Queuine Micronutrient Deficiency Promotes Warburg Metabolism and Reversal of the Mitochondrial ATP Synthase in Hela Cells
Abstract
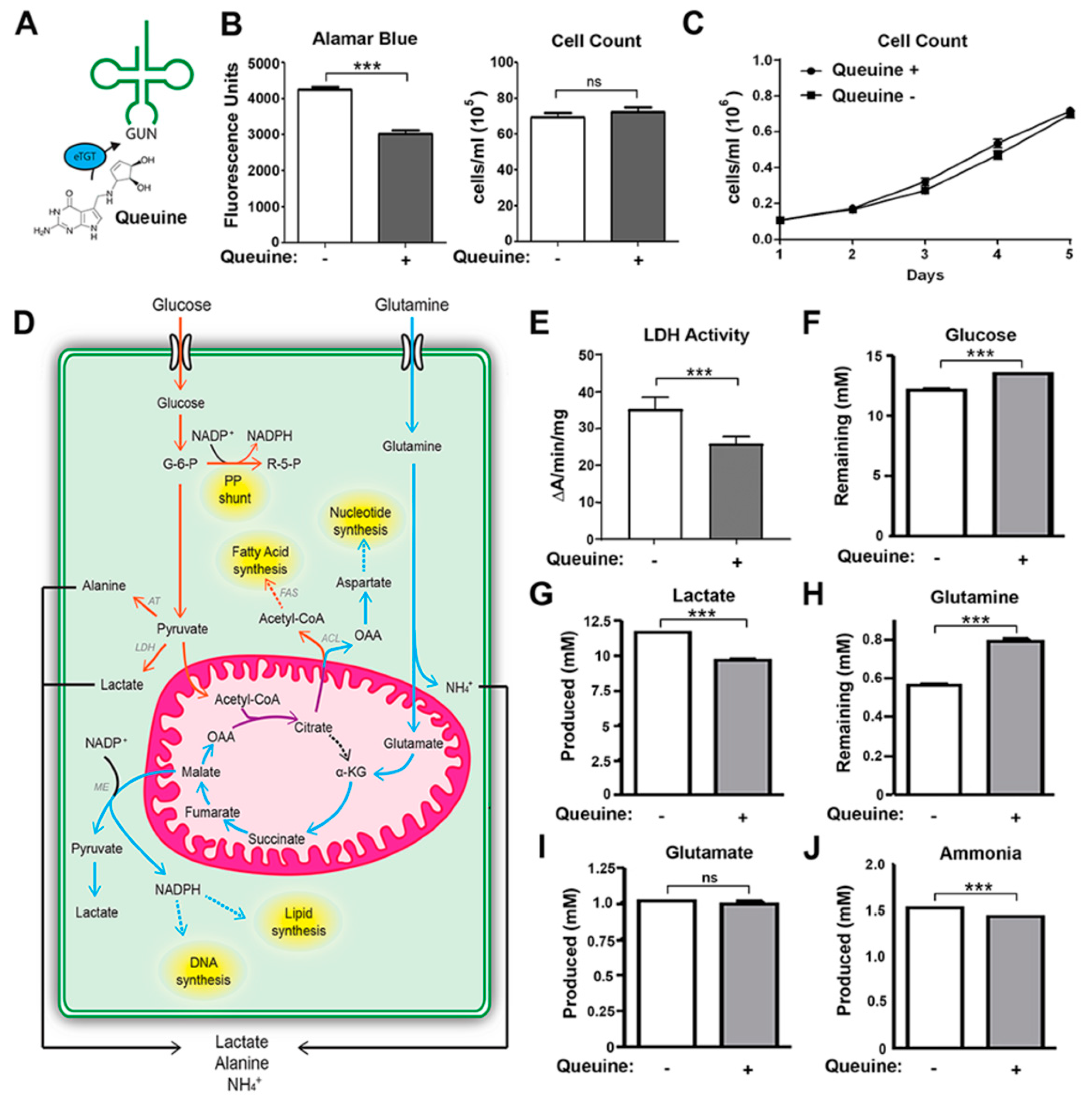
:1. Introduction
2. Materials and Methods
2.1. Hela Cell Culture
2.2. Detection of Q-modified tRNA Using Acryloyl Aminophenylboronic Acid Gels
2.3. Enzyme Activity Assays
2.4. ATP Measurement
2.5. Metabolite Analysis
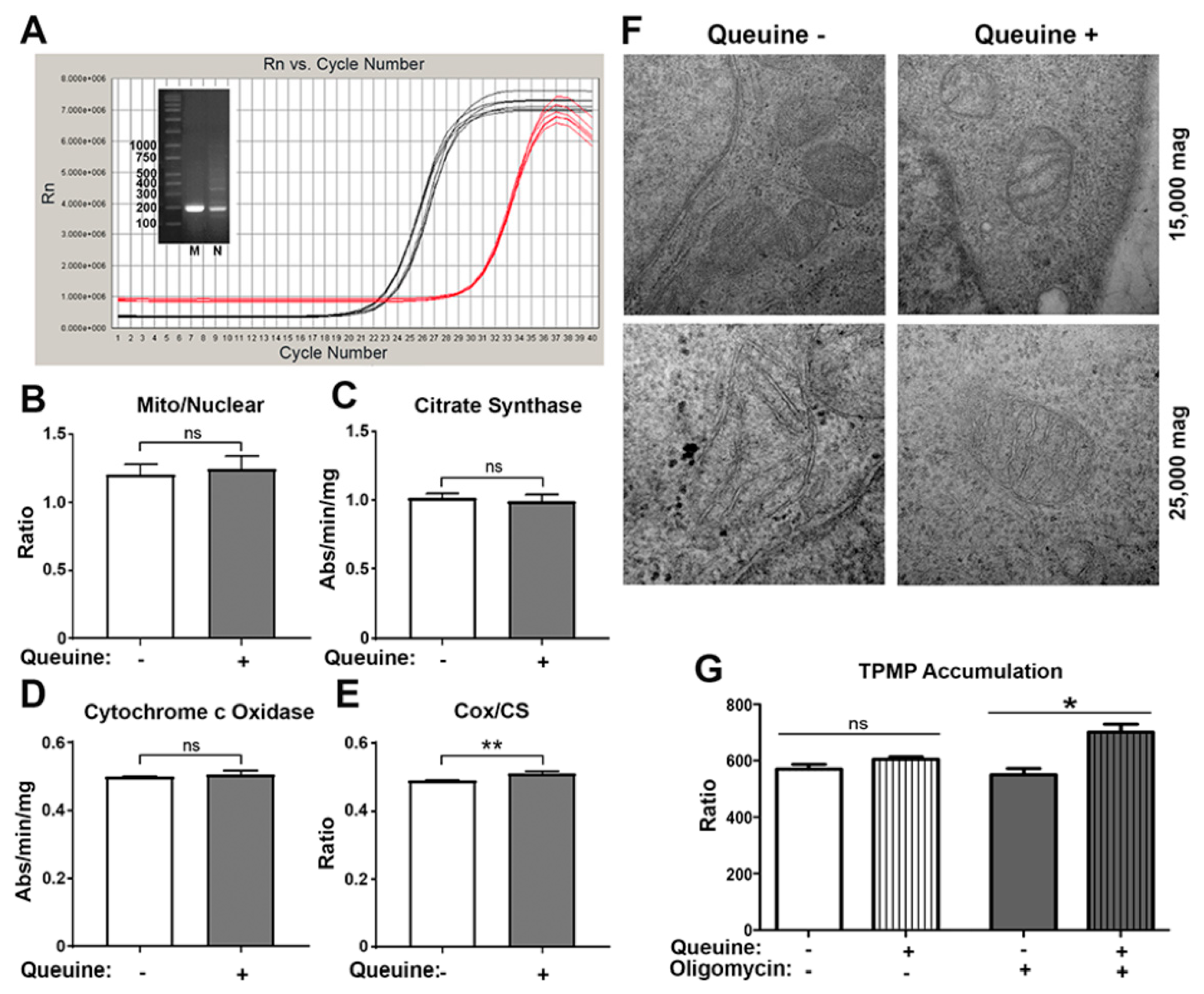
2.6. Mitochondrial and Nuclear DNA Quantitation
2.7. Mitochondrial Membrane Potential Analysis
2.8. Transmission Electron Microscopy
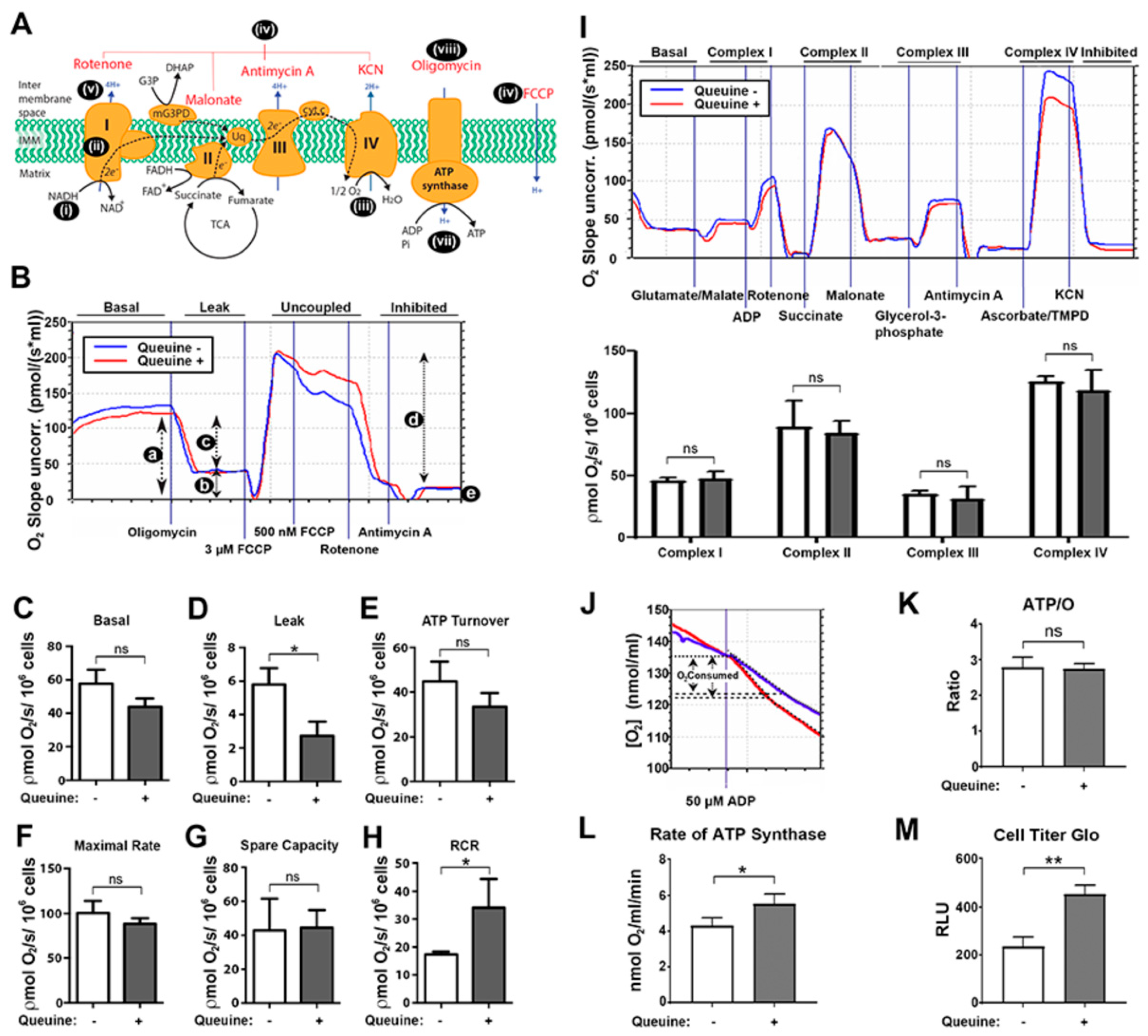
2.9. Mitochondrial Function Analysis
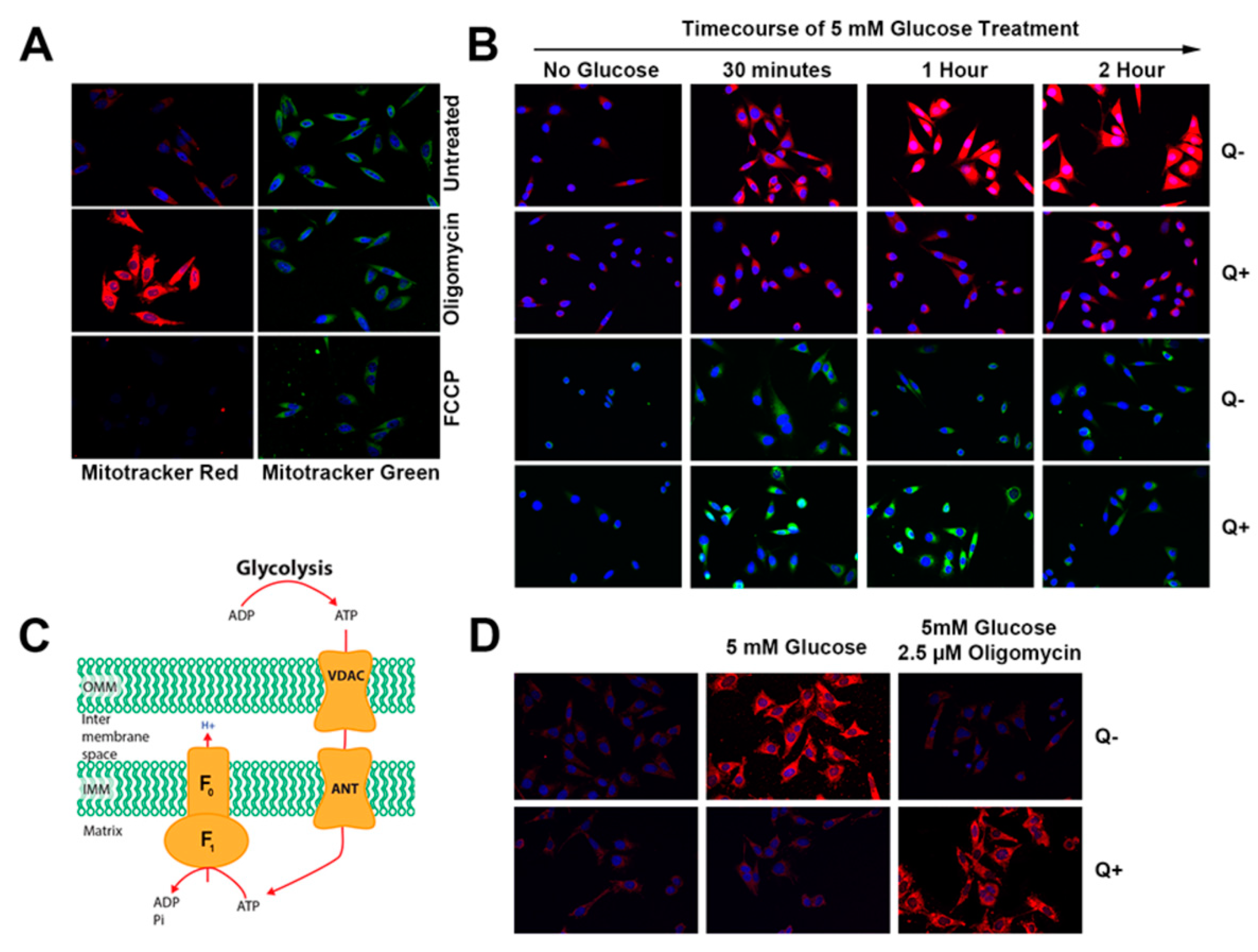
2.10. Confocal Microscopy
3. Results
3.1. Queuine Deficiency Induces Aerobic Glycolysis and Glutaminolysis in Hela Cells
3.2. Queuine Deficiency Affects Mitochondrial Membrane Hyperpolarisation by Oligomycin
3.3. Queuine Deficient Cells Display an Increased Proton Leak and Reduced Rate of ATP Synthesis
3.4. Queuine Deficiency Permits the F1F0 ATP Synthase to Operate in the Reverse Mode
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, W.Q.; Sun, X.; Xing, L.; Li, M.; Yue, J.; Qu, W.; Kong, L.; Yu, J. Oesophageal squamous cell carcinoma: Relationship between fluorine-18 fludeoxyglucose positron emission tomography ct maximum standardised uptake value, metabolic tumour volume, and tumour, node and metastasis classification. Br. J. Radiol. 2012, 85, e383–e387. [Google Scholar] [CrossRef] [Green Version]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, A.J.; Puzio-Kuter, A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fergus, C.; Barnes, D.; Alqasem, M.A.; Kelly, V.P. The queuine micronutrient: Charting a course from microbe to man. Nutrients 2015, 7, 2897–2929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, F.; Nishimura, S. Possible anticodon sequences of tRNA His, tRNA Asm, and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry 1972, 11, 301–308. [Google Scholar] [CrossRef]
- Randerath, E.; Agrawal, H.P.; Randerath, K. Specific lack of the hypermodified nucleoside, queuosine, in hepatoma mitochondrial aspartate transfer RNA and its possible biological significance. Cancer Res. 1984, 44, 1167–1171. [Google Scholar]
- Morl, M.; Dorner, M.; Paabo, S. C to U editing and modifications during the maturation of the mitochondrial tRNA(Asp) in marsupials. Nucleic Acids Res. 1995, 23, 3380–3384. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014, 42, 7346–7357. [Google Scholar] [CrossRef] [Green Version]
- Baranowski, W.; Dirheimer, G.; Jakowicki, J.A.; Keith, G. Deficiency of queuine, a highly modified purine base, in transfer RNAs from primary and metastatic ovarian malignant tumors in women. Cancer Res. 1994, 54, 4468–4471. [Google Scholar] [PubMed]
- Huang, B.S.; Wu, R.T.; Chien, K.Y. Relationship of the queuine content of transfer ribonucleic acids to histopathological grading and survival in human lung cancer. Cancer Res. 1992, 52, 4696–4700. [Google Scholar] [PubMed]
- Emmerich, B.; Zubrod, E.; Weber, H.; Maubach, P.A.; Kersten, H.; Kersten, W. Relationship of queuine-lacking transfer RNA to the grade of malignancy in human leukemias and lymphomas. Cancer Res. 1985, 45, 4308–4314. [Google Scholar] [PubMed]
- Dirheimer, G.; Baranowski, W.; Keith, G. Variations in trna modifications, particularly of their queuine content in higher eukaryotes. Its relation to malignancy grading. Biochimie 1995, 77, 99–103. [Google Scholar] [CrossRef]
- Aytac, U.; Gunduz, U. Q-modification of tRNAs in human brain tumors. Cancer Biochem. Biophys. 1994, 14, 93–98. [Google Scholar]
- Langgut, W.; Kersten, H. The deazaguanine-derivative, queuine, affects cell proliferation, protein phosphorylation and the expression of the proto oncogenes c-fos and c-myc in HeLa cells. FEBS Lett. 1990, 265, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Reisser, T.; Langgut, W.; Kersten, H. The nutrient factor queuine protects HeLa cells from hypoxic stress and improves metabolic adaptation to oxygen availability. Eur. J. Biochem./FEBS 1994, 221, 979–986. [Google Scholar] [CrossRef]
- Langgut, W.; Reisser, T.; Kersten, H.; Nishimura, S. Modulation of epidermal growth factor receptor activity and related responses by the 7-deazaguanine derivative, queuine. Oncogene 1993, 8, 3141–3147. [Google Scholar]
- Pathak, C.; Vinayak, M. Modulation of lactate dehydrogenase isozymes by modified base queuine. Mol. Biol. Rep. 2005, 32, 191–196. [Google Scholar] [CrossRef]
- Shindo-Okada, N.; Terada, M.; Nishimura, S. Changes in amount of hypo-modified tRNA having guanine in place of queuine during erythroid differentiation of murine erythroleukemia cells. Eur. J. Biochem. / FEBS 1981, 115, 423–428. [Google Scholar] [CrossRef]
- Chen, Y.L.; Wu, R.T. Altered queuine modification of transfer RNA involved in the differentiation of human K562 erythroleukemia cells in the presence of distinct differentiation inducers. Cancer Res. 1994, 54, 2192–2198. [Google Scholar]
- Morgan, C.J.; Merrill, F.L.; Trewyn, R.W. Defective transfer RNA-queuine modification in C3H10T1/2 murine fibroblasts transfected with oncogenic ras. Cancer Res. 1996, 56, 594–598. [Google Scholar] [PubMed]
- Muralidhar, G.; Trewyn, R.W. Enhancement of the chemical transformation of chinese hamster embryo cells in vitro by 7-methylguanine. Cancer Res. 1987, 47, 2440–2444. [Google Scholar] [PubMed]
- Muralidhar, G.; Ochieng, J.; Trewyn, R.W. Altered queuine modification of transfer RNA involved in the in vitro transformation of chinese hamster embryo cells. Cancer Res. 1989, 49, 7110–7114. [Google Scholar] [PubMed]
- Ishiguro, K.; Sartorelli, A.C. Enhancement of the differentiation-inducing properties of 6-thioguanine by hypoxanthine and its nucleosides in HL-60 promyelocytic leukemia cells. Cancer Res. 1985, 45, 91–95. [Google Scholar]
- Kretz, K.A.; Katze, J.R.; Trewyn, R.W. Guanine analog-induced differentiation of human promyelocytic leukemia cells and changes in queuine modification of tRNA. Mol. Cell. Biol. 1987, 7, 3613–3619. [Google Scholar] [CrossRef] [Green Version]
- French, B.T.; Patrick, D.E.; Grever, M.R.; Trewyn, R.W. Queuine, a tRNA anticodon wobble base, maintains the proliferative and pluripotent potential of HL-60 cells in the presence of the differentiating agent 6-thioguanine. Proc. Natl. Acad. Sci. USA 1991, 88, 370–374. [Google Scholar] [CrossRef] [Green Version]
- Morgan, C.J.; Chawdry, R.N.; Smith, A.R.; Siravo-Sagraves, G.; Trewyn, R.W. 6-thioguanine-induced growth arrest in 6-mercaptopurine-resistant human leukemia cells. Cancer Res. 1994, 54, 5387–5393. [Google Scholar]
- Zaborske, J.M.; Bauer DuMont, V.L.; Wallace, E.W.J.; Pan, T.; Aquadro, C.F.; Drummond, D.A. A Nutrient-Driven tRNA Modification Alters Translational Fidelity and Genome-wide Protein Coding across an Animal Genus. PLoS Biol. 2014, 12, e1002015. [Google Scholar] [CrossRef] [Green Version]
- Hyslop, S.J.; James, A.M.; Maw, M.; Fischel-Ghodsian, N.; Murphy, M.P. The effect on mitochondrial function of the tRNA Ser(UCN)/COI A7445G mtDNA point mutation associated with maternally-inherited sensorineural deafness. Biochem. Mol. Biol. Int. 1997, 42, 567–575. [Google Scholar] [CrossRef]
- Appleby, R.D.; Porteous, W.K.; Hughes, G.; James, A.M.; Shannon, D.; Wei, Y.H.; Murphy, M.P. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. / FEBS 1999, 262, 108–116. [Google Scholar] [CrossRef]
- Langgut, W. Changes of the phosphorylation of membrane-associated proteins following treatment of HeLa cells with the guanine analogue, queuine. Biofactors 1993, 4, 117–122. [Google Scholar] [PubMed]
- Rakovich, T.; Boland, C.; Bernstein, I.; Chikwana, V.M.; Iwata-Reuyl, D.; Kelly, V.P. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J. Biol. Chem. 2011, 286, 19354–19363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, P.; Sheng, S.; Sun, X.; Liu, J.; Huang, G. Lactate dehydrogenase a in cancer: A promising target for diagnosis and therapy. IUBMB Life 2013, 65, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Page, B.; Page, M.; Noel, C. A new fluorometric assay for cytotoxicity measurements in-vitro. Int. J. Oncol. 1993, 3, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Rampersad, S.N. Multiple applications of alamar blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors (Basel) 2012, 12, 12347–12360. [Google Scholar] [CrossRef]
- Trounce, I.A.; Kim, Y.L.; Jun, A.S.; Wallace, D.C. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996, 264, 484–509. [Google Scholar]
- Alirol, E.; Martinou, J.C. Mitochondria and cancer: Is there a morphological connection? Oncogene 2006, 25, 4706–4716. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, R.; Gilkerson, R.; Aggeler, R.; Yamagata, K.; Remington, S.J.; Capaldi, R.A. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004, 64, 985–993. [Google Scholar] [CrossRef] [Green Version]
- Ross, M.F.; Prime, T.A.; Abakumova, I.; James, A.M.; Porteous, C.M.; Smith, R.A.; Murphy, M.P. Rapid and extensive uptake and activation of hydrophobic triphenylphosphonium cations within cells. Biochem. J. 2008, 411, 633–645. [Google Scholar] [CrossRef] [Green Version]
- Brand, M.D.; Nicholls, D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011, 435, 297–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eagle, H.; Barban, S.; Levy, M.; Schulze, H.O. The utilization of carbohydrates by human cell cultures. J. Biol. Chem. 1958, 233, 551–558. [Google Scholar] [PubMed]
- Hall, A.; Larsen, A.K.; Parhamifar, L.; Meyle, K.D.; Wu, L.P.; Moghimi, S.M. High resolution respirometry analysis of polyethylenimine-mediated mitochondrial energy crisis and cellular stress: Mitochondrial proton leak and inhibition of the electron transport system. Biochim. Biophys. Acta 2013, 1827, 1213–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reitzer, L.J.; Wice, B.M.; Kennell, D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 1979, 254, 2669–2676. [Google Scholar] [PubMed]
- Pendergrass, W.; Wolf, N.; Poot, M. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytom. Part A J. Int. Soc. Anal. Cytol. 2004, 61, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Heerdt, B.G.; Houston, M.A.; Augenlicht, L.H. The intrinsic mitochondrial membrane potential of colonic carcinoma cells is linked to the probability of tumor progression. Cancer Res. 2005, 65, 9861–9867. [Google Scholar] [CrossRef] [Green Version]
- Heerdt, B.G.; Houston, M.A.; Augenlicht, L.H. Growth properties of colonic tumor cells are a function of the intrinsic mitochondrial membrane potential. Cancer Res. 2006, 66, 1591–1596. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Shi, X.; Johnson, R.H.; Kelbauskas, L.; Zhang, W.; Meldrum, D.R. Single-cell analysis reveals early manifestation of cancerous phenotype in pre-malignant esophageal cells. PLoS ONE 2013, 8, e75365. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.J.; Qiu, W.; Xu, S.L.; Wang, B.; Ye, X.Z.; Ping, Y.F.; Zhang, X.; Bian, X.W.; Yu, S.C. Strategies for isolating and enriching cancer stem cells: Well begun is half done. Stem Cells Dev. 2013, 22, 2221–2239. [Google Scholar] [CrossRef] [Green Version]
- Forrest, M.D. Why Cancer Cells Have a More Hyperpolarised Mitochondrial Membrane Potential and Emergent Prospects for Therapy. 2015. Available online: http://biorxiv.org/content/early/2015/08/21/025197 (accessed on 15 January 2018).
- Cetinbas, N.M.; Sudderth, J.; Harris, R.C.; Cebeci, A.; Negri, G.L.; Yilmaz, O.H.; DeBerardinis, R.J.; Sorensen, P.H. Glucose-dependent anaplerosis in cancer cells is required for cellular redox balance in the absence of glutamine. Sci. Rep. 2016, 6, 32606. [Google Scholar] [CrossRef] [Green Version]
- San-Millan, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the warburg effect. Carcinogenesis 2016, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lehuede, C.; Dupuy, F.; Rabinovitch, R.; Jones, R.G.; Siegel, P.M. Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 2016, 76, 5201–5208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molon, B.; Cali, B.; Viola, A. T cells and cancer: How metabolism shapes immunity. Front. Immunol. 2016, 7, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Campanella, M.; Parker, N.; Tan, C.H.; Hall, A.M.; Duchen, M.R. IF(1): Setting the pace of the F(1)F(o)-ATP synthase. Trends Biochem. Sci. 2009, 34, 343–350. [Google Scholar] [CrossRef]
- Akerman, K.E. Qualitative measurements of the mitochondrial membrane potential in situ in Ehrlich ascites tumour cells using the safranine method. Biochim Biophys Acta 1979, 546, 341–347. [Google Scholar] [CrossRef]
- Nolan, D.P.; Voorheis, H.P. The mitochondrion in Trypanosoma brucei is energised by the electrogenic translocation of H+ catalysed by the F1F0-ATPase. Eur. J. Biochem. 1992, 209, 207–216. [Google Scholar] [CrossRef]
- Jacobson, K.B.; Farkas, W.R.; Katze, J.R. Presence of queuine in drosophila melanogaster: Correlation of free pool with queuosine content of tRNA and effect of mutations in pteridine metabolism. Nucleic Acids Res. 1981, 9, 2351–2366. [Google Scholar] [CrossRef] [Green Version]
- Morris, R.C.; Galicia, M.C.; Clase, K.L.; Elliott, M.S. Determination of queuosine modification system deficiencies in cultured human cells. Mol. Genet. Metab. 1999, 68, 56–67. [Google Scholar] [CrossRef]
- Singhal, R.P.; Kopper, R.A.; Nishimura, S.; Shindo-Okada, N. Modification of guanine to queuine in transfer RNAs during development and aging. Biochem. Biophys. Res. Commun. 1981, 99, 120–126. [Google Scholar] [CrossRef]
- Frazer, J.M.; Yang, W.K. Isoaccepting transfer ribonucleic acids in liver and brain of young and old BC3F 1 mice. Arch. Biochem. Biophys. 1972, 153, 610–618. [Google Scholar] [CrossRef]
- Costa, A.; Pais de Barros, J.P.; Keith, G.; Baranowski, W.; Desgres, J. Determination of queuosine derivatives by reverse-phase liquid chromatography for the hypomodification study of Q-bearing tRNAs from various mammal liver cells. J. Chromatogr. BAnal. Technol. Biomed. Life Sci. 2004, 801, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Landin, R.M.; Boisnard, M.; Petrissant, G. Correlation between the presence of tRNA His GUG and the erythropoietic function in foetal sheep liver. Nucleic Acids Res. 1979, 7, 1635–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayes, P.; Fergus, C.; Ghanim, M.; Cirzi, C.; Burtnyak, L.; McGrenaghan, C.J.; Tuorto, F.; Nolan, D.P.; Kelly, V.P. Queuine Micronutrient Deficiency Promotes Warburg Metabolism and Reversal of the Mitochondrial ATP Synthase in Hela Cells. Nutrients 2020, 12, 871. https://doi.org/10.3390/nu12030871
Hayes P, Fergus C, Ghanim M, Cirzi C, Burtnyak L, McGrenaghan CJ, Tuorto F, Nolan DP, Kelly VP. Queuine Micronutrient Deficiency Promotes Warburg Metabolism and Reversal of the Mitochondrial ATP Synthase in Hela Cells. Nutrients. 2020; 12(3):871. https://doi.org/10.3390/nu12030871
Chicago/Turabian StyleHayes, Patti, Claire Fergus, Magda Ghanim, Cansu Cirzi, Lyubomyr Burtnyak, Callum J. McGrenaghan, Francesca Tuorto, Derek P. Nolan, and Vincent P. Kelly. 2020. "Queuine Micronutrient Deficiency Promotes Warburg Metabolism and Reversal of the Mitochondrial ATP Synthase in Hela Cells" Nutrients 12, no. 3: 871. https://doi.org/10.3390/nu12030871
APA StyleHayes, P., Fergus, C., Ghanim, M., Cirzi, C., Burtnyak, L., McGrenaghan, C. J., Tuorto, F., Nolan, D. P., & Kelly, V. P. (2020). Queuine Micronutrient Deficiency Promotes Warburg Metabolism and Reversal of the Mitochondrial ATP Synthase in Hela Cells. Nutrients, 12(3), 871. https://doi.org/10.3390/nu12030871




