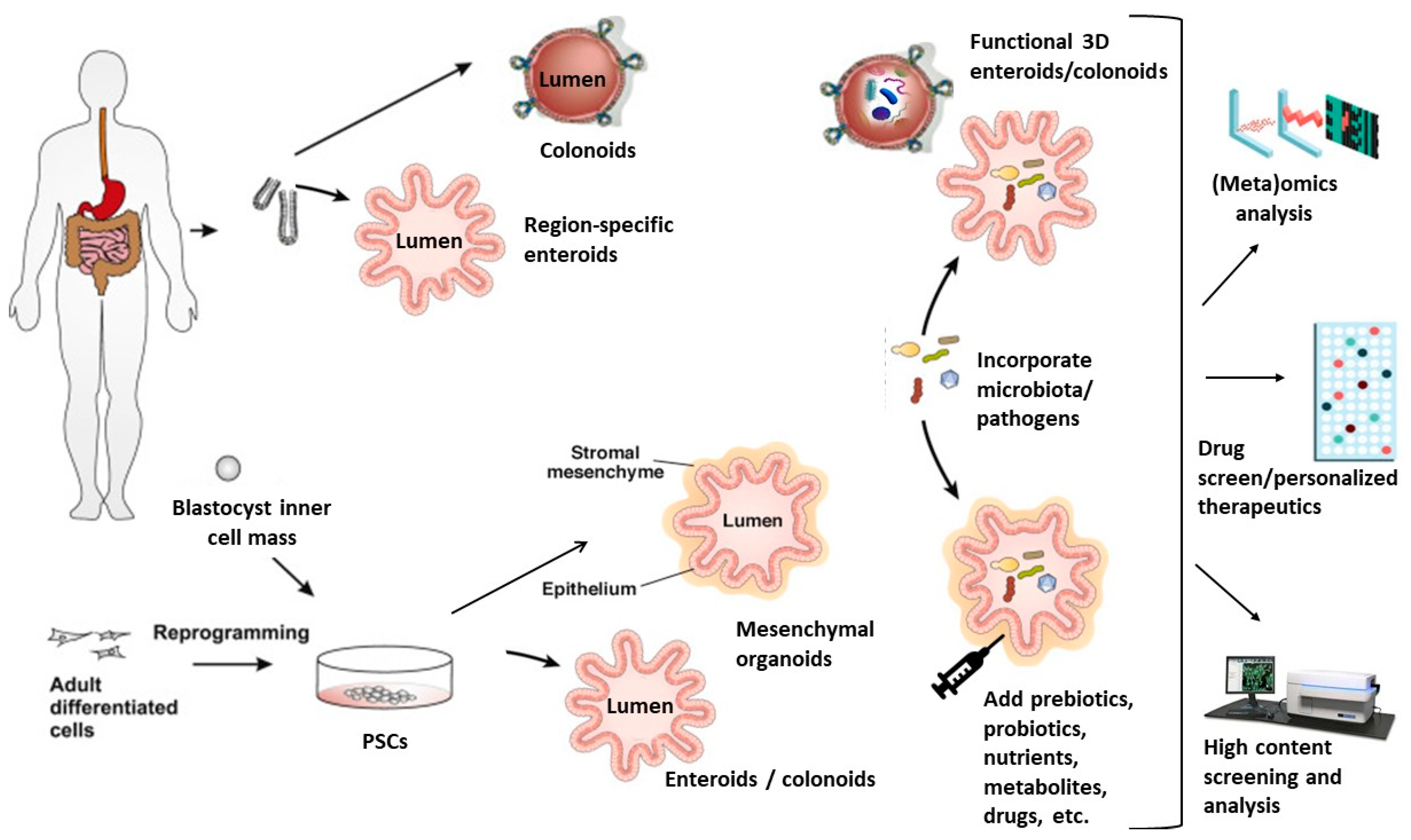
3.2. Human Intestinal Organoids
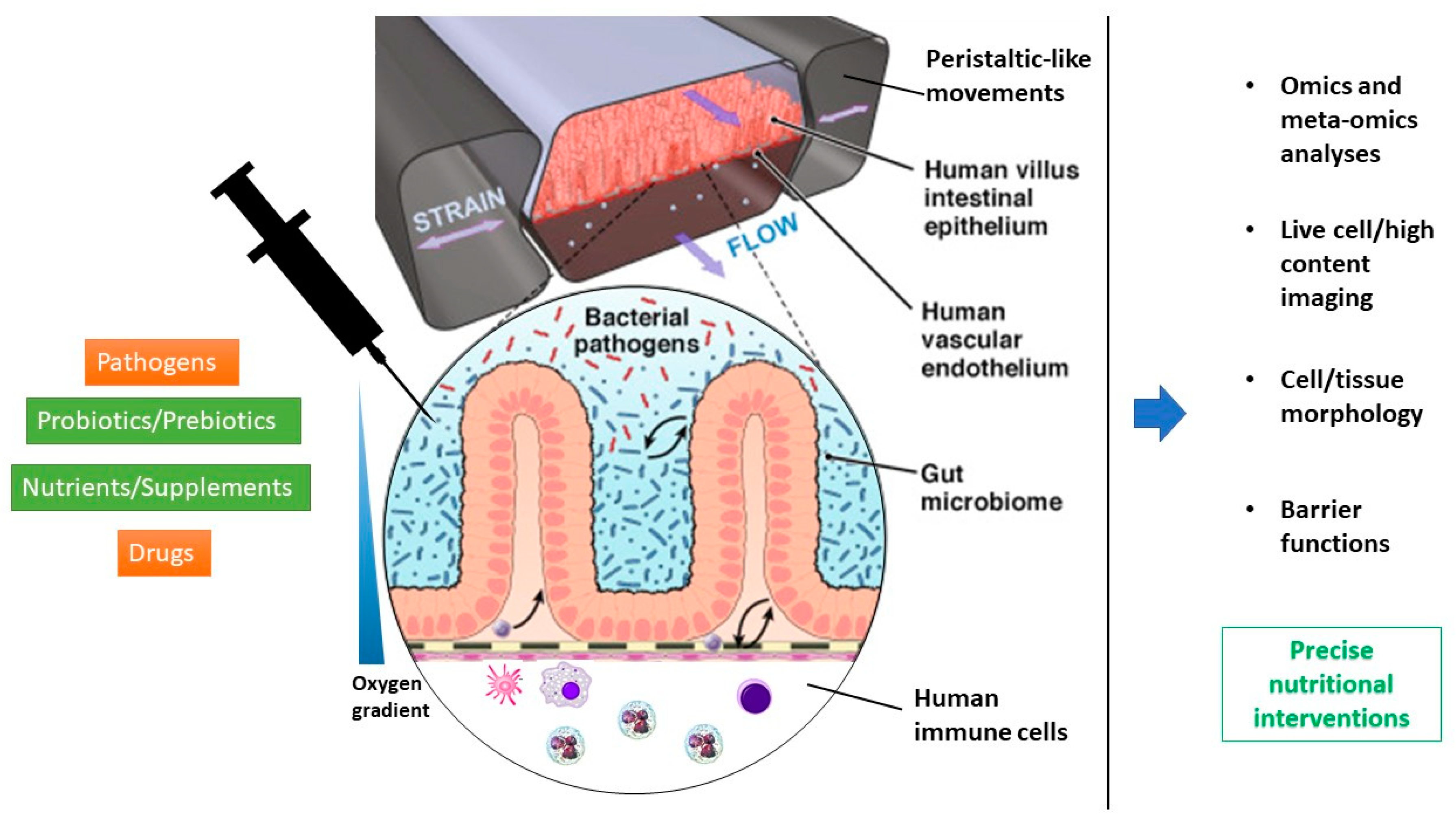
3.3. Organs-On-a-Chip/Microphysiological Systems
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Krawinkel, M.B. Interaction of nutrition and infections globally: An overview. Ann. Nutr. Metab. 2012, 61, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.M.; Morris, L.S.; Marchesi, J.R. The gut microbiome: The role of a virtual organ in the endocrinology of the host. J. Endocrinol. 2013, 218, R37–R47. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Jaiswal, S.K.; Chaudhary, N.; Sharma, V.K. A novel approach for the prediction of species-specific biotransformation of xenobiotic/drug molecules by the human gut microbiota. Sci. Rep. 2017, 7, 9751. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, Y.M.; Tomicic, S.; Lundberg, A.; Böttcher, M.F.; Björkstén, B.; Sverremark-Ekström, E.; Jenmalm, M.C. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2009, 39, 1842–1851. [Google Scholar] [CrossRef]
- Graf, K.; Last, A.; Gratz, R.; Allert, S.; Linde, S.; Westermann, M.; Gröger, M.; Mosig, A.S.; Gresnigt, M.S.; Hube, B. Keeping Candida commensal: How lactobacilli antagonize pathogenicity of Candida albicans in an in vitro gut model. Dis. Model. Mech. 2019, 12. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Pistollato, F.; Sumalla Cano, S.; Elio, I.; Masias Vergara, M.; Giampieri, F.; Battino, M. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr. Rev. 2016, 74, 624–634. [Google Scholar] [CrossRef]
- Bekkering, P.; Jafri, I.; van Overveld, F.J.; Rijkers, G.T. The intricate association between gut microbiota and development of type 1, type 2 and type 3 diabetes. Expert Rev. Clin. Immunol. 2013, 9, 1031–1041. [Google Scholar] [CrossRef]
- Esteve, E.; Ricart, W.; Fernández-Real, J.-M. Gut microbiota interactions with obesity, insulin resistance and type 2 diabetes: Did gut microbiote co-evolve with insulin resistance? Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 483–490. [Google Scholar] [CrossRef]
- Horta-Baas, G.; Romero-Figueroa, M.D.S.; Montiel-Jarquín, A.J.; Pizano-Zárate, M.L.; García-Mena, J.; Ramírez-Durán, N. Intestinal Dysbiosis and Rheumatoid Arthritis: A Link between Gut Microbiota and the Pathogenesis of Rheumatoid Arthritis. J. Immunol. Res. 2017, 2017, 4835189. [Google Scholar] [CrossRef]
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef] [PubMed]
- van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Yooseph, S.; Kirkness, E.F.; Tran, T.M.; Harkins, D.M.; Jones, M.B.; Torralba, M.G.; O’Connell, E.; Nutman, T.B.; Doumbo, S.; Doumbo, O.K.; et al. Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genom. 2015, 16, 631. [Google Scholar] [CrossRef] [PubMed]
- Libertucci, J.; Young, V.B. The role of the microbiota in infectious diseases. Nat. Microbiol. 2019, 4, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Di Segni, A.; BenShoshan, M.; Asaf, R.; Squires, J.E.; Farage Barhom, S.; Glick Saar, E.; Cesarkas, K.; Smollan, G.; Weiss, B.; et al. Fecal microbial characterization of hospitalized patients with suspected infectious diarrhea shows significant dysbiosis. Sci. Rep. 2017, 7, 1088. [Google Scholar] [CrossRef]
- Stecher, B. The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Kampmann, C.; Dicksved, J.; Engstrand, L.; Rautelin, H. Composition of human faecal microbiota in resistance to Campylobacter infection. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016, 22, 61.e61–61.e68. [Google Scholar] [CrossRef]
- Sun, Y.; O’Riordan, M.X.D. Regulation of bacterial pathogenesis by intestinal short-chain Fatty acids. Adv. Appl. Microbiol. 2013, 85, 93–118. [Google Scholar] [CrossRef]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.A.; Zheng, D.; Elinav, E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef]
- Preidis, G.A.; Versalovic, J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology 2009, 136, 2015–2031. [Google Scholar] [CrossRef] [PubMed]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Markowiak, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Segre, J.A. Microbial growth dynamics and human disease. Science 2015, 349, 1058. [Google Scholar] [CrossRef]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, R89. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne pathogens. Aims Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Zhang, P.; Dong, J.; Bai, L.; Qiu, Z. [Clinical analysis of 53 patients with Clostridium botulinum food poisoning]. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2017, 29, 459–464. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef]
- Dadar, M.; Shahali, Y.; Whatmore, A.M. Human brucellosis caused by raw dairy products: A review on the occurrence, major risk factors and prevention. Int. J. Food Microbiol. 2019, 292, 39–47. [Google Scholar] [CrossRef]
- Bandera, A.; De Benedetto, I.; Bozzi, G.; Gori, A. Altered gut microbiome composition in HIV infection: Causes, effects and potential intervention. Curr. Opin. HIV Aids 2018, 13, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Fois, C.A.M.; Le, T.Y.L.; Schindeler, A.; Naficy, S.; McClure, D.D.; Read, M.N.; Valtchev, P.; Khademhosseini, A.; Dehghani, F. Models of the Gut for Analyzing the Impact of Food and Drugs. Adv. Healthc. Mater. 2019, 8, e1900968. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- George, N.S.; Cheung, L.; Luthria, D.L.; Santin, M.; Dawson, H.D.; Bhagwat, A.A.; Smith, A.D. Pomegranate peel extract alters the microbiome in mice and dysbiosis caused by Citrobacter rodentium infection. Food Sci. Nutr. 2019, 7, 2565–2576. [Google Scholar] [CrossRef] [PubMed]
- Greek, R.; Hansen, L.A. Questions regarding the predictive value of one evolved complex adaptive system for a second: Exemplified by the SOD1 mouse. Prog. Biophys. Mol. Biol. 2013, 113, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Greek, R.; Shanks, N. Complex systems, evolution, and animal models. Stud. Hist. Philos. Biol. Biomed. Sci. 2011, 42, 542–544. [Google Scholar] [CrossRef]
- Van Norman, G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is it Time to Rethink Our Current Approach? JACC Basic Transl. Sci. 2019, 4, 845–854. [Google Scholar] [CrossRef]
- Leist, M.; Hartung, T. Inflammatory findings on species extrapolations: Humans are definitely no 70-kg mice. Arch. Toxicol. 2013, 87, 563–567. [Google Scholar] [CrossRef]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C.W. Of Mice and Not Men: Differences between Mouse and Human Immunology. J. Immunol. 2004, 172, 2731. [Google Scholar] [CrossRef]
- Treuting, P.M.; Arends, M.J.; Dintzis, S.M. 12—Lower Gastrointestinal Tract. In Comparative Anatomy and Histology, 2nd ed.; Treuting, P.M., Dintzis, S.M., Montine, K.S., Eds.; Academic Press: San Diego, CA, USA, 2018; pp. 213–228. [Google Scholar] [CrossRef]
- Casteleyn, C.; Rekecki, A.; Van der Aa, A.; Simoens, P.; Van den Broeck, W. Surface area assessment of the murine intestinal tract as a prerequisite for oral dose translation from mouse to man. Lab. Anim. 2010, 44, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, F.; de Vos, W.M. Mouse models for human intestinal microbiota research: A critical evaluation. Cell. Mol. Life Sci. CMLS 2018, 75, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Cunliffe, R.N.; Rose, F.R.A.J.; Keyte, J.; Abberley, L.; Chan, W.C.; Mahida, Y.R. Human defensin 5 is stored in precursor form in normal Paneth cells and is expressed by some villous epithelial cells and by metaplastic Paneth cells in the colon in inflammatory bowel disease. Gut 2001, 48, 176. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Porter, E.; Shen, B.; Lee, S.K.; Wilk, D.; Drazba, J.; Yadav, S.P.; Crabb, J.W.; Ganz, T.; Bevins, C.L. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat. Immunol. 2002, 3, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, A.J.; Selsted, M.E. Paneth cell defensins: Endogenous peptide components of intestinal host defense. FASEB J. 1996, 10, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, E. Digestive strategies of small hindgut fermenters. Anim. Sci. J. 2003, 74, 327–337. [Google Scholar] [CrossRef]
- Hildebrand, F.; Nguyen, T.L.; Brinkman, B.; Yunta, R.G.; Cauwe, B.; Vandenabeele, P.; Liston, A.; Raes, J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013, 14, R4. [Google Scholar] [CrossRef]
- Wang, J.; Linnenbrink, M.; Kunzel, S.; Fernandes, R.; Nadeau, M.J.; Rosenstiel, P.; Baines, J.F. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc. Natl. Acad. Sci. USA 2014, 111, E2703–E2710. [Google Scholar] [CrossRef]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Cheng, Y.L.; Song, L.Q.; Huang, Y.M.; Xiong, Y.W.; Zhang, X.A.; Sun, H.; Zhu, X.P.; Meng, G.X.; Xu, J.G.; Ren, Z.H. Effect of enterohaemorrhagic Escherichia coli O157:H7-specific enterohaemolysin on interleukin-1beta production differs between human and mouse macrophages due to the different sensitivity of NLRP3 activation. Immunology 2015, 145, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Perlman, R.L. Mouse models of human disease: An evolutionary perspective. Evol. Med. Public Health 2016, 2016, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Pamp, S.J.; Hill, J.A.; Surana, N.K.; Edelman, S.M.; Troy, E.B.; Reading, N.C.; Villablanca, E.J.; Wang, S.; Mora, J.R.; et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012, 149, 1578–1593. [Google Scholar] [CrossRef] [PubMed]
- Eppinger, M.; Cebula, T.A. Future perspectives, applications and challenges of genomic epidemiology studies for food-borne pathogens: A case study of Enterohemorrhagic Escherichia coli (EHEC) of the O157:H7 serotype. Gut Microbes 2015, 6, 194–201. [Google Scholar] [CrossRef][Green Version]
- Arrieta, M.-C.; Walter, J.; Finlay, B.B. Human Microbiota-Associated Mice: A Model with Challenges. Cell Host Microbe 2016, 19, 575–578. [Google Scholar] [CrossRef]
- van Nuenen, M.H.; de Ligt, R.A.; Doornbos, R.P.; van der Woude, J.C.; Kuipers, E.J.; Venema, K. The influence of microbial metabolites on human intestinal epithelial cells and macrophages in vitro. FEMS Immunol. Med. Microbiol. 2005, 45, 183–189. [Google Scholar] [CrossRef]
- Parlesak, A.; Haller, D.; Brinz, S.; Baeuerlein, A.; Bode, C. Modulation of cytokine release by differentiated CACO-2 cells in a compartmentalized coculture model with mononuclear leucocytes and nonpathogenic bacteria. Scand. J. Immunol. 2004, 60, 477–485. [Google Scholar] [CrossRef]
- Toki, S.; Kagaya, S.; Shinohara, M.; Wakiguchi, H.; Matsumoto, T.; Takahata, Y.; Morimatsu, F.; Saito, H.; Matsumoto, K. Lactobacillus rhamnosus GG and Lactobacillus casei suppress Escherichia coli-induced chemokine expression in intestinal epithelial cells. Int. Arch. Allergy Immunol. 2009, 148, 45–58. [Google Scholar] [CrossRef]
- Furrie, E.; Macfarlane, S.; Thomson, G.; Macfarlane, G.T. Toll-like receptors-2, -3 and -4 expression patterns on human colon and their regulation by mucosal-associated bacteria. Immunology 2005, 115, 565–574. [Google Scholar] [CrossRef]
- Zenhom, M.; Hyder, A.; de Vrese, M.; Heller, K.J.; Roeder, T.; Schrezenmeir, J. Prebiotic oligosaccharides reduce proinflammatory cytokines in intestinal Caco-2 cells via activation of PPARgamma and peptidoglycan recognition protein 3. J. Nutr. 2011, 141, 971–977. [Google Scholar] [CrossRef]
- Altamimi, M.; Abdelhay, O.; Rastall, R.A. Effect of oligosaccharides on the adhesion of gut bacteria to human HT-29 cells. Anaerobe 2016, 39, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Resta-Lenert, S.; Barrett, K.E. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 2003, 52, 988–997. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Bahrami, B.; Macfarlane, S.; Gueimonde, M.; Macfarlane, G.T.; de los Reyes-Gavilan, C.G. Production of immune response mediators by HT-29 intestinal cell-lines in the presence of Bifidobacterium-treated infant microbiota. Benef. Microbes 2015, 6, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, P.; Minieri, M.; Ahluwalia, A. Engineering the Stem Cell Niche and the Differentiative Micro- and Macroenvironment: Technologies and Tools for Applying Biochemical, Physical and Structural Stimuli and Their Effects on Stem Cells. In Stem Cell Engineering: Principles and Applications; Artmann, G.M., Minger, S., Hescheler, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 41–59. [Google Scholar] [CrossRef]
- Pamies, D.; Hartung, T. 21st Century Cell Culture for 21st Century Toxicology. Chem. Res. Toxicol. 2017, 30, 43–52. [Google Scholar] [CrossRef]
- Nandakumar, N.S.; Pugazhendhi, S.; Ramakrishna, B.S. Effects of enteropathogenic bacteria & lactobacilli on chemokine secretion & Toll like receptor gene expression in two human colonic epithelial cell lines. Indian J. Med. Res. 2009, 130, 170–178. [Google Scholar]
- Raja, S.B.; Murali, M.R.; Devaraj, H.; Devaraj, S.N. Differential expression of gastric MUC5AC in colonic epithelial cells: TFF3-wired IL1 β/Akt crosstalk-induced mucosal immune response against Shigella dysenteriae infection. J. Cell Sci. 2012, 125, 703–713. [Google Scholar] [CrossRef]
- Rausell, A.; Muñoz, M.; Martinez, R.; Roger, T.; Telenti, A.; Ciuffi, A. Innate immune defects in HIV permissive cell lines. Retrovirology 2016, 13, 43. [Google Scholar] [CrossRef]
- Kolawole, A.O.; Mirabelli, C.; Hill, D.R.; Svoboda, S.A.; Janowski, A.B.; Passalacqua, K.D.; Rodriguez, B.N.; Dame, M.K.; Freiden, P.; Berger, R.P.; et al. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 2019, 15, e1008057. [Google Scholar] [CrossRef]
- Kolawole, A.O.; Wobus, C.E. Gastrointestinal organoid technology advances studies of enteric virus biology. PLoS Pathog. 2020, 16, e1008212. [Google Scholar] [CrossRef]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human Intestinal Enteroids: A New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef]
- Ginis, I.; Luo, Y.; Miura, T.; Thies, S.; Brandenberger, R.; Gerecht-Nir, S.; Amit, M.; Hoke, A.; Carpenter, M.K.; Itskovitz-Eldor, J.; et al. Differences between human and mouse embryonic stem cells. Dev. Biol. 2004, 269, 360–380. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.L.; Karlsson-Sjöberg, J.M.T.; Pütsep, K.L.A. CRS-peptides: Unique defense peptides of mouse Paneth cells. Mucosal Immunol. 2012, 5, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, V.; White, J.H. Species-specific regulation of innate immunity by vitamin D signaling. J. Steroid Biochem. Mol. Biol. 2016, 164, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Park, G.S.; Park, M.H.; Shin, W.; Zhao, C.; Sheikh, S.; Oh, S.J.; Kim, H.J. Emulating Host-Microbiome Ecosystem of Human Gastrointestinal Tract in Vitro. Stem Cell Rev. Rep. 2017, 13, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A. Lost in translation: The valley of death across preclinical and clinical divide–identification of problems and overcoming obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
- Hartung, T. Look back in anger—What clinical studies tell us about preclinical work. Altex 2013, 30, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A. Efficacy and safety of new medicines: A human focus. Cell Tissue Bank. 2011, 12, 3–5. [Google Scholar] [CrossRef]
- Herrmann, K.; Pistollato, F.; Stephens, M.L. Beyond the 3Rs: Expanding the use of human-relevant replacement methods in biomedical research. Altex 2019, 36, 343–352. [Google Scholar] [CrossRef]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis. Evol. Bioinform. Online 2016, 12, 5–16. [Google Scholar] [CrossRef]
- Schloissnig, S.; Arumugam, M.; Sunagawa, S.; Mitreva, M.; Tap, J.; Zhu, A.; Waller, A.; Mende, D.R.; Kultima, J.R.; Martin, J.; et al. Genomic variation landscape of the human gut microbiome. Nature 2013, 493, 45–50. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Huang, K.; Meadow, J.F.; Gevers, D.; Lemon, K.P.; Bohannan, B.J.M.; Huttenhower, C. Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. USA 2015, 112, 2930. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971. [Google Scholar] [CrossRef] [PubMed]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662.e620. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Vázquez-Castellanos, J.F.; Serrano-Villar, S.; Jiménez-Hernández, N.; Soto Del Rio, M.D.; Gayo, S.; Rojo, D.; Ferrer, M.; Barbas, C.; Moreno, S.; Estrada, V.; et al. Interplay between gut microbiota metabolism and inflammation in HIV infection. ISME J. 2018, 12, 1964–1976. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Salipante, S.J.; Kawashima, T.; Rosenthal, C.; Hoogestraat, D.R.; Cummings, L.A.; Sengupta, D.J.; Harkins, T.T.; Cookson, B.T.; Hoffman, N.G. Performance comparison of Illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl. Environ. Microbiol. 2014, 80, 7583–7591. [Google Scholar] [CrossRef] [PubMed]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2019, 9, 2868. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; De Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N.; et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe 2019, 25, 444–453.e443. [Google Scholar] [CrossRef]
- Wu, G.; Zhang, C.; Wu, H.; Wang, R.; Shen, J.; Wang, L.; Zhao, Y.; Pang, X.; Zhang, X.; Zhao, L.; et al. Genomic Microdiversity of Bifidobacterium pseudocatenulatum Underlying Differential Strain-Level Responses to Dietary Carbohydrate Intervention. mBio 2017, 8. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. TIG 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, V.; Jiang, C.; Zhou, W.; Jahanbani, F.; Batzoglou, S.; Snyder, M. Synthetic long-read sequencing reveals intraspecies diversity in the human microbiome. Nat. Biotechnol. 2016, 34, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, A.C.; Xavier, R.J. Dissecting the human microbiome with single-cell genomics. Genome Med. 2017, 9, 56. [Google Scholar] [CrossRef]
- Sadowsky, M.J.; Staley, C.; Heiner, C.; Hall, R.; Kelly, C.R.; Brandt, L.; Khoruts, A. Analysis of gut microbiota—An ever changing landscape. Gut Microbes 2017, 8, 268–275. [Google Scholar] [CrossRef]
- Zhao, J.; Nian, L.; Kwok, L.Y.; Sun, T.; Zhao, J. Reduction in fecal microbiota diversity and short-chain fatty acid producers in Methicillin-resistant Staphylococcus aureus infected individuals as revealed by PacBio single molecule, real-time sequencing technology. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2017, 36, 1463–1472. [Google Scholar] [CrossRef]
- Baumgartner, M.; Bayer, F.; Pfrunder-Cardozo, K.R.; Buckling, A.; Hall, A.R. Resident microbial communities inhibit growth and antibiotic-resistance evolution of Escherichia coli in human gut microbiome samples. PLoS Biol. 2020, 18, e3000465. [Google Scholar] [CrossRef]
- Leggett, R.M.; Alcon-Giner, C.; Heavens, D.; Caim, S.; Brook, T.C.; Kujawska, M.; Martin, S.; Peel, N.; Acford-Palmer, H.; Hoyles, L.; et al. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat. Microbiol. 2020, 5, 430–442. [Google Scholar] [CrossRef]
- Hou, Q.; Zhao, F.; Liu, W.; Lv, R.; Khine, W.W.T.; Han, J.; Sun, Z.; Lee, Y.-K.; Zhang, H. Probiotic-directed modulation of gut microbiota is basal microbiome dependent. Gut Microbes 2020, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Neville, B.A.; Forster, S.C.; Lawley, T.D. Transmission of the gut microbiota: Spreading of health. Nat. Rev. Microbiol. 2017, 15, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Hirata, S.I.; Kunisawa, J. Gut microbiome, metabolome, and allergic diseases. Allergol. Int. Off. J. Jpn. Soc. Allergol. 2017, 66, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Quan, G.; Jiang, X.; Yang, Y.; Ding, X.; Zhang, D.; Wang, X.; Hardwidge, P.R.; Ren, W.; Zhu, G. Effects of Metabolites Derived From Gut Microbiota and Hosts on Pathogens. Front. Cell. Infect. Microbiol. 2018, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.; Barbas, C. Gas chromatography-mass spectrometry (GC-MS)-based metabolomics. Methods Mol. Biol. 2011, 708, 191–204. [Google Scholar] [CrossRef]
- Larive, C.K.; Barding, G.A., Jr.; Dinges, M.M. NMR spectroscopy for metabolomics and metabolic profiling. Anal. Chem. 2015, 87, 133–146. [Google Scholar] [CrossRef]
- Duncan, K.D.; Fyrestam, J.; Lanekoff, I. Advances in mass spectrometry based single-cell metabolomics. Analyst 2019, 144, 782–793. [Google Scholar] [CrossRef]
- Junot, C.; Fenaille, F.; Colsch, B.; Bécher, F. High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass Spectrom. Rev. 2014, 33, 471–500. [Google Scholar] [CrossRef]
- Dunham, S.J.B.; Ellis, J.F.; Li, B.; Sweedler, J.V. Mass Spectrometry Imaging of Complex Microbial Communities. Acc. Chem. Res. 2017, 50, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Wang, K.; Huang, C.; Nice, E.C. Mining the fecal proteome: From biomarkers to personalised medicine. Expert Rev. Proteom. 2017, 14, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, W.; Ning, Z.; Mayne, J.; Mack, D.; Stintzi, A.; Tian, R.; Figeys, D. Deep Metaproteomics Approach for the Study of Human Microbiomes. Anal. Chem. 2017, 89, 9407–9415. [Google Scholar] [CrossRef] [PubMed]
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; de Wouters, T.; Kasendra, M.; Super, M.; et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Efremova, M.; Teichmann, S.A. Computational methods for single-cell omics across modalities. Nat. Methods 2020, 17, 14–17. [Google Scholar] [CrossRef]
- Jiang, D.; Armour, C.R.; Hu, C.; Mei, M.; Tian, C.; Sharpton, T.J.; Jiang, Y. Microbiome Multi-Omics Network Analysis: Statistical Considerations, Limitations, and Opportunities. Front. Genet. 2019, 10, 995. [Google Scholar] [CrossRef]
- Benítez-Páez, A.; Kjølbæk, L.; Gómez del Pulgar, E.M.; Brahe, L.K.; Astrup, A.; Matysik, S.; Schött, H.-F.; Krautbauer, S.; Liebisch, G.; Boberska, J.; et al. A Multi-omics Approach to Unraveling the Microbiome-Mediated Effects of Arabinoxylan Oligosaccharides in Overweight Humans. mSystems 2019, 4, e00209-19. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Tang, Z.-Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-Omic Analysis of the Microbiome and Metabolome in Healthy Subjects Reveals Microbiome-Dependent Relationships Between Diet and Metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.M.; Van de Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Pires, E.S.; Hardoim, C.C.P.; Miranda, K.R.; Secco, D.A.; Lobo, L.A.; de Carvalho, D.P.; Han, J.; Borchers, C.H.; Ferreira, R.B.R.; Salles, J.F.; et al. The Gut Microbiome and Metabolome of Two Riparian Communities in the Amazon. Front. Microbiol. 2019, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Abdelwareth, A.; Sallam, I.E.; El Shorbagi, M.; Jehmlich, N.; Fritz-Wallace, K.; Serena Schäpe, S.; Rolle-Kampczyk, U.; Ehrlich, A.; Wessjohann, L.A.; et al. Metabolomics reveals impact of seven functional foods on metabolic pathways in a gut microbiota model. J. Adv. Res. 2020, 23, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Visconti, A.; Le Roy, C.I.; Rosa, F.; Rossi, N.; Martin, T.C.; Mohney, R.P.; Li, W.; de Rinaldis, E.; Bell, J.T.; Venter, J.C.; et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019, 10, 4505. [Google Scholar] [CrossRef]
- Xiong, W.; Giannone, R.J.; Morowitz, M.J.; Banfield, J.F.; Hettich, R.L. Development of an enhanced metaproteomic approach for deepening the microbiome characterization of the human infant gut. J. Proteome Res. 2015, 14, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Forbester, J.L.; Goulding, D.; Vallier, L.; Hannan, N.; Hale, C.; Pickard, D.; Mukhopadhyay, S.; Dougan, G. Interaction of Salmonella Enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells. Infect. Immun. 2015, 83, 2926. [Google Scholar] [CrossRef]
- Dimitrov, D.V. The Human Gutome: Nutrigenomics of the Host–Microbiome Interactions. Omics A J. Integr. Biol. 2010, 15, 419–430. [Google Scholar] [CrossRef]
- Aruoma, O.I.; Hausman-Cohen, S.; Pizano, J.; Schmidt, M.A.; Minich, D.M.; Joffe, Y.; Brandhorst, S.; Evans, S.J.; Brady, D.M. Personalized Nutrition: Translating the Science of NutriGenomics Into Practice: Proceedings From the 2018 American College of Nutrition Meeting. J. Am. Coll. Nutr. 2019, 38, 287–301. [Google Scholar] [CrossRef]
- Haller, D. Nutrigenomics and IBD: The Intestinal Microbiota at the Cross-road Between Inflammation and Metabolism. J. Clin. Gastroenterol. 2010, 44, S6–S9. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Heinken, A.; Kutt, L.; Ravcheev, D.A.; Bauer, E.; Noronha, A.; Greenhalgh, K.; Jäger, C.; Baginska, J.; Wilmes, P.; et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 2017, 35, 81–89. [Google Scholar] [CrossRef]
- Shoaie, S.; Ghaffari, P.; Kovatcheva-Datchary, P.; Mardinoglu, A.; Sen, P.; Pujos-Guillot, E.; de Wouters, T.; Juste, C.; Rizkalla, S.; Chilloux, J.; et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015, 22, 320–331. [Google Scholar] [CrossRef]
- Noecker, C.; Eng, A.; Srinivasan, S.; Theriot, C.M.; Young, V.B.; Jansson, J.K.; Fredricks, D.N.; Borenstein, E. Metabolic Model-Based Integration of Microbiome Taxonomic and Metabolomic Profiles Elucidates Mechanistic Links between Ecological and Metabolic Variation. mSystems 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.; Kim, S.; Cabatbat, J.J.T.; Jang, S.; Jin, Y.S.; Jung, G.Y.; Chia, N.; Kim, P.J. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat. Commun. 2017, 8, 15393. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, M.; Taylor, B.C.; Debelius, J.W.; Li, W.; Knight, R.; Smarr, L. Using machine learning to identify major shifts in human gut microbiome protein family abundance in disease. In Proceedings of the 2016 IEEE International Conference on Big Data (Big Data), Washington, DC, USA, 5–8 December 2016; pp. 1272–1280. [Google Scholar]
- Eetemadi, A.; Rai, N.; Pereira, B.M.P.; Kim, M.; Schmitz, H.; Tagkopoulos, I. The Computational Diet: A Review of Computational Methods Across Diet, Microbiome, and Health. Front. Microbiol. 2020, 11, 393. [Google Scholar] [CrossRef] [PubMed]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Kim, M.; Tagkopoulos, I. Data integration and predictive modeling methods for multi-omics datasets. Mol. Omics 2018, 14, 8–25. [Google Scholar] [CrossRef]
- Wainberg, M.; Merico, D.; Delong, A.; Frey, B.J. Deep learning in biomedicine. Nat. Biotechnol. 2018, 36, 829. [Google Scholar] [CrossRef]
- Ghosh, S.; Matsuoka, Y.; Asai, Y.; Hsin, K.-Y.; Kitano, H. Software for systems biology: From tools to integrated platforms. Nat. Rev. Genet. 2011, 12, 821–832. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef]
- Lehmann, R.; Lee, C.M.; Shugart, E.C.; Benedetti, M.; Charo, R.A.; Gartner, Z.; Hogan, B.; Knoblich, J.; Nelson, C.M.; Wilson, K.M. Human organoids: A new dimension in cell biology. Mol. Biol. Cell 2019, 30, 1129–1137. [Google Scholar] [CrossRef]
- Korytnikov, R.; Nostro, M.C. Generation of polyhormonal and multipotent pancreatic progenitor lineages from human pluripotent stem cells. Methods 2016, 101, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Fiorotto, R.; Amenduni, M.; Mariotti, V.; Fabris, L.; Spirli, C.; Strazzabosco, M. Liver diseases in the dish: IPSC and organoids as a new approach to modeling liver diseases. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 920–928. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Cata, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef]
- Yoo, J.-H.; Donowitz, M. Intestinal enteroids/organoids: A novel platform for drug discovery in inflammatory bowel diseases. World J. Gastroenterol. 2019, 25, 4125–4147. [Google Scholar] [CrossRef]
- Jung, K.B.; Lee, H.; Son, Y.S.; Lee, M.-O.; Kim, Y.-D.; Oh, S.J.; Kwon, O.; Cho, S.; Cho, H.-S.; Kim, D.-S.; et al. Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nat. Commun. 2018, 9, 3039. [Google Scholar] [CrossRef]
- Zachos, N.C.; Kovbasnjuk, O.; Foulke-Abel, J.; In, J.; Blutt, S.E.; de Jonge, H.R.; Estes, M.K.; Donowitz, M. Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology. J. Biol. Chem. 2016, 291, 3759–3766. [Google Scholar] [CrossRef]
- Mithal, A.; Capilla, A.; Heinze, D.; Berical, A.; Villacorta-Martin, C.; Vedaie, M.; Jacob, A.; Abo, K.; Szymaniak, A.; Peasley, M.; et al. Generation of mesenchyme free intestinal organoids from human induced pluripotent stem cells. Nat. Commun. 2020, 11, 215. [Google Scholar] [CrossRef]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell 2018, 23, 787–793.e786. [Google Scholar] [CrossRef]
- Dedhia, P.H.; Bertaux-Skeirik, N.; Zavros, Y.; Spence, J.R. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology 2016, 150, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.L.; Huang, S.; Opp, J.S.; Nagy, M.S.; Kobayashi, M.; Young, V.B.; Spence, J.R. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 2015, 83, 138–145. [Google Scholar] [CrossRef]
- Pearce, S.C.; Coia, H.G.; Karl, J.P.; Pantoja-Feliciano, I.G.; Zachos, N.C.; Racicot, K. Intestinal in vitro and ex vivo Models to Study Host-Microbiome Interactions and Acute Stressors. Front. Physiol. 2018, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Foulke-Abel, J.; In, J.; Yin, J.; Zachos, N.C.; Kovbasnjuk, O.; Estes, M.K.; de Jonge, H.; Donowitz, M. Human Enteroids as a Model of Upper Small Intestinal Ion Transport Physiology and Pathophysiology. Gastroenterology 2016, 150, 638–649.e638. [Google Scholar] [CrossRef] [PubMed]
- Freire, R.; Ingano, L.; Serena, G.; Cetinbas, M.; Anselmo, A.; Sapone, A.; Sadreyev, R.I.; Fasano, A.; Senger, S. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci. Rep. 2019, 9, 7029. [Google Scholar] [CrossRef] [PubMed]
- Angus, H.C.K.; Butt, A.G.; Schultz, M.; Kemp, R.A. Intestinal Organoids as a Tool for Inflammatory Bowel Disease Research. Front. Med. 2020, 6, 334. [Google Scholar] [CrossRef] [PubMed]
- Michels, B.E.; Mosa, M.H.; Grebbin, B.M.; Yepes, D.; Darvishi, T.; Hausmann, J.; Urlaub, H.; Zeuzem, S.; Kvasnicka, H.M.; Oellerich, T.; et al. Human colon organoids reveal distinct physiologic and oncogenic Wnt responses. J. Exp. Med. 2019, 216, 704–720. [Google Scholar] [CrossRef]
- Fernández-Barral, A.; Costales-Carrera, A.; Buira, S.P.; Jung, P.; Ferrer-Mayorga, G.; Larriba, M.J.; Bustamante-Madrid, P.; Domínguez, O.; Real, F.X.; Guerra-Pastrián, L.; et al. Vitamin D differentially regulates colon stem cells in patient-derived normal and tumor organoids. FEBS J. 2020, 287, 53–72. [Google Scholar] [CrossRef]
- Engel, R.M.; Chan, W.H.; Nickless, D.; Hlavca, S.; Richards, E.; Kerr, G.; Oliva, K.; McMurrick, P.J.; Jardé, T.; Abud, H.E. Patient-Derived Colorectal Cancer Organoids Upregulate Revival Stem Cell Marker Genes following Chemotherapeutic Treatment. J. Clin. Med. 2020, 9, 128. [Google Scholar] [CrossRef]
- Cristobal, A.; van den Toorn, H.W.P.; van de Wetering, M.; Clevers, H.; Heck, A.J.R.; Mohammed, S. Personalized Proteome Profiles of Healthy and Tumor Human Colon Organoids Reveal Both Individual Diversity and Basic Features of Colorectal Cancer. Cell Rep. 2017, 18, 263–274. [Google Scholar] [CrossRef]
- Rubert, J.; Schweiger, P.J.; Mattivi, F.; Tuohy, K.; Jensen, K.B.; Lunardi, A. Intestinal Organoids: A Tool for Modelling Diet–Microbiome–Host Interactions. Trends Endocrinol. Metab. 2020. [Google Scholar] [CrossRef]
- Yin, Y.-B.; de Jonge, H.R.; Wu, X.; Yin, Y.-L. Enteroids for Nutritional Studies. Mol. Nutr. Food Res. 2019, 63, 1801143. [Google Scholar] [CrossRef] [PubMed]
- Schlaermann, P.; Toelle, B.; Berger, H.; Schmidt, S.C.; Glanemann, M.; Ordemann, J.; Bartfeld, S.; Mollenkopf, H.J.; Meyer, T.F. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 2016, 65, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Bijvelds, M.; Dang, W.; Xu, L.; van der Eijk, A.A.; Knipping, K.; Tuysuz, N.; Dekkers, J.F.; Wang, Y.; de Jonge, J.; et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antivir. Res. 2015, 123, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Bartfeld, S. Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev. Biol. 2016, 420, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zhou, D. Organoid and Enteroid Modeling of Salmonella Infection. Front. Cell Infect. Microbiol. 2018, 8, 102. [Google Scholar] [CrossRef]
- Kovbasnjuk, O.; Zachos, N.C.; In, J.; Foulke-Abel, J.; Ettayebi, K.; Hyser, J.M.; Broughman, J.R.; Zeng, X.L.; Middendorp, S.; de Jonge, H.R.; et al. Human enteroids: Preclinical models of non-inflammatory diarrhea. Stem Cell Res. Ther. 2013, 4, S3. [Google Scholar] [CrossRef]
- Karve, S.S.; Pradhan, S.; Ward, D.V.; Weiss, A.A. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS ONE 2017, 12, e0178966. [Google Scholar] [CrossRef]
- In, J.; Foulke-Abel, J.; Zachos, N.C.; Hansen, A.M.; Kaper, J.B.; Bernstein, H.D.; Halushka, M.; Blutt, S.; Estes, M.K.; Donowitz, M.; et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 48–62.e43. [Google Scholar] [CrossRef]
- Blutt, S.E.; Crawford, S.E.; Ramani, S.; Zou, W.Y.; Estes, M.K. Engineered Human Gastrointestinal Cultures to Study the Microbiome and Infectious Diseases. Cell. Mol. Gastroenterol. Hepatol. 2017, 5, 241–251. [Google Scholar] [CrossRef]
- Foulke-Abel, J.; In, J.; Kovbasnjuk, O.; Zachos, N.C.; Ettayebi, K.; Blutt, S.E.; Hyser, J.M.; Zeng, X.L.; Crawford, S.E.; Broughman, J.R.; et al. Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Exp. Biol. Med. 2014, 239, 1124–1134. [Google Scholar] [CrossRef]
- Finkbeiner, S.R.; Zeng, X.L.; Utama, B.; Atmar, R.L.; Shroyer, N.F.; Estes, M.K. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 2012, 3, e00159-12. [Google Scholar] [CrossRef] [PubMed]
- Williamson, I.A.; Arnold, J.W.; Samsa, L.A.; Gaynor, L.; DiSalvo, M.; Cocchiaro, J.L.; Carroll, I.; Azcarate-Peril, M.A.; Rawls, J.F.; Allbritton, N.L.; et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Simon, L.M.; Zeng, X.L.; Blutt, S.E.; Crawford, S.E.; Sastri, N.P.; Karandikar, U.C.; Ajami, N.J.; Zachos, N.C.; Kovbasnjuk, O.; et al. A paradox of transcriptional and functional innate interferon responses of human intestinal enteroids to enteric virus infection. Proc. Natl. Acad. Sci. USA 2017, 114, E570–E579. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.-L.; Qu, L.; et al. Replication of human noroviruses in stem cell–derived human enteroids. Science 2016, 353, 1387. [Google Scholar] [CrossRef]
- Drummond, C.G.; Bolock, A.M.; Ma, C.; Luke, C.J.; Good, M.; Coyne, C.B. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc. Natl. Acad. Sci. USA 2017, 114, 1672. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, F.M.; Santhanam, S.; Kumar, P.; Luo, Q.; Ciorba, M.A.; Fleckenstein, J.M. Blood Group O–Dependent Cellular Responses to Cholera Toxin: Parallel Clinical and Epidemiological Links to Severe Cholera. Am. J. Trop. Med. Hyg. 2016, 95, 440–443. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koestler, B.J.; Ward, C.M.; Fisher, C.R.; Rajan, A.; Maresso, A.W.; Payne, S.M. Human Intestinal Enteroids as a Model System of Shigella Pathogenesis. Infect. Immun. 2019, 87, e00733-18. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Dutta, D.; Schaefer, D.A.; Iakobachvili, N.; Artegiani, B.; Sachs, N.; Boonekamp, K.E.; Bowden, G.; Hendrickx, A.P.A.; Willems, R.J.L.; et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018, 3, 814–823. [Google Scholar] [CrossRef]
- Au-Dutta, D.; Au-Heo, I.; Au-O’Connor, R. Studying Cryptosporidium Infection in 3D Tissue-derived Human Organoid Culture Systems by Microinjection. JoVE 2019, e59610. [Google Scholar] [CrossRef]
- Conlan, J.W.; Bardy, S.L.; KuoLee, R.; Webb, A.; Perry, M.B. Ability of Escherichia coli O157:H7 isolates to colonize the intestinal tract of conventional adult CD1 mice is transient. Can. J. Microbiol. 2001, 47, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.Y.; Chen, Y.X.; Fang, J.Y. 2019 Novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020, 21, 125–126. [Google Scholar] [CrossRef] [PubMed]
- De Weirdt, R.; Crabbé, A.; Roos, S.; Vollenweider, S.; Lacroix, C.; van Pijkeren, J.P.; Britton, R.A.; Sarker, S.; Van de Wiele, T.; Nickerson, C.A. Glycerol supplementation enhances L. reuteri’s protective effect against S. Typhimurium colonization in a 3-D model of colonic epithelium. PLoS ONE 2012, 7, e37116. [Google Scholar] [CrossRef]
- Forbester, J.L.; Hannan, N.; Vallier, L.; Dougan, G. Derivation of Intestinal Organoids from Human Induced Pluripotent Stem Cells for Use as an Infection System. Methods Mol. Biol. 2019, 1576, 157–169. [Google Scholar] [CrossRef]
- Wallach, T.E.; Bayrer, J.R. Intestinal Organoids: New Frontiers in the Study of Intestinal Disease and Physiology. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 180–185. [Google Scholar] [CrossRef]
- Rajan, A.; Vela, L.; Zeng, X.L.; Yu, X.; Shroyer, N.; Blutt, S.E.; Poole, N.M.; Carlin, L.G.; Nataro, J.P.; Estes, M.K.; et al. Novel Segment- and Host-Specific Patterns of Enteroaggregative Escherichia coli Adherence to Human Intestinal Enteroids. mBio 2018, 9. [Google Scholar] [CrossRef]
- Holloway, E.M.; Capeling, M.M.; Spence, J.R. Biologically inspired approaches to enhance human organoid complexity. Development 2019, 146, dev166173. [Google Scholar] [CrossRef]
- Grebenyuk, S.; Ranga, A. Engineering Organoid Vascularization. Front. Bioeng. Biotechnol. 2019, 7, 39. [Google Scholar] [CrossRef]
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 45270. [Google Scholar] [CrossRef]
- Sidar, B.; Jenkins, B.R.; Huang, S.; Spence, J.R.; Walk, S.T.; Wilking, J.N. Long-term flow through human intestinal organoids with the gut organoid flow chip (GOFlowChip). Lab Chip 2019, 19, 3552–3562. [Google Scholar] [CrossRef] [PubMed]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer-Phelps, A.; Chou, D.B.; Tovaglieri, A.; Ferrante, T.C.; Duckworth, T.; Fadel, C.; Frismantas, V.; Sutherland, A.D.; Jalili-Firoozinezhad, S.; Kasendra, M.; et al. Human Colon-on-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 507–526. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Aleman, J.; Arneri, A.; Bersini, S.; Piraino, F.; Shin, S.R.; Dokmeci, M.R.; Khademhosseini, A. From cardiac tissue engineering to heart-on-a-chip: Beating challenges. Biomed. Mater. 2015, 10, 034006. [Google Scholar] [CrossRef]
- Lee, J.; Kim, S. Kidney-on-a-Chip: A New Technology for Predicting Drug Efficacy, Interactions, and Drug-induced Nephrotoxicity. Curr. Drug Metab. 2018, 19, 577–583. [Google Scholar] [CrossRef]
- Jahromi, M.A.M.; Abdoli, A.; Rahmanian, M.; Bardania, H.; Bayandori, M.; Moosavi Basri, S.M.M.; Kalbasi, A.; Aref, A.R.; Karimi, M.; Hamblin, M.R. Microfluidic Brain-on-a-Chip: Perspectives for Mimicking Neural System Disorders. Mol. Neurobiol. 2019, 56, 8489–8512. [Google Scholar] [CrossRef]
- Knowlton, S.; Tasoglu, S. A Bioprinted Liver-on-a-Chip for Drug Screening Applications. Trends Biotechnol. 2016, 34, 681–682. [Google Scholar] [CrossRef]
- de Graaf, M.N.S.; Cochrane, A.; van den Hil, F.E.; Buijsman, W.; van der Meer, A.D.; van den Berg, A.; Mummery, C.L.; Orlova, V.V. Scalable microphysiological system to model three-dimensional blood vessels. APL Bioeng. 2019, 3, 026105. [Google Scholar] [CrossRef]
- Goyal, G.; Long, J.; Levy, O.; Ingber, D.E. Biologically Inspired, iterative engineering of a Human Lymphoid Follicle Chip. J. Immunol. 2018, 200, 120–134. [Google Scholar]
- Poceviciute, R.; Ismagilov, R.F. Human-gut-microbiome on a chip. Nat. Biomed. Eng. 2019, 3, 500–501. [Google Scholar] [CrossRef]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Workman, M.J.; Gleeson, J.P.; Troisi, E.J.; Estrada, H.Q.; Kerns, S.J.; Hinojosa, C.D.; Hamilton, G.A.; Targan, S.R.; Svendsen, C.N.; Barrett, R.J. Enhanced Utilization of Induced Pluripotent Stem Cell-Derived Human Intestinal Organoids Using Microengineered Chips. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 669–677.e662. [Google Scholar] [CrossRef]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. Quant. Biosci. Nano Macro 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- de Haan, P.; Ianovska, M.A.; Mathwig, K.; van Lieshout, G.A.A.; Triantis, V.; Bouwmeester, H.; Verpoorte, E. Digestion-on-a-chip: A continuous-flow modular microsystem recreating enzymatic digestion in the gastrointestinal tract. Lab Chip 2019, 19, 1599–1609. [Google Scholar] [CrossRef]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef]
- Kim, J.; Hegde, M.; Jayaraman, A. Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions. Lab Chip 2010, 10, 43–50. [Google Scholar] [CrossRef]
- Costello, C.M.; Sorna, R.M.; Goh, Y.L.; Cengic, I.; Jain, N.K.; March, J.C. 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol. Pharm. 2014, 11, 2030–2039. [Google Scholar] [CrossRef]
- Costello, C.M.; Hongpeng, J.; Shaffiey, S.; Yu, J.; Jain, N.K.; Hackam, D.; March, J.C. Synthetic small intestinal scaffolds for improved studies of intestinal differentiation. Biotechnol. Bioeng. 2014, 111, 1222–1232. [Google Scholar] [CrossRef]
- Grassart, A.; Malardé, V.; Gobaa, S.; Sartori-Rupp, A.; Kerns, J.; Karalis, K.; Marteyn, B.; Sansonetti, P.; Sauvonnet, N. Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection. Cell Host Microbe 2019, 26, 565. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, M.R.; Isabella, V.M.; Li, N.; Kurtz, C.B. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 2020, 11, 1738. [Google Scholar] [CrossRef] [PubMed]
- Lycke, N. Recent progress in mucosal vaccine development: Potential and limitations. Nat. Rev. Immunol. 2012, 12, 592–605. [Google Scholar] [CrossRef]
- Villenave, R.; Wales, S.Q.; Hamkins-Indik, T.; Papafragkou, E.; Weaver, J.C.; Ferrante, T.C.; Bahinski, A.; Elkins, C.A.; Kulka, M.; Ingber, D.E. Human Gut-On-A-Chip Supports Polarized Infection of Coxsackie B1 Virus In Vitro. PLoS ONE 2017, 12, e0169412. [Google Scholar] [CrossRef]
- Kratz, S.R.A.; Höll, G.; Schuller, P.; Ertl, P.; Rothbauer, M. Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors 2019, 9, 110. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Maurer, M.; Gresnigt, M.S.; Last, A.; Wollny, T.; Berlinghof, F.; Pospich, R.; Cseresnyes, Z.; Medyukhina, A.; Graf, K.; Gröger, M.; et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 2019, 220, 119396. [Google Scholar] [CrossRef]
- Wikswo, J.P. The relevance and potential roles of microphysiological systems in biology and medicine. Exp. Biol. Med. 2014, 239, 1061–1072. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef]
- Raimondi, M.T.; Albani, D.; Giordano, C. An Organ-On-A-Chip Engineered Platform to Study the Microbiota–Gut–Brain Axis in Neurodegeneration. Trends Mol. Med. 2019, 25, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Choe, A.; Ha, S.K.; Choi, I.; Choi, N.; Sung, J.H. Microfluidic Gut-liver chip for reproducing the first pass metabolism. Biomed. Microdevices 2017, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Philips, C.A.; Pande, A.; Shasthry, S.M.; Jamwal, K.D.; Khillan, V.; Chandel, S.S.; Kumar, G.; Sharma, M.K.; Maiwall, R.; Jindal, A.; et al. Healthy Donor Fecal Microbiota Transplantation in Steroid-Ineligible Severe Alcoholic Hepatitis: A Pilot Study. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2017, 15, 600–602. [Google Scholar] [CrossRef] [PubMed]
- Vernetti, L.; Gough, A.; Baetz, N.; Blutt, S.; Broughman, J.R.; Brown, J.A.; Foulke-Abel, J.; Hasan, N.; In, J.; Kelly, E.; et al. Functional Coupling of Human Microphysiology Systems: Intestine, Liver, Kidney Proximal Tubule, Blood-Brain Barrier and Skeletal Muscle. Sci. Rep. 2017, 7, 42296. [Google Scholar] [CrossRef] [PubMed]
- Pound, P.; Ritskes-Hoitinga, M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J. Transl. Med. 2018, 16, 304. [Google Scholar] [CrossRef]
- National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy; The National Academies Press: Washington, DC, USA, 2007; p. 216. [CrossRef]
- Langley, G.R. Considering a new paradigm for Alzheimer’s disease research. Drug Discov. Today 2014, 19, 1114–1124. [Google Scholar] [CrossRef]
- van der Worp, H.B.; Howells, D.W.; Sena, E.S.; Porritt, M.J.; Rewell, S.; O’Collins, V.; Macleod, M.R. Can animal models of disease reliably inform human studies? PLoS Med. 2010, 7, e1000245. [Google Scholar] [CrossRef]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar]
- Begley, C.G.; Ellis, L.M. Drug development: Raise standards for preclinical cancer research. Nature 2012, 483, 531–533. [Google Scholar] [CrossRef]
- Geerts, H. Of mice and men: Bridging the translational disconnect in CNS drug discovery. CNS Drugs 2009, 23, 915–926. [Google Scholar] [CrossRef]
- van de Stolpe, A.; Kauffmann, R.H. Innovative human-specific investigational approaches to autoimmune disease. RSC Adv. 2015, 5, 18451–18463. [Google Scholar] [CrossRef]
- Pistollato, F.; Cavanaugh, S.E.; Chandrasekera, P.C. A Human-Based Integrated Framework for Alzheimer’s Disease Research. J. Alzheimers Dis. 2015, 47, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Archibald, K.; Tsaioun, K.; Kenna, J.G.; Pound, P. Better science for safer medicines: The human imperative. J. R. Soc. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Burrows, S.R. Human immunology: A case for the ascent of non-furry immunology. Immunol. Cell Biol. 2011, 89, 330–331. [Google Scholar] [CrossRef]
- Langley, G.R.; Adcock, I.M.; Busquet, F.; Crofton, K.M.; Csernok, E.; Giese, C.; Heinonen, T.; Herrmann, K.; Hofmann-Apitius, M.; Landesmann, B.; et al. Towards a 21st-century roadmap for biomedical research and drug discovery: Consensus report and recommendations. Drug Discov. Today 2017, 22, 327–339. [Google Scholar] [CrossRef]
- Chandrasekera, P.C.; Pippin, J.J. The human subject: An integrative animal model for 21(st) century heart failure research. Am. J. Transl. Res. 2015, 7, 1636–1647. [Google Scholar]
- Pistollato, F.; Iglesias, R.C.; Ruiz, R.; Aparicio, S.; Crespo, J.; Lopez, L.D.; Manna, P.P.; Giampieri, F.; Battino, M. Nutritional patterns associated with the maintenance of neurocognitive functions and the risk of dementia and Alzheimer’s disease: A focus on human studies. Pharmacol. Res. 2018, 131, 32–43. [Google Scholar] [CrossRef]
- Scientific Committee on Health and Environmental Risks (SCHER); Scientific Committee on Consumer Safety (SCCS); Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR). Addressing the New Challenges for Risk Assessment; European Commission: Brussels, Belgium, 2014. [Google Scholar]
- Collins, F.S. Reengineering translational science: The time is right. Sci. Transl. Med. 2011, 3, 90cm17. [Google Scholar] [CrossRef]
- Ben-Zvi, D.; Melton, D.A. Modeling Human Nutrition Using Human Embryonic Stem Cells. Cell 2015, 161, 12–17. [Google Scholar] [CrossRef][Green Version]
- Parfrey, L.W.; Knight, R. Spatial and temporal variability of the human microbiota. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 8–11. [Google Scholar] [CrossRef]
- Nie, J.; Hashino, E. Organoid technologies meet genome engineering. EMBO Rep. 2017, 18, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Ahluwalia, A. Advances and Current Challenges in Intestinal in vitro Model Engineering: A Digest. Front. Bioeng. Biotechnol. 2019, 7, 144. [Google Scholar] [CrossRef] [PubMed]
| Type of Omic Approach | Type of Model | Description/Major Findings | References |
|---|---|---|---|
| Multi-omics | hGoC | Identification of specific human microbiome metabolites modulating EHEC pathogenesis | [116] |
| Metagenomics/Metabolomics | HITChip/M-SHIME | In-depth microbial characterization of luminal and mucosal gut microbes | [122] |
| Metagenomics/Metabolomics | Human subjects/stool samples | Characterization of the gut microbiome of individuals living in the Amazon showed striking differences in the microbial communities from these two types of populations | [123] |
| Metabolomics | In vitro SIHUMIx | Analysis of the impact of functional food on the microbic metabolic pathways | [124] |
| Multi-omics | Human subjects/stool and plasma samples | Investigation of the interplay between the human gut microbiome and the host metabolism | [125] |
| Meta-proteomics | Human subjects/stool samples | Extensive microbiome comparison between infants and the identification of previously undetected microbial functional categories | [126] |
| Transcriptomics/Metatranscriptomics | hiOs | Exploration of the interaction of Salmonella enterica serovar Typhimurium with hiOs; clear changes in transcriptional signatures were detected | [127] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cassotta, M.; Forbes-Hernández, T.Y.; Calderón Iglesias, R.; Ruiz, R.; Elexpuru Zabaleta, M.; Giampieri, F.; Battino, M. Links between Nutrition, Infectious Diseases, and Microbiota: Emerging Technologies and Opportunities for Human-Focused Research. Nutrients 2020, 12, 1827. https://doi.org/10.3390/nu12061827
Cassotta M, Forbes-Hernández TY, Calderón Iglesias R, Ruiz R, Elexpuru Zabaleta M, Giampieri F, Battino M. Links between Nutrition, Infectious Diseases, and Microbiota: Emerging Technologies and Opportunities for Human-Focused Research. Nutrients. 2020; 12(6):1827. https://doi.org/10.3390/nu12061827
Chicago/Turabian StyleCassotta, Manuela, Tamara Yuliett Forbes-Hernández, Ruben Calderón Iglesias, Roberto Ruiz, Maria Elexpuru Zabaleta, Francesca Giampieri, and Maurizio Battino. 2020. "Links between Nutrition, Infectious Diseases, and Microbiota: Emerging Technologies and Opportunities for Human-Focused Research" Nutrients 12, no. 6: 1827. https://doi.org/10.3390/nu12061827
APA StyleCassotta, M., Forbes-Hernández, T. Y., Calderón Iglesias, R., Ruiz, R., Elexpuru Zabaleta, M., Giampieri, F., & Battino, M. (2020). Links between Nutrition, Infectious Diseases, and Microbiota: Emerging Technologies and Opportunities for Human-Focused Research. Nutrients, 12(6), 1827. https://doi.org/10.3390/nu12061827