Bifidobacterium animalis subsp. lactis 420 for Metabolic Health: Review of the Research
Abstract
1. Introduction
2. B420 and Health Benefits
2.1. Gut Microbiota Composition in Obesity
2.2. Influence of B420 on Weight Management
2.3. Influence of B420 on Glycemia, Lipidemia, Insulin Sensitivity, and Cardiovascular Disease Risk
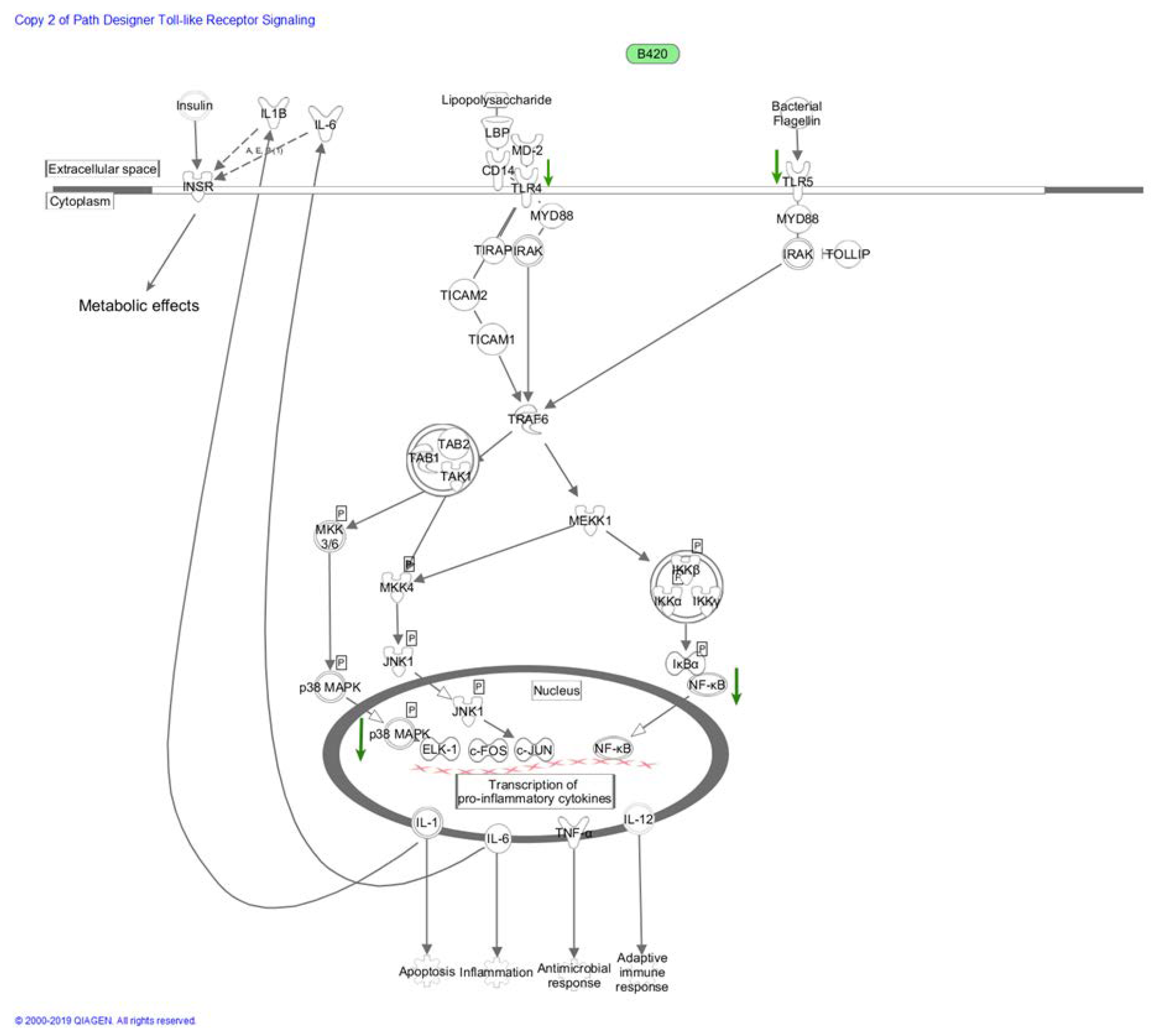
2.4. Metabolic Endotoxemia and Chronic Low-Grade Inflammation in Gut Dysbiosis
2.5. Immunomodulatory Pathways in Metabolic Endotoxemia
2.5.1. Cyclooxygenase and Nitric Oxide Synthase Pathways
2.5.2. NF-κB and MAPK Pathways
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sanchez-Tapia, M.; Tovar, A.R.; Torres, N. Diet as regulator of gut microbiota and its role in health and disease. Arch. Med. Res 2019, 50, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.P.; Antoine, J.M.; Midtvedt, T.; van Hemert, S. Manipulating the gut microbiota to maintain health and treat disease. Microb. Ecol. Health Dis. 2015, 26, 25877. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Chang, C.J.; Lu, C.C.; Martel, J.; Ojcius, D.M.; Ko, Y.F.; Young, J.D.; Lai, H.C. Impact of the gut microbiota, prebiotics, and probiotics on human health and disease. Biomed. J. 2014, 37, 259–268. [Google Scholar] [CrossRef]
- Vandenplas, Y.; Huys, G.; Daube, G. Probiotics: An update. J. Pediatr. 2015, 91, 6–21. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Kirjavainen, P.; Shortt, C.; Salminen, S. Probiotics: Mechanisms and established effects. Int. Dairy J. 1999, 9, 43–52. [Google Scholar] [CrossRef]
- Guarner, F.; Malagelada, J.R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. MMBR 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Turroni, F.; Foroni, E.; Pizzetti, P.; Giubellini, V.; Ribbera, A.; Merusi, P.; Cagnasso, P.; Bizzarri, B.; de’Angelis, G.L.; Shanahan, F.; et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009, 75, 1534–1545. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Stark, P.L.; Lee, A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J. Med. Microbiol. 1982, 15, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; van den Brandt, P.A.; Stobberingh, E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Benno, Y.; Sawada, K.; Mitsuoka, T. The intestinal microflora of infants: Composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 1984, 28, 975–986. [Google Scholar] [CrossRef]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef]
- Meile, L.; Ludwig, W.; Rueger, U.; Gut, C.; Kaufmann, P.; Dasen, G.; Wenger, S.; Teuber, M. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 1997, 20, 57–64. [Google Scholar] [CrossRef]
- Masco, L.; Ventura, M.; Zink, R.; Huys, G.; Swings, J. Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: Reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1137–1143. [Google Scholar] [CrossRef]
- Stahl, B.; Barrangou, R. Complete Genome Sequences of Probiotic Strains Bifidobacterium animalis subsp. lactis B420 and Bi-07. J. Bacteriol. 2012, 62, 584–593. [Google Scholar] [CrossRef]
- Stenman, L.K.; Lehtinen, M.J.; Meland, N.; Christensen, J.E.; Yeung, N.; Saarinen, M.T.; Courtney, M.; Burcelin, R.; Lahdeaho, M.L.; Linros, J.; et al. Probiotic with or without fiber controls body fat mass, associated with serum zonulin, in overweight and obese adults-randomized controlled trial. EBioMedicine 2016, 190–200. [Google Scholar] [CrossRef]
- Mokkala, K.; Laitinen, K.; Roytio, H. Bifidobacterium lactis 420 and fish oil enhance intestinal epithelial integrity in Caco-2 cells. Nutr. Res. (N. Y.) 2016, 36, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Putaala, H.; Salusjarvi, T.; Nordstrom, M.; Saarinen, M.; Ouwehand, A.C.; Bech-Hansen, E.; Rautonen, N. Effect of four probiotic strains and Escherichia coli O157:H7 on tight junction integrity and cyclo-oxygenase expression. Res. Microbiol. 2008, 159, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Stenman, L.K.; Waget, A.; Garret, C.; Klopp, P.; Burcelin, R.; Lahtinen, S. Potential probiotic Bifidobacterium animalis ssp. lactis 420 prevents weight gain and glucose intolerance in diet-induced obese mice. Benef. Microbes 2014, 5, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Surono, I.; Meriluoto, J.; Salminen, S. Indigenous dadih lactic acid bacteria: Cell-Surface properties and interactions with pathogens. J. Food Sci. 2007, 72, M89–M93. [Google Scholar] [CrossRef]
- Amar, J.; Chabo, C.; Waget, A.; Klopp, P.; Vachoux, C.; Bermudez-Humaran, L.G.; Smirnova, N.; Berge, M.; Sulpice, T.; Lahtinen, S.; et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: Molecular mechanisms and probiotic treatment. EMBO Mol. Med. 2011, 3, 559–572. [Google Scholar] [CrossRef]
- Stenman, L.K.; Waget, A.; Garret, C.; Briand, F.; Burcelin, R.; Sulpice, T.; Lahtinen, S. Probiotic B420 and prebiotic polydextrose improve efficacy of antidiabetic drugs in mice. Diabetol. Metab. Syndr. 2015, 7, 75. [Google Scholar] [CrossRef]
- Lyra, A.; Saarinen, M.; Putaala, H.; Olli, K.; Lahtinen, S.J.; Ouwehand, A.C.; Madetoja, M.; Tiihonen, K. Bifidobacterium animalis ssp. lactis 420 protects against indomethacin-induced gastric permeability in rats. Gastroenterol. Res. Pract. 2012, 2012, 615051. [Google Scholar] [CrossRef][Green Version]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Riva, A.; Borgo, F.; Lassandro, C.; Verduci, E.; Morace, G.; Borghi, E.; Berry, D. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. 2017, 19, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Bervoets, L.; Van Hoorenbeeck, K.; Kortleven, I.; Van Noten, C.; Hens, N.; Vael, C.; Goossens, H.; Desager, K.N.; Vankerckhoven, V. Differences in gut microbiota composition between obese and lean children: A cross-sectional study. Gut Pathog. 2013, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-Analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef] [PubMed]
- Maukonen, J.; Simoes, C.; Saarela, M. The currently used commercial DNA-extraction methods give different results of clostridial and actinobacterial populations derived from human fecal samples. FEMS Microbiol. Ecol. 2012, 79, 697–708. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- Simoes, C.D.; Maukonen, J.; Kaprio, J.; Rissanen, A.; Pietilainen, K.H.; Saarela, M. Habitual dietary intake is associated with stool microbiota composition in monozygotic twins. J. Nutr. 2013, 143, 417–423. [Google Scholar] [CrossRef]
- Jumpertz, R.; Le, D.S.; Turnbaugh, P.J.; Trinidad, C.; Bogardus, C.; Gordon, J.I.; Krakoff, J. Energy-Balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011, 94, 58–65. [Google Scholar] [CrossRef]
- Falcinelli, S.; Rodiles, A.; Hatef, A.; Picchietti, S.; Cossignani, L.; Merrifield, D.L.; Unniappan, S.; Carnevali, O. Influence of probiotics administration on gut microbiota core: A review on the effects on appetite control, glucose, and lipid metabolism. J. Clin. Gastroenterol. 2018, S50–S56. [Google Scholar] [CrossRef]
- Hibberd, A.A.; Yde, C.C.; Ziegler, M.L.; Honore, A.H.; Saarinen, M.T.; Lahtinen, S.; Stahl, B.; Jensen, H.M.; Stenman, L.K. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Benef. Microbes 2018, 1–16. [Google Scholar] [CrossRef]
- Heimann, E.; Nyman, M.; Pålbrink, A.-K.; Lindkvist-Petersson, K.; Degerman, E. Branched short-chain fatty acids modulate glucose and lipid metabolism in primary adipocytes. Adipocyte 2016, 5, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Makelainen, H.; Saarinen, M.; Stowell, J.; Rautonen, N.; Ouwehand, A.C. Xylo-Oligosaccharides and lactitol promote the growth of Bifidobacterium lactis and Lactobacillus species in pure cultures. Benef. Microbes 2010, 1, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Rautonen, N.; Heli, P.; Ouwehand, A.; Tiihonen, K.; Zdzislawa, M.; Wouter, K.; Noordman, H. Use of Probiotic Microorganisms for the Treatment and Prevention of Obesity and Related Disorders. U.S. Patent WO2007085970A2, 2 August 2007. [Google Scholar]
- Cryan, J.F.; Dinan, T.G. Mind-Altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- van de Wouw, M.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. Microbiota-Gut-Brain axis: Modulator of host metabolism and appetite. J. Nutr. 2017, 147, 727–745. [Google Scholar] [CrossRef]
- Catrysse, L.; van Loo, G. Inflammation and the metabolic syndrome: The tissue-specific functions of NF-κB. Trends Cell Biol. 2017, 27, 417–429. [Google Scholar] [CrossRef]
- Quon, M.J.; Butte, A.J.; Zarnowski, M.J.; Sesti, G.; Cushman, S.W.; Taylor, S.I. Insulin receptor substrate 1 mediates the stimulatory effect of insulin on GLUT4 translocation in transfected rat adipose cells. J. Biol. Chem. 1994, 269, 27920–27924. [Google Scholar]
- Foufelle, F.; Ferre, P. New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: A role for the transcription factor sterol regulatory element binding protein-1c. Biochem. J. 2002, 366, 377–391. [Google Scholar] [CrossRef]
- Boden, G.; Shulman, G.I. Free fatty acids in obesity and type 2 diabetes: Defining their role in the development of insulin resistance and beta-cell dysfunction. Eur. J. Clin. Investig. 2002, 32 (Suppl. 3), 14–23. [Google Scholar] [CrossRef]
- Weidinger, C.; Ziegler, J.F.; Letizia, M.; Schmidt, F.; Siegmund, B. Adipokines and their role in intestinal inflammation. Front. Immunol. 2018, 9, 1974. [Google Scholar] [CrossRef]
- Unger, R.H. Lipid overload and overflow: Metabolic trauma and the metabolic syndrome. Trends Endocrinol. Metab. TEM 2003, 14, 398–403. [Google Scholar] [CrossRef]
- Unger, R.H. Minireview: Weapons of lean body mass destruction: The role of ectopic lipids in the metabolic syndrome. Endocrinology 2003, 144, 5159–5165. [Google Scholar] [CrossRef] [PubMed]
- McGarry, J.D.; Mannaerts, G.P.; Foster, D.W. A possible role for malonyl-CoA in the regulation of hepatic fatty acid oxidation and ketogenesis. J. Clin. Investig. 1977, 60, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Shiga, T.; Moriyoshi, Y.; Nagahara, H.; Shiratori, K. Nonalcoholic fatty liver is a risk factor for postprandial hyperglycemia, but not for impaired fasting glucose. J. Gastroenterol. 2009, 44, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojarvi, J.; Kootte, R.S.; Bartelsman, J.F.; Dallinga-Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012, 143, 913–916, e917. [Google Scholar] [CrossRef]
- Lam, V.; Su, J.; Koprowski, S.; Hsu, A.; Tweddell, J.S.; Rafiee, P.; Gross, G.J.; Salzman, N.H.; Baker, J.E. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 1727–1735. [Google Scholar] [CrossRef]
- Danilo, C.A.; Constantopoulos, E.; McKee, L.A.; Chen, H.; Regan, J.A.; Lipovka, Y.; Lahtinen, S.; Stenman, L.K.; Nguyen, T.V.; Doyle, K.P.; et al. Bifidobacterium animalis subsp. lactis 420 mitigates the pathological impact of myocardial infarction in the mouse. Benef. Microbes 2017, 8, 257–269. [Google Scholar] [CrossRef]
- Lam, V.; Su, J.; Hsu, A.; Gross, G.J.; Salzman, N.H.; Baker, J.E. Intestinal microbial metabolites are linked to severity of myocardial infarction in rats. PLoS ONE 2016, 11, e0160840. [Google Scholar] [CrossRef]
- Garidou, L.; Pomie, C.; Klopp, P.; Waget, A.; Charpentier, J.; Aloulou, M.; Giry, A.; Serino, M.; Stenman, L.; Lahtinen, S.; et al. The gut microbiota regulates intestinal CD4 T cells expressing RORgammat and controls metabolic disease. Cell Metab. 2015, 22, 100–112. [Google Scholar] [CrossRef]
- Klein, A.; Friedrich, U.; Vogelsang, H.; Jahreis, G. Lactobacillus acidophilus 74-2 and Bifidobacterium animalis subsp lactis DGCC 420 modulate unspecific cellular immune response in healthy adults. Eur. J. Clin. Nutr. 2008, 62, 584–593. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Hooks, K.B.; O’Malley, M.A. Dysbiosis and its discontents. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Esser, N.; Legrand-Poels, S.; Piette, J.; Scheen, A.J.; Paquot, N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 2014, 105, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Burcelin, R. Regulation of metabolism: A cross talk between gut microbiota and its human host. Physiology 2012, 27, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Brun, P.; Castagliuolo, I.; Di Leo, V.; Buda, A.; Pinzani, M.; Palu, G.; Martines, D. Increased intestinal permeability in obese mice: New evidence in the pathogenesis of nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G518–G525. [Google Scholar] [CrossRef] [PubMed]
- Damms-Machado, A.; Louis, S.; Schnitzer, A.; Volynets, V.; Rings, A.; Basrai, M.; Bischoff, S.C. Gut permeability is related to body weight, fatty liver disease, and insulin resistance in obese individuals undergoing weight reduction. Am. J. Clin. Nutr. 2017, 105, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Chi, W.; Dao, D.; Lau, T.C.; Henriksbo, B.D.; Cavallari, J.F.; Foley, K.P.; Schertzer, J.D. Bacterial peptidoglycan stimulates adipocyte lipolysis via NOD1. PLoS ONE 2014, 9, e97675. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef]
- Neal, M.D.; Leaphart, C.; Levy, R.; Prince, J.; Billiar, T.R.; Watkins, S.; Li, J.; Cetin, S.; Ford, H.; Schreiber, A.; et al. Enterocyte TLR4 mediates phagocytosis and translocation of bacteria across the intestinal barrier. J. Immunol. 2006, 176, 3070–3079. [Google Scholar] [CrossRef]
- Kanczkowski, W.; Ziegler, C.G.; Zacharowski, K.; Bornstein, S.R. Toll-Like receptors in endocrine disease and diabetes. Neuroimmunomodulation 2008, 15, 54–60. [Google Scholar] [CrossRef]
- Allen, J.A.; Diemer, T.; Janus, P.; Hales, K.H.; Hales, D.B. Bacterial endotoxin lipopolysaccharide and reactive oxygen species inhibit Leydig cell steroidogenesis via perturbation of mitochondria. Endocrine 2004, 25, 265–275. [Google Scholar] [CrossRef]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Possemiers, S.; Van de, W.T.; Guiot, Y.; Everard, A.; Rottier, O.; Geurts, L.; Naslain, D.; Neyrinck, A.; Lambert, D.M.; et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009, 58, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, D.S. The glycemic index: Physiological mechanisms relating to obesity, diabetes, and cardiovascular disease. JAMA 2002, 287, 2414–2423. [Google Scholar] [CrossRef]
- Ruderman, N.B.; Xu, X.J.; Nelson, L.; Cacicedo, J.M.; Saha, A.K.; Lan, F.; Ido, Y. AMPK and SIRT1: A long-standing partnership? Am. J. Physiol. Endocrinol. Metab. 2010, 298, E751–E760. [Google Scholar] [CrossRef]
- Sorokin, A. Nitric oxide synthase and cyclooxygenase pathways: A complex interplay in cellular signaling. Curr. Med. Chem. 2016, 23, 2559–2578. [Google Scholar] [CrossRef]
- Simmons, D.L.; Botting, R.M.; Hla, T. Cyclooxygenase isozymes: The biology of prostaglandin synthesis and inhibition. Pharmacol. Rev. 2004, 56, 387–437. [Google Scholar] [CrossRef]
- Needleman, P.; Turk, J.; Jakschik, B.A.; Morrison, A.R.; Lefkowith, J.B. Arachidonic acid metabolism. Annu. Rev. Biochem. 1986, 55, 69–102. [Google Scholar] [CrossRef]
- Laughton, M.J.; Evans, P.J.; Moroney, M.A.; Hoult, J.R.; Halliwell, B. Inhibition of mammalian 5-lipoxygenase and cyclo-oxygenase by flavonoids and phenolic dietary additives: Relationship to antioxidant activity and to iron ion-reducing ability. Biochem. Pharmacol. 1991, 42, 1673–1681. [Google Scholar] [CrossRef]
- Peran, L.; Camuesco, D.; Comalada, M.; Nieto, A.; Concha, A.; Diaz-Ropero, M.P.; Olivares, M.; Xaus, J.; Zarzuelo, A.; Galvez, J. Preventative effects of a probiotic, Lactobacillus salivarius ssp. salivarius, in the TNBS model of rat colitis. World J. Gastroenterol. 2005, 11, 5185–5192. [Google Scholar]
- Nurmi, J.T.; Puolakkainen, P.A.; Rautonen, N.E. Bifidobacterium lactis sp. 420 up-regulates cyclooxygenase (COX)-1 and down-regulates COX-2 gene expression in a Caco-2 cell culture model. Nutr. Cancer 2005, 51, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Mahkonen, A.; Putaala, H.; Mustonen, H.; Rautonen, N.; Puolakkainen, P. Lactobacillus acidophilus 74-2 and butyrate induce cyclooxygenase (COX)-1 expression in gastric cancer cells. Immunopharmacol. Immunotoxicol. 2008, 30, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Nitric oxide as a secretory product of mammalian cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1992, 6, 3051–3064. [Google Scholar] [CrossRef]
- Kolios, G.; Valatas, V.; Ward, S.G. Nitric oxide in inflammatory bowel disease: A universal messenger in an unsolved puzzle. Immunology 2004, 113, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Toumi, R.; Soufli, I.; Rafa, H.; Belkhelfa, M.; Biad, A.; Touil-Boukoffa, C. Probiotic bacteria Lactobacillus and Bifidobacterium attenuate inflammation in dextran sulfate sodium-induced experimental colitis in mice. Int. J. Immunopathol. Pharmacol. 2014, 27, 615–627. [Google Scholar] [CrossRef]
- Tse, J.K.Y. Gut microbiota, nitric oxide, and microglia as prerequisites for neurodegenerative disorders. ACS Chem. Neurosci. 2017, 8, 1438–1447. [Google Scholar] [CrossRef]
- Lim, S.M.; Jang, H.M.; Jang, S.E.; Han, M.J.; Kim, D.H. Lactobacillus fermentum IM12 attenuates inflammation in mice by inhibiting NF-kappaB-STAT3 signalling pathway. Benef. Microbes 2017, 8, 407–419. [Google Scholar] [CrossRef]
- Mauricio, M.D.; Serna, E.; Fernandez-Murga, M.L.; Portero, J.; Aldasoro, M.; Valles, S.L.; Sanz, Y.; Vila, J.M. Bifidobacterium pseudocatenulatum CECT 7765 supplementation restores altered vascular function in an experimental model of obese mice. Int. J. Med Sci. 2017, 14, 444–451. [Google Scholar] [CrossRef]
- Putaala, H.; Barrangou, R.; Leyer, G.J.; Ouwehand, A.C.; Hansen, E.B.; Romero, D.A.; Rautonen, N. Analysis of the human intestinal epithelial cell transcriptional response to Lactobacillus acidophilus, Lactobacillus salivarius, Bifidobacterium lactis and Escherichia coli. Benef. Microbes 2010, 1, 283–295. [Google Scholar] [CrossRef]
- Peran, L.; Camuesco, D.; Comalada, M.; Bailon, E.; Henriksson, A.; Xaus, J.; Zarzuelo, A.; Galvez, J. A comparative study of the preventative effects exerted by three probiotics, Bifidobacterium lactis, Lactobacillus casei and Lactobacillus acidophilus, in the TNBS model of rat colitis. J. Appl. Microbiol. 2007, 103, 836–844. [Google Scholar] [CrossRef]
- Konner, A.C.; Bruning, J.C. Toll-Like receptors: Linking inflammation to metabolism. Trends Endocrinol. Metab. TEM 2011, 22, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Yamamoto, Y.; Wang, Q.M. The IKK NF-kappa B system: A treasure trove for drug development. Nat. Rev. Drug Discov. 2004, 3, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Ho, F.M.; Pei-Dawn Lee, C.; Chen, C.P.; Jeng, K.C.; Hsu, H.B.; Lee, S.T.; Wen Tung, W.; Lin, W.W. Inhibition of iNOS gene expression by quercetin is mediated by the inhibition of IkappaB kinase, nuclear factor-kappa B and STAT1, and depends on heme oxygenase-1 induction in mouse BV-2 microglia. Eur. J. Pharmacol. 2005, 521, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, A.; Foey, A. Probiotic modulation of innate cell pathogen sensing and signaling events. Nutrients 2017, 9. [Google Scholar] [CrossRef]
- Karin, M. The regulation of AP-1 activity by mitogen-activated protein kinases. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1996, 351, 127–134. [Google Scholar] [CrossRef]
- Mayor, F., Jr.; Jurado-Pueyo, M.; Campos, P.M.; Murga, C. Interfering with MAP kinase docking interactions: Implications and perspective for the p38 route. Cell Cycle (Georget. Tex.) 2007, 6, 528–533. [Google Scholar] [CrossRef][Green Version]
- Boutagy, N.E.; McMillan, R.P.; Frisard, M.I.; Hulver, M.W. Metabolic endotoxemia with obesity: Is it real and is it relevant? Biochimie 2016, 124, 11–20. [Google Scholar] [CrossRef]
- Davis, J.E.; Gabler, N.K.; Walker-Daniels, J.; Spurlock, M.E. Tlr-4 deficiency selectively protects against obesity induced by diets high in saturated fat. Obesity 2008, 16, 1248–1255. [Google Scholar] [CrossRef]
- Takanashi, N.; Tomosada, Y.; Villena, J.; Murata, K.; Takahashi, T.; Chiba, E.; Tohno, M.; Shimazu, T.; Aso, H.; Suda, Y.; et al. Advanced application of bovine intestinal epithelial cell line for evaluating regulatory effect of Lactobacilli against heat-killed enterotoxigenic Escherichia coli-mediated inflammation. BMC Microbiol. 2013, 13, 54. [Google Scholar] [CrossRef]
- Finamore, A.; Roselli, M.; Imbinto, A.; Seeboth, J.; Oswald, I.P.; Mengheri, E. Lactobacillus amylovorus inhibits the TLR4 inflammatory signaling triggered by enterotoxigenic Escherichia coli via modulation of the negative regulators and involvement of TLR2 in intestinal Caco-2 cells and pig explants. PLoS ONE 2014, 9, e94891. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uusitupa, H.-M.; Rasinkangas, P.; Lehtinen, M.J.; Mäkelä, S.M.; Airaksinen, K.; Anglenius, H.; Ouwehand, A.C.; Maukonen, J. Bifidobacterium animalis subsp. lactis 420 for Metabolic Health: Review of the Research. Nutrients 2020, 12, 892. https://doi.org/10.3390/nu12040892
Uusitupa H-M, Rasinkangas P, Lehtinen MJ, Mäkelä SM, Airaksinen K, Anglenius H, Ouwehand AC, Maukonen J. Bifidobacterium animalis subsp. lactis 420 for Metabolic Health: Review of the Research. Nutrients. 2020; 12(4):892. https://doi.org/10.3390/nu12040892
Chicago/Turabian StyleUusitupa, Henna-Maria, Pia Rasinkangas, Markus J. Lehtinen, Sanna M. Mäkelä, Kaisa Airaksinen, Heli Anglenius, Arthur C. Ouwehand, and Johanna Maukonen. 2020. "Bifidobacterium animalis subsp. lactis 420 for Metabolic Health: Review of the Research" Nutrients 12, no. 4: 892. https://doi.org/10.3390/nu12040892
APA StyleUusitupa, H.-M., Rasinkangas, P., Lehtinen, M. J., Mäkelä, S. M., Airaksinen, K., Anglenius, H., Ouwehand, A. C., & Maukonen, J. (2020). Bifidobacterium animalis subsp. lactis 420 for Metabolic Health: Review of the Research. Nutrients, 12(4), 892. https://doi.org/10.3390/nu12040892




