Saturated Fatty Acids Promote GDF15 Expression in Human Macrophages through the PERK/eIF2/CHOP Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of FFA Solutions
2.2. Cell Culture and Treatments
2.3. siRNA Transfection
2.4. Cell Viability Assay
2.5. Luminex Assay
2.6. RT-qPCR Analysis
2.7. Western Blot Analysis
2.8. ChIP-qPCR Analysis
2.9. Statistical Analyses
3. Results
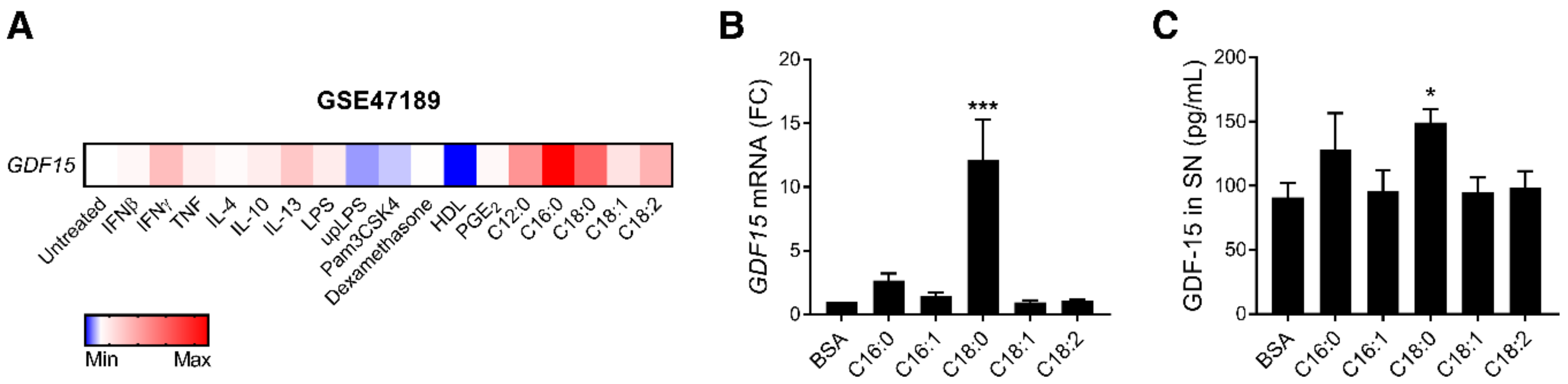
3.1. SFAs Promote GDF15 Expression and Secretion in Macrophages
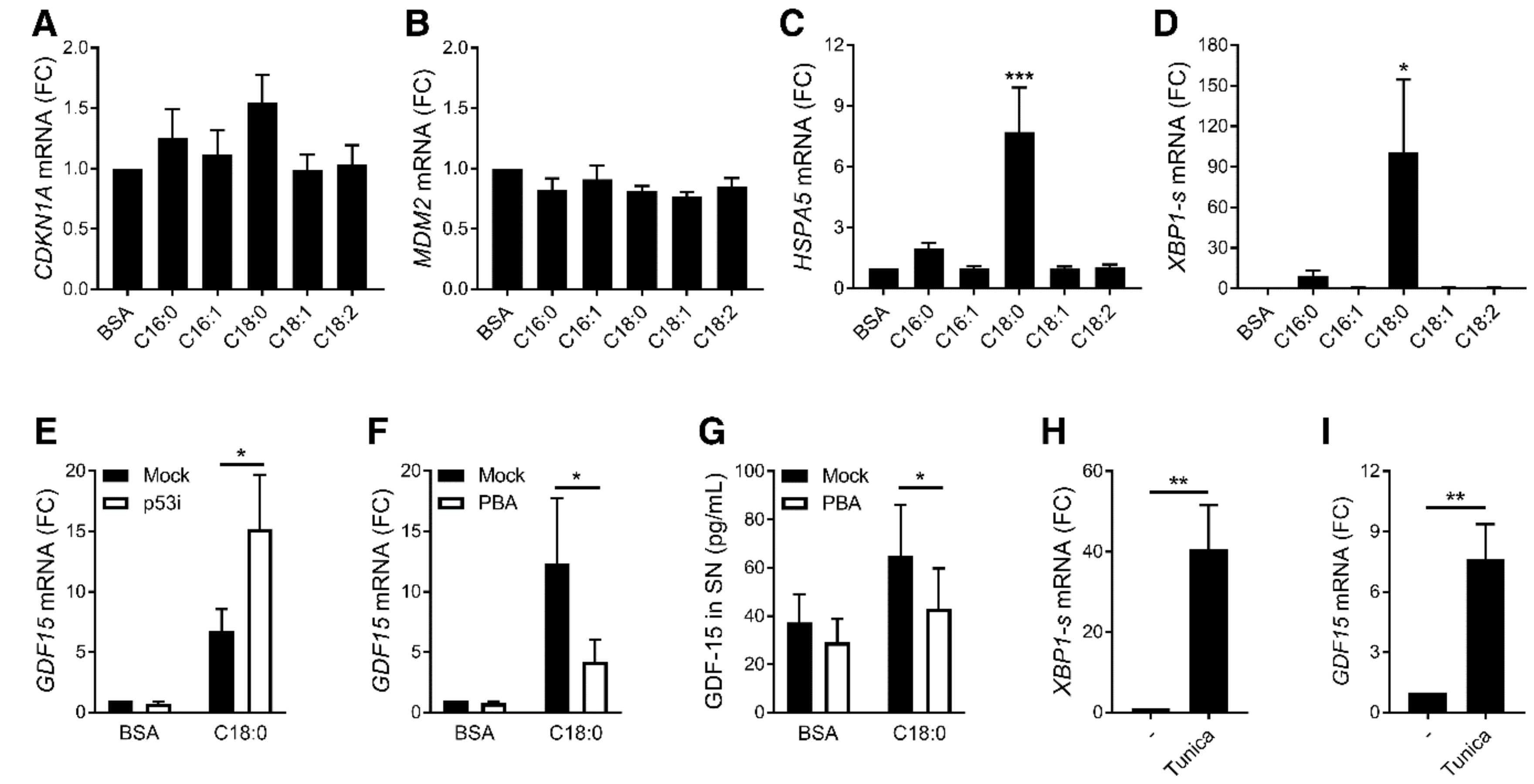
3.2. ER Stress, but Not p53, Is Involved in SFA-Induced GDF15 Expression
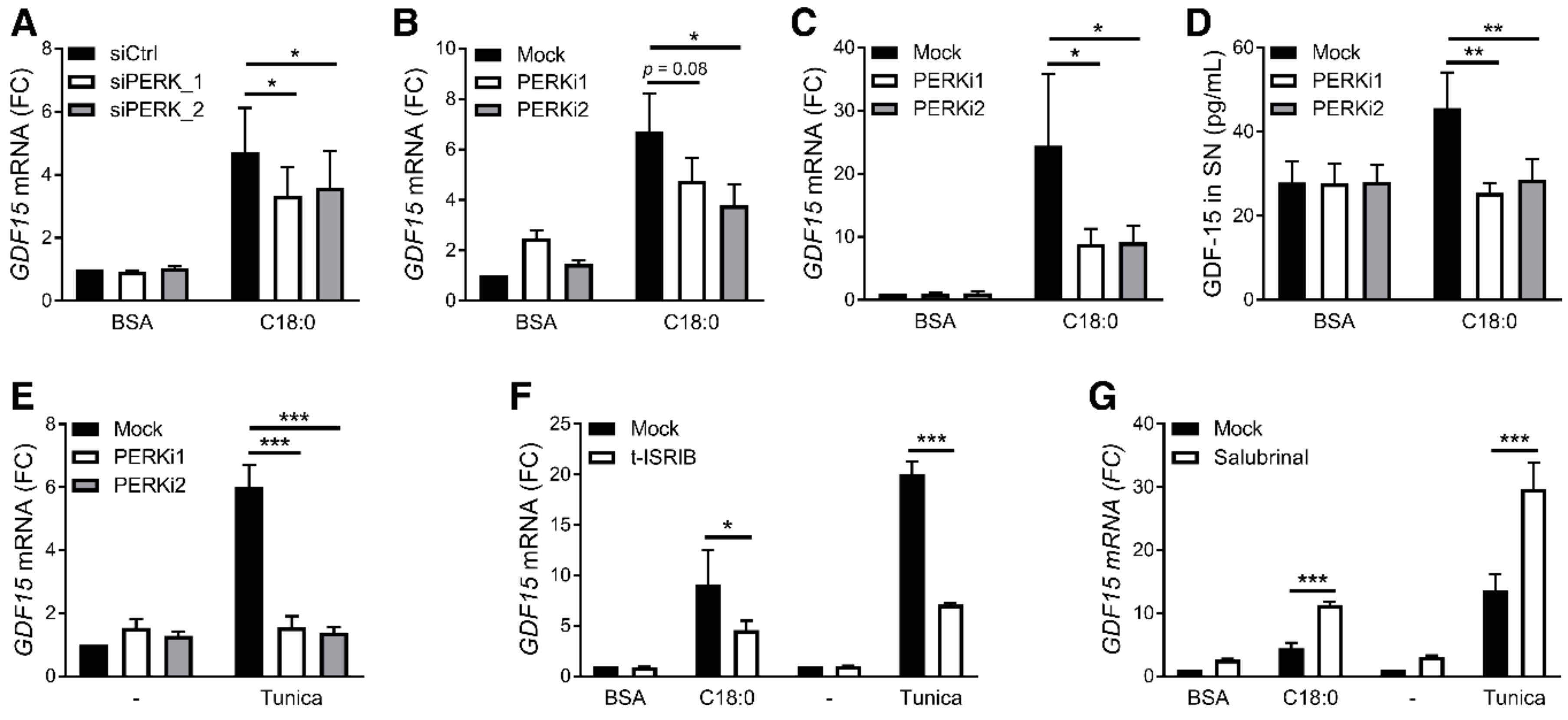
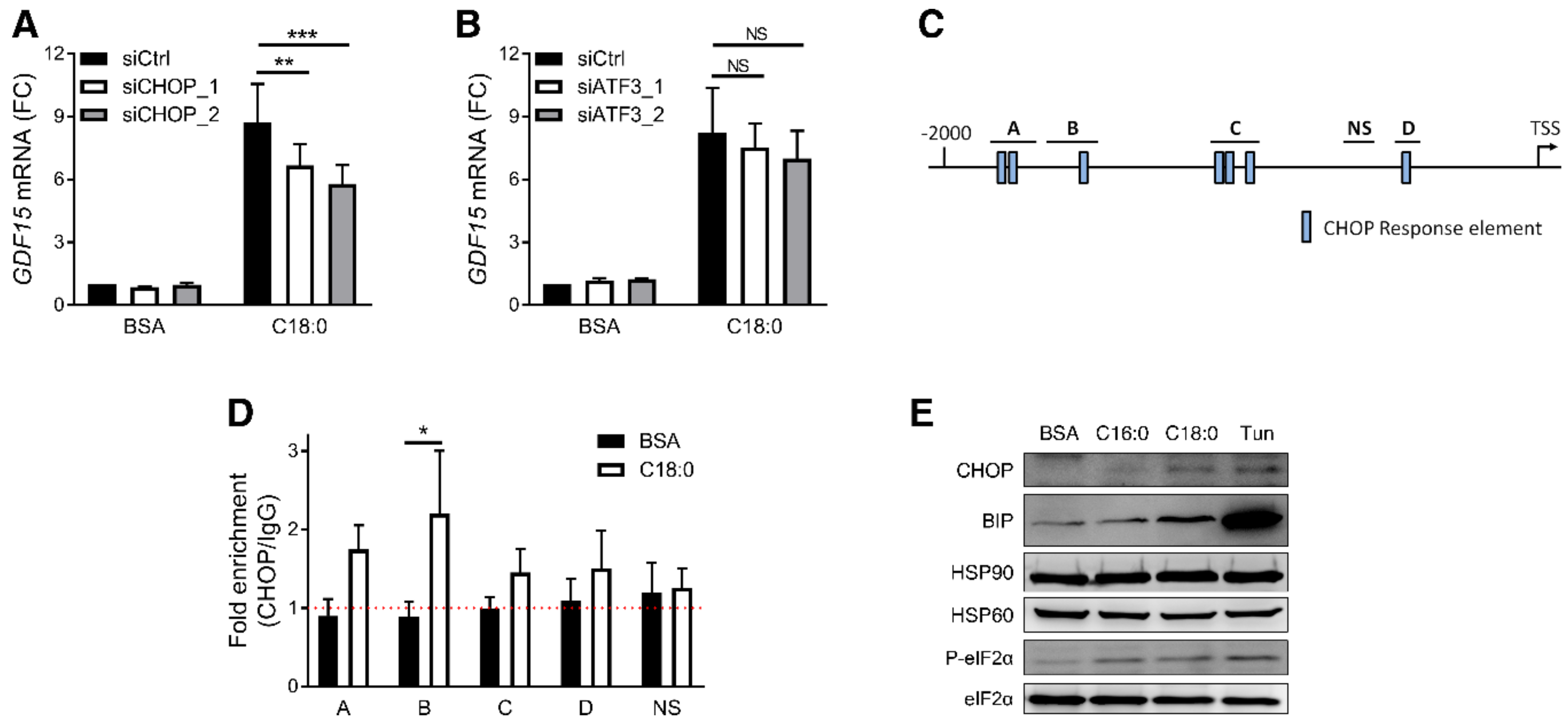
3.3. PERK/eIF2/CHOP Pathway Regulates SFA-Induced GDF15 Expression
3.4. UFAs Inhibit SFA-Induced ER Stress and GDF15 Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dostálová, I.; Roubícek, T.; Bártlová, M.; Mráz, M.; Lacinová, Z.; Haluzíková, D.; Kaválková, P.; Matoulek, M.; Kasalicky, M.; Haluzík, M. Increased serum concentrations of macrophage inhibitory cytokine-1 in patients with obesity and type 2 diabetes mellitus: The influence of very low calorie diet. Eur. J. Endocrinol. 2009, 161, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila, G.; Riedl, M.; Anderwald, C.; Resl, M.; Handisurya, A.; Clodi, M.; Prager, G.; Ludvik, B.; Krebs, M.; Luger, A. The relationship between insulin resistance and the cardiovascular biomarker growth differentiation factor-15 in obese patients. Clin. Chem. 2011, 57, 309–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, X.; Borné, Y.; Muhammad, I.F.; Nilsson, J.; Lind, L.; Melander, O.; Niu, K.; Orho-Melander, M.; Engström, G. Growth differentiation factor 15 is positively associated with incidence of diabetes mellitus: The Malmö Diet and Cancer-Cardiovascular Cohort. Diabetologia 2019, 62, 78–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kempf, T.; Guba-Quint, A.; Torgerson, J.; Magnone, M.C.; Haefliger, C.; Bobadilla, M.; Wollert, K.C. Growth differentiation factor 15 predicts future insulin resistance and impaired glucose control in obese nondiabetic individuals: Results from the XENDOS trial. Eur. J. Endocrinol. 2012, 167, 671–678. [Google Scholar] [CrossRef]
- Koo, B.K.; Um, S.H.; Seo, D.S.; Joo, S.K.; Bae, J.M.; Park, J.H.; Chang, M.S.; Kim, J.H.; Lee, J.; Jeong, W.-I.; et al. Growth differentiation factor 15 predicts advanced fibrosis in biopsy-proven non-alcoholic fatty liver disease. Liver Int. 2018, 38, 695–705. [Google Scholar] [CrossRef]
- Brown, D.A.; Breit, S.N.; Buring, J.; Fairlie, W.D.; Bauskin, A.R.; Liu, T.; Ridker, P.M. Concentration in plasma of macrophage inhibitory cytokine-1 and risk of cardiovascular events in women: A nested case-control study. Lancet 2002, 359, 2159–2163. [Google Scholar] [CrossRef]
- Brown, D.A.; Ward, R.L.; Buckhaults, P.; Liu, T.; Romans, K.E.; Hawkins, N.J.; Bauskin, A.R.; Kinzler, K.W.; Vogelstein, B.; Breit, S.N. MIC-1 serum level and genotype: Associations with progress and prognosis of colorectal carcinoma. Clin. Cancer Res. 2003, 9, 2642–2650. [Google Scholar]
- Koopmann, J.; Buckhaults, P.; Brown, D.A.; Zahurak, M.L.; Sato, N.; Fukushima, N.; Sokoll, L.J.; Chan, D.W.; Yeo, C.J.; Hruban, R.H.; et al. Serum macrophage inhibitory cytokine 1 as a marker of pancreatic and other periampullary cancers. Clin. Cancer Res. 2004, 10, 2386–2392. [Google Scholar] [CrossRef] [Green Version]
- Johnen, H.; Lin, S.; Kuffner, T.; Brown, D.A.; Tsai, V.W.-W.; Bauskin, A.R.; Wu, L.; Pankhurst, G.; Jiang, L.; Junankar, S.; et al. Tumor-induced anorexia and weight loss are mediated by the TGF-beta superfamily cytokine MIC-1. Nat. Med. 2007, 13, 1333–1340. [Google Scholar] [CrossRef]
- Osada, M.; Park, H.L.; Park, M.J.; Liu, J.-W.; Wu, G.; Trink, B.; Sidransky, D. A p53-type response element in the GDF15 promoter confers high specificity for p53 activation. Biochem. Biophys. Res. Commun. 2007, 354, 913–918. [Google Scholar] [CrossRef] [Green Version]
- Tsai, V.W.W.; Husaini, Y.; Sainsbury, A.; Brown, D.A.; Breit, S.N. The MIC-1/GDF15-GFRAL Pathway in Energy Homeostasis: Implications for Obesity, Cachexia, and Other Associated Diseases. Cell Metab. 2018, 28, 353–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chrysovergis, K.; Wang, X.; Kosak, J.; Lee, S.-H.; Kim, J.S.; Foley, J.F.; Travlos, G.; Singh, S.; Baek, S.J.; Eling, T.E. NAG-1/GDF-15 prevents obesity by increasing thermogenesis, lipolysis and oxidative metabolism. Int. J. Obes. 2014, 38, 1555–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, H.K.; Ryu, D.; Kim, K.S.; Chang, J.Y.; Kim, Y.K.; Yi, H.-S.; Kang, S.G.; Choi, M.J.; Lee, S.E.; Jung, S.-B.; et al. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J. Cell Biol. 2017, 216, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, P.J.; Wang, F.; Du, Y.; Liu, Q.; Pickard, R.T.; Gonciarz, M.D.; Coskun, T.; Hamang, M.J.; Sindelar, D.K.; Ballman, K.K.; et al. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat. Med. 2017, 23, 1215–1219. [Google Scholar] [CrossRef]
- Jung, S.-B.; Choi, M.J.; Ryu, D.; Yi, H.-S.; Lee, S.E.; Chang, J.Y.; Chung, H.K.; Kim, Y.K.; Kang, S.G.; Lee, J.H.; et al. Reduced oxidative capacity in macrophages results in systemic insulin resistance. Nat.Commun. 2018, 9, 1551. [Google Scholar] [CrossRef] [Green Version]
- Macia, L.; Tsai, V.W.-W.; Nguyen, A.D.; Johnen, H.; Kuffner, T.; Shi, Y.-C.; Lin, S.; Herzog, H.; Brown, D.A.; Breit, S.N.; et al. Macrophage inhibitory cytokine 1 (MIC-1/GDF15) decreases food intake, body weight and improves glucose tolerance in mice on normal & obesogenic diets. PLoS ONE 2012, 7, e34868. [Google Scholar] [CrossRef] [Green Version]
- Mullican, S.E.; Lin-Schmidt, X.; Chin, C.-N.; Chavez, J.A.; Furman, J.L.; Armstrong, A.A.; Beck, S.C.; South, V.J.; Dinh, T.Q.; Cash-Mason, T.D.; et al. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat. Med. 2017, 23, 1150–1157. [Google Scholar] [CrossRef]
- Tran, T.; Yang, J.; Gardner, J.; Xiong, Y. GDF15 deficiency promotes high fat diet-induced obesity in mice. PLoS ONE 2018, 13, e0201584. [Google Scholar] [CrossRef] [Green Version]
- Hsu, J.-Y.; Crawley, S.; Chen, M.; Ayupova, D.A.; Lindhout, D.A.; Higbee, J.; Kutach, A.; Joo, W.; Gao, Z.; Fu, D.; et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 2017, 550, 255–259. [Google Scholar] [CrossRef]
- Yang, L.; Chang, C.-C.; Sun, Z.; Madsen, D.; Zhu, H.; Padkjær, S.B.; Wu, X.; Huang, T.; Hultman, K.; Paulsen, S.J.; et al. GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat. Med. 2017, 23, 1158–1166. [Google Scholar] [CrossRef]
- Tsai, V.W.-W.; Manandhar, R.; Jørgensen, S.B.; Lee-Ng, K.K.M.; Zhang, H.P.; Marquis, C.P.; Jiang, L.; Husaini, Y.; Lin, S.; Sainsbury, A.; et al. The anorectic actions of the TGFβ cytokine MIC-1/GDF15 require an intact brainstem area postrema and nucleus of the solitary tract. PLoS ONE 2014, 9, e100370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Q.; Mracek, T.; Gonzalez-Muniesa, P.; Kos, K.; Wilding, J.; Trayhurn, P.; Bing, C. Identification of macrophage inhibitory cytokine-1 in adipose tissue and its secretion as an adipokine by human adipocytes. Endocrinology 2009, 150, 1688–1696. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, H.; Zhong, Y. Hepatic GDF15 is regulated by CHOP of the unfolded protein response and alleviates NAFLD progression in obese mice. Biochem. Biophys. Res. Commun. 2018, 498, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-H.; Yu, M.; Kim, J.; Moon, Y. C/EBP homologous protein promotes NSAID-activated gene 1-linked pro-inflammatory signals and enterocyte invasion by enteropathogenic Escherichia coli. Microbes Infect. 2017, 19, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Bootcov, M.R.; Bauskin, A.R.; Valenzuela, S.M.; Moore, A.G.; Bansal, M.; He, X.Y.; Zhang, H.P.; Donnellan, M.; Mahler, S.; Pryor, K.; et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc. Natl. Acad. Sci. USA 1997, 94, 11514–11519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- Perdiguero, E.G.; Geissmann, F. The development and maintenance of resident macrophages. Nat. Immunol. 2016, 17, 2–8. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Alvarez-Guaita, A.; Melvin, A.; Rimmington, D.; Dattilo, A.; Miedzybrodzka, E.L.; Cimino, I.; Maurin, A.-C.; Roberts, G.P.; Meek, C.L.; et al. GDF15 Provides an Endocrine Signal of Nutritional Stress in Mice and Humans. Cell Metab. 2019, 29, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Park, S.H.; Choi, H.J.; Moon, Y. The integrated stress response-associated signals modulates intestinal tumor cell growth by NSAID-activated gene 1 (NAG-1/MIC-1/PTGF-beta). Carcinogenesis 2010, 31, 703–711. [Google Scholar] [CrossRef] [Green Version]
- Boden, G.; Duan, X.; Homko, C.; Molina, E.J.; Song, W.; Perez, O.; Cheung, P.; Merali, S. Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes 2008, 57, 2438–2444. [Google Scholar] [CrossRef] [Green Version]
- Laybutt, D.R.; Preston, A.M.; Akerfeldt, M.C.; Kench, J.G.; Busch, A.K.; Biankin, A.V.; Biden, T.J. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia 2007, 50, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, U.; Cao, Q.; Yilmaz, E.; Lee, A.-H.; Iwakoshi, N.N.; Ozdelen, E.; Tuncman, G.; Görgün, C.; Glimcher, L.H.; Hotamisligil, G.S. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 2004, 306, 457–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Murthy, R.; Wood, B.; Song, B.; Wang, S.; Sun, B.; Malhi, H.; Kaufman, R.J. ER stress signalling through eIF2α and CHOP, but not IRE1α, attenuates adipogenesis in mice. Diabetologia 2013, 56, 911–924. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Gao, J.; Ishigaki, Y.; Kondo, K.; Sawada, S.; Izumi, T.; Uno, K.; Kaneko, K.; Tsukita, S.; Takahashi, K.; et al. ER Stress Protein CHOP Mediates Insulin Resistance by Modulating Adipose Tissue Macrophage Polarity. Cell Rep. 2017, 18, 2045–2057. [Google Scholar] [CrossRef] [Green Version]
- L’homme, L.; Esser, N.; Riva, L.; Scheen, A.; Paquot, N.; Piette, J.; Legrand-Poels, S. Unsaturated fatty acids prevent activation of NLRP3 inflammasome in human monocytes/macrophages. J. Lipid Res. 2013, 54, 2998–3008. [Google Scholar] [CrossRef] [Green Version]
- De Jonge, H.J.M.; Fehrmann, R.S.N.; de Bont, E.S.J.M.; Hofstra, R.M.W.; Gerbens, F.; Kamps, W.A.; de Vries, E.G.E.; van der Zee, A.G.J.; te Meerman, G.J.; ter Elst, A. Evidence Based Selection of Housekeeping Genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef] [Green Version]
- Bergman, R.N.; Ader, M. Free fatty acids and pathogenesis of type 2 diabetes mellitus. Trends Endocrinol. Metab. 2000, 11, 351–356. [Google Scholar] [CrossRef]
- Hodson, L.; Skeaff, C.M.; Fielding, B.A. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog. Lipid Res. 2008, 47, 348–380. [Google Scholar] [CrossRef]
- Gianfrancesco, M.A.; Dehairs, J.; L’homme, L.; Herinckx, G.; Esser, N.; Jansen, O.; Habraken, Y.; Lassence, C.; Swinnen, J.V.; Rider, M.H.; et al. Saturated fatty acids induce NLRP3 activation in human macrophages through K+ efflux resulting from phospholipid saturation and Na, K-ATPase disruption. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Macrae, K.; Stretton, C.; Lipina, C.; Blachnio-Zabielska, A.; Baranowski, M.; Gorski, J.; Marley, A.; Hundal, H.S. Defining the role of DAG, mitochondrial function, and lipid deposition in palmitate-induced proinflammatory signaling and its counter-modulation by palmitoleate. J. Lipid Res. 2013, 54, 2366–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein-Platat, C.; Drai, J.; Oujaa, M.; Schlienger, J.-L.; Simon, C. Plasma fatty acid composition is associated with the metabolic syndrome and low-grade inflammation in overweight adolescents. Am. J. Clin. Nutr. 2005, 82, 1178–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Wu, J.H.Y.; Wang, Q.; Lemaitre, R.N.; Mukamal, K.J.; Djoussé, L.; King, I.B.; Song, X.; Biggs, M.L.; Delaney, J.A.; et al. Prospective association of fatty acids in the de novo lipogenesis pathway with risk of type 2 diabetes: The Cardiovascular Health Study. Am. J. Clin. Nutr. 2015, 101, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Haversen, L.; Danielsson, K.N.; Fogelstrand, L.; Wiklund, O. Induction of proinflammatory cytokines by long-chain saturated fatty acids in human macrophages. Atherosclerosis 2009, 202, 382–393. [Google Scholar] [CrossRef]
- Kratz, M.; Coats, B.R.; Hisert, K.B.; Hagman, D.; Mutskov, V.; Peris, E.; Schoenfelt, K.Q.; Kuzma, J.N.; Larson, I.; Billing, P.S.; et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014, 20, 614–625. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
L’homme, L.; Sermikli, B.P.; Staels, B.; Piette, J.; Legrand-Poels, S.; Dombrowicz, D. Saturated Fatty Acids Promote GDF15 Expression in Human Macrophages through the PERK/eIF2/CHOP Signaling Pathway. Nutrients 2020, 12, 3771. https://doi.org/10.3390/nu12123771
L’homme L, Sermikli BP, Staels B, Piette J, Legrand-Poels S, Dombrowicz D. Saturated Fatty Acids Promote GDF15 Expression in Human Macrophages through the PERK/eIF2/CHOP Signaling Pathway. Nutrients. 2020; 12(12):3771. https://doi.org/10.3390/nu12123771
Chicago/Turabian StyleL’homme, Laurent, Benan Pelin Sermikli, Bart Staels, Jacques Piette, Sylvie Legrand-Poels, and David Dombrowicz. 2020. "Saturated Fatty Acids Promote GDF15 Expression in Human Macrophages through the PERK/eIF2/CHOP Signaling Pathway" Nutrients 12, no. 12: 3771. https://doi.org/10.3390/nu12123771






