A High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.2. Oral Glucose Tolerance Test
2.3. Biochemical Measurements
2.4. Metabolic Rates Analysis
2.5. Gene Expression Analysis
2.6. Gut Microbiota Sequencing
2.7. Detection of Short-Chain Fatty Acids (SCFAs) from Intestinal Contents
2.8. Statistical Analysis
3. Results
3.1. High-Fat Diet (HFD) Triggered Obesity and Disrupted Glucose Homeostasis
3.2. HFD Enhanced Energy Expenditure and Lipid Metabolism
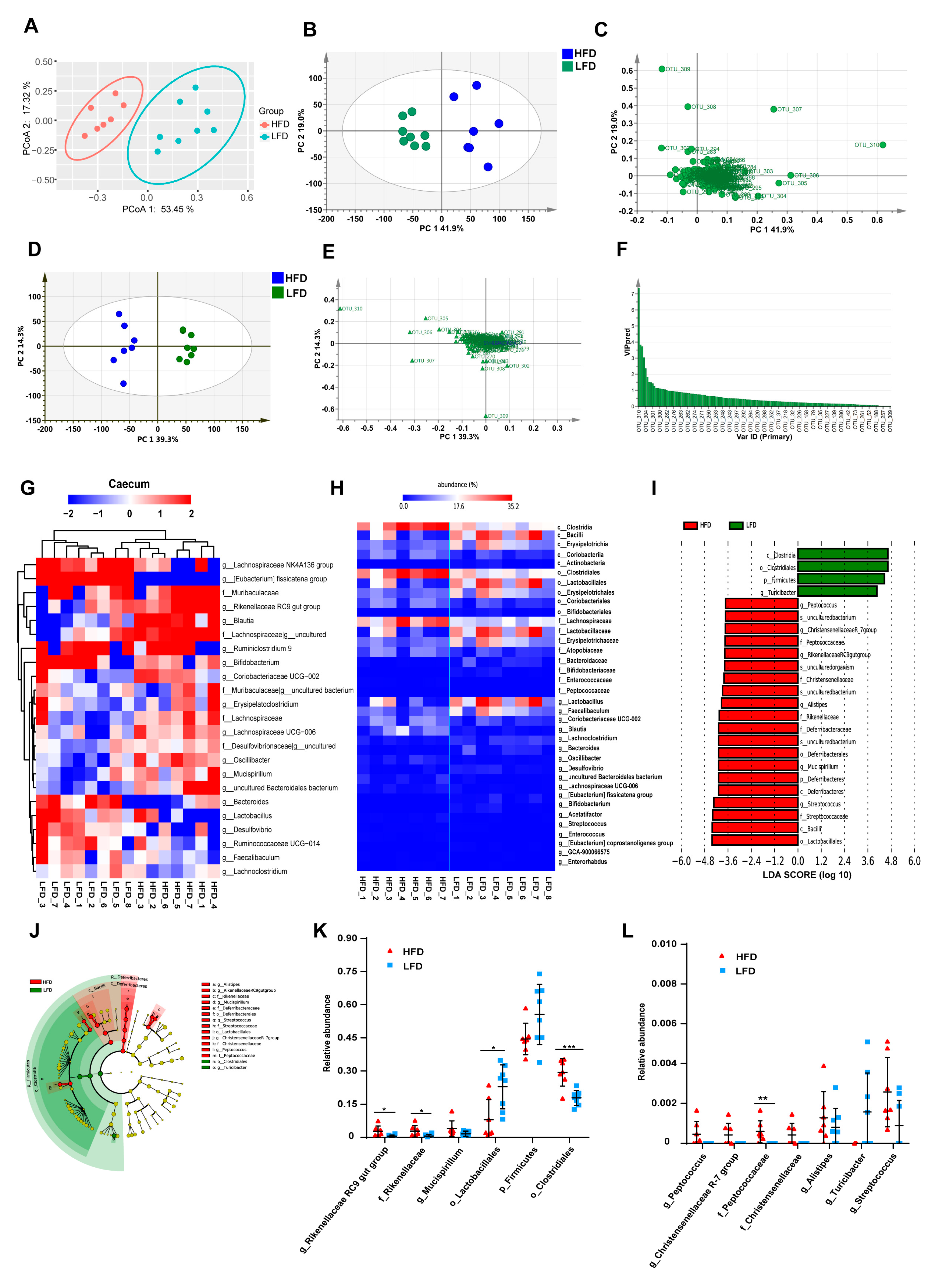
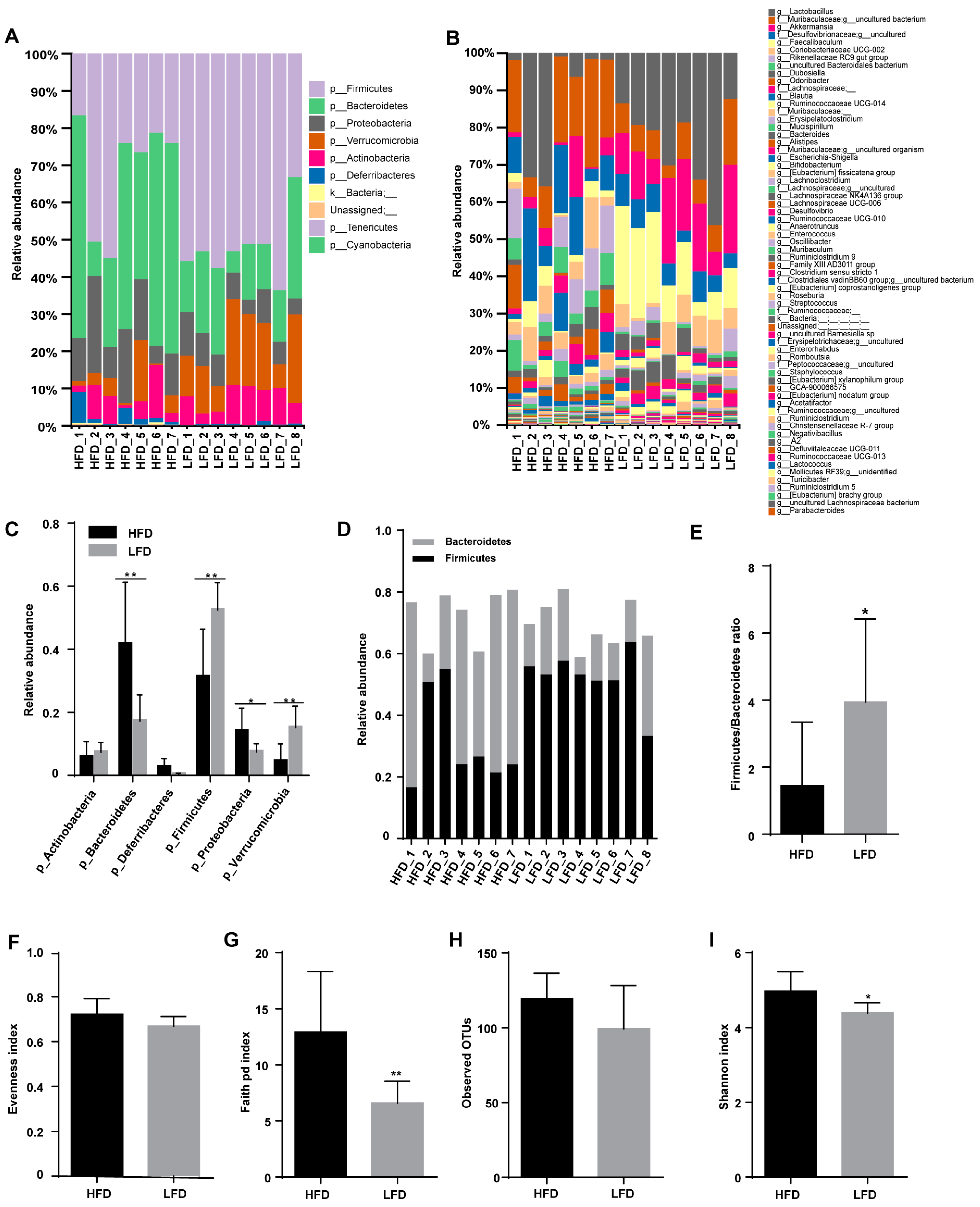
3.3. HFD Increased Biodiversity and Influenced Gut Microbial Composition in the Cecum
3.4. Low-Fat Diet (LFD) Reduced Colonic Gut Microbial Biodiversity But Increased the Retention of Beneficial Bacteria
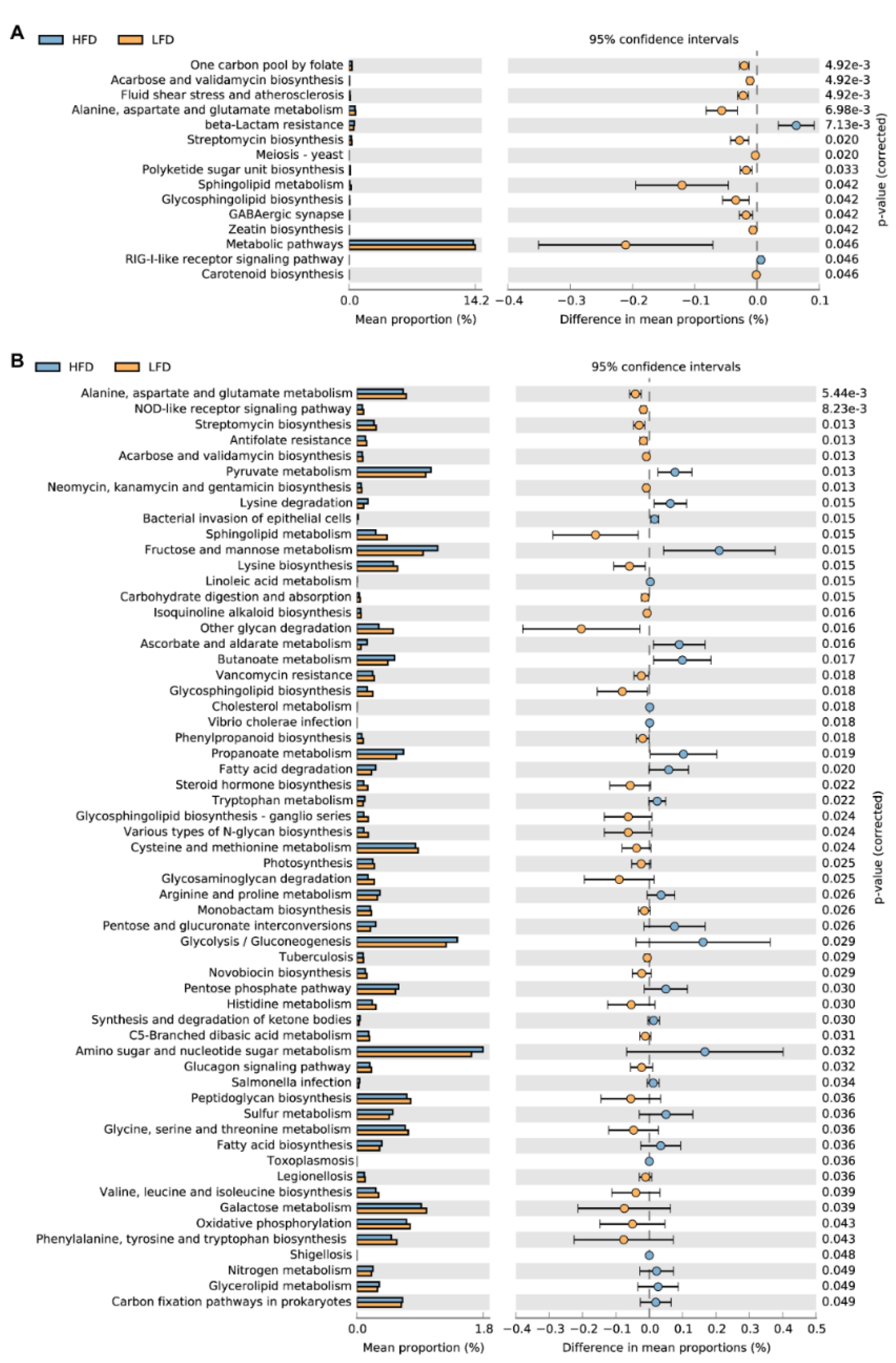
3.5. Predicted Metabolic Pathways
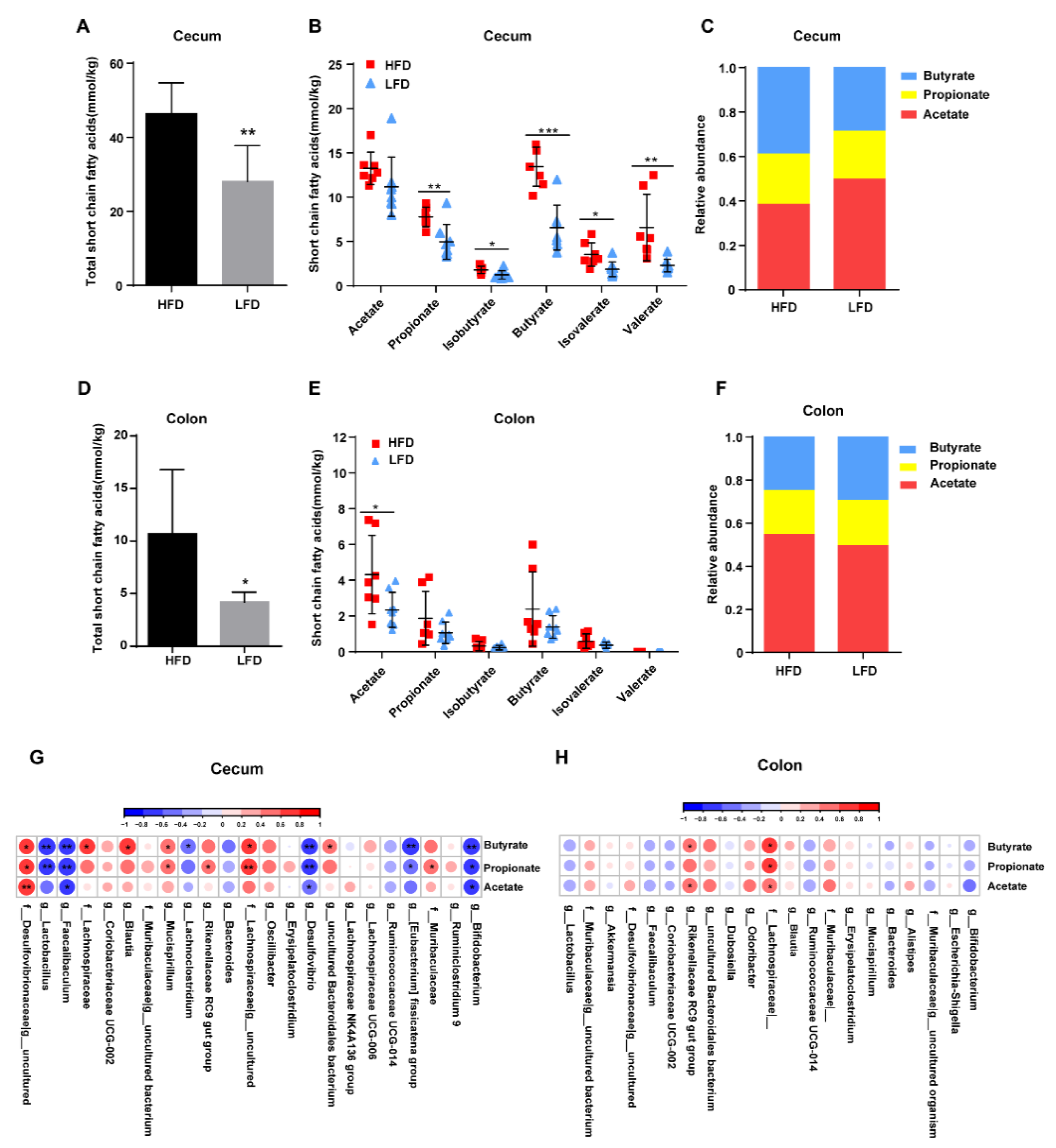
3.6. HFD Promoted the Production of Intestinal SCFAs Dependent on Specific Bacteria Colonization
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gregg, E.W.; Shaw, J.E. Global Health Effects of Overweight and Obesity. N. Engl. J. Med. 2017, 377, 80–81. [Google Scholar] [CrossRef] [PubMed]
- Bjorge, T.; Haggstrom, C.; Ghaderi, S.; Nagel, G.; Manjer, J.; Tretli, S.; Ulmer, H.; Harlid, S.; Rosendahl, A.H.; Lang, A.; et al. BMI and weight changes and risk of obesity-related cancers: A pooled European cohort study. Int. J. Epidemiol. 2019, 48, 1872–1885. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Engineer, A. Denis Burkitt and the origins of the dietary fibre hypothesis. Nutr. Res. Rev. 2018, 31, 1–15. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.J. The association between dietary fibre deficiency and high-income lifestyle-associated diseases: Burkitt’s hypothesis revisited. Lancet Gastroenterol. Hepatol. 2019, 4, 984–996. [Google Scholar] [CrossRef]
- King, D.E.; Mainous, A.G., III; Lambourne, C.A. Trends in dietary fiber intake in the United States, 1999–2008. J. Acad. Nutr. Diet. 2012, 112, 642–648. [Google Scholar] [CrossRef]
- Slavin, J.L. Dietary fiber and body weight. Nutrition 2005, 21, 411–418. [Google Scholar] [CrossRef]
- Howarth, N.C.; Huang, T.T.; Roberts, S.B.; McCrory, M.A. Dietary fiber and fat are associated with excess weight in young and middle-aged U.S. adults. J. Am. Diet. Assoc. 2005, 105, 1365–1372. [Google Scholar] [CrossRef]
- Du, H.; van der, A.D.; Boshuizen, H.C.; Forouhi, N.G.; Wareham, N.J.; Halkjaer, J.; Tjonneland, A.; Overvad, K.; Jakobsen, M.U.; Boeing, H.; et al. Dietary fiber and subsequent changes in body weight and waist circumference in European men and women. Am. J. Clin. Nutr. 2010, 91, 329–336. [Google Scholar] [CrossRef]
- Zhao, L. The gut microbiota and obesity: From correlation to causality. Nat. Rev. Microbiol. 2013, 11, 639–647. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Greenhill, C. Obesity: Gut microbiota, host genetics and diet interact to affect the risk of developing obesity and the metabolic syndrome. Nat. Rev. Endocrinol. 2015, 11, 630. [Google Scholar] [CrossRef]
- Parks, B.W.; Nam, E.; Org, E.; Kostem, E.; Norheim, F.; Hui, S.T.; Pan, C.; Civelek, M.; Rau, C.D.; Bennett, B.J.; et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013, 17, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Bouter, K.E.; van Raalte, D.H.; Groen, A.K.; Nieuwdorp, M. Role of the gut microbiome in the pathogenesis of obesity and obesity-related metabolic dysfunction. Gastroenterology 2017, 152, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Osto, M.; Geurts, L.; Everard, A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes 2012, 3, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Vangay, P.; Johnson, A.J.; Ward, T.L.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Hillmann, B.M.; Lucas, S.K.; Beura, L.K.; Thompson, E.A.; Till, L.M.; et al. U.S. immigration westernizes the human gut microbiome. Cell 2018, 175, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Suh, J.M.; Reilly, S.M.; Yu, E.; Osborn, O.; Lackey, D.; Yoshihara, E.; Perino, A.; Jacinto, S.; Lukasheva, Y.; et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat. Med. 2015, 21, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Bifidobacterium with the role of 5-hydroxytryptophan synthesis regulation alleviates the symptom of depression and related microbiota dysbiosis. J. Nutr. Biochem. 2019, 66, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef]
- Wang, G.; Yu, Y.; Garcia-Gutierrez, E.; Jin, X.; He, Y.; Wang, L.; Tian, P.; Liu, Z.; Zhao, J.; Zhang, H.; et al. Lactobacillus acidophilus JCM 1132 strain and its mutant with different bacteriocin-producing behaviour have various in situ effects on the gut microbiota of healthy mice. Microorganisms 2019, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Neyrinck, A.M.; Schüppel, V.L.; Lockett, T.; Haller, D.; Delzenne, N.M. Microbiome and metabolic disorders related to obesity: Which lessons to learn from experimental models? Trends Food Sci. Technol. 2016, 57, 256–264. [Google Scholar] [CrossRef]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; Gonzalez, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef]
- Chambers, E.S.; Byrne, C.S.; Morrison, D.J.; Murphy, K.G.; Preston, T.; Tedford, C.; Garcia-Perez, I.; Fountana, S.; Serrano-Contreras, J.I.; Holmes, E.; et al. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: A randomised cross-over trial. Gut 2019, 68, 1430–1438. [Google Scholar] [CrossRef]
- Dalby, M.J.; Ross, A.W.; Walker, A.W.; Morgan, P.J. Dietary uncoupling of gut microbiota and energy harvesting from obesity and glucose tolerance in mice. Cell Rep. 2017, 21, 1521–1533. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Petersen, C.; Bell, R.; Klag, K.A.; Lee, S.-H.; Soto, R.; Ghazaryan, A.; Buhrke, K.; Ekiz, H.A.; Ost, K.S.; Boudina, S.; et al. T cell–mediated regulation of the microbiota protects against obesity. Science 2019, 365, eaat9351. [Google Scholar] [CrossRef]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.Y.; Ha, C.W.; Campbell, C.R.; Mitchell, A.J.; Dinudom, A.; Oscarsson, J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE 2012, 7, e34233. [Google Scholar] [CrossRef]
- Everard, A.; Lazarevic, V.; Gaïa, N.; Johansson, M.; Ståhlman, M.; Backhed, F.; Delzenne, N.M.; Schrenzel, J.; François, P.; Cani, P.D. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014, 8, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xiao, X.; Zhang, Q.; Zheng, J.; Li, M.; Yu, M.; Wang, X.; Deng, M.; Zhai, X.; Li, R. Improved glucose and lipid metabolism in the early life of female offspring by maternal dietary genistein is associated with alterations in the gut microbiota. Front. Endocrinol. 2018, 9, 516. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Hu, H.; Yao, Y.; Zhao, D.; Wang, S.; Pan, C.; Duan, X.; Gao, Y.; Liu, J.; Zhang, Y.; et al. Coordinated changes of gut microbiome and lipidome differentiates nonalcoholic steatohepatitis (NASH) from isolated steatosis. Liver Int. 2020, 40, 622–637. [Google Scholar] [CrossRef]
- Seedorf, H.; Griffin, N.W.; Ridaura, V.K.; Reyes, A.; Cheng, J.; Rey, F.E.; Smith, M.I.; Simon, G.M.; Scheffrahn, R.H.; Woebken, D.; et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 2014, 159, 253–266. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Galvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef]
- Barouei, J.; Bendiks, Z.; Martinic, A.; Mishchuk, D.; Heeney, D.; Hsieh, Y.H.; Kieffer, D.; Zaragoza, J.; Martin, R.; Slupsky, C.; et al. Microbiota, metabolome, and immune alterations in obese mice fed a high-fat diet containing type 2 resistant starch. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef]
- Obanda, D.; Page, R.; Guice, J.; Raggio, A.M.; Husseneder, C.; Marx, B.; Stout, R.W.; Welsh, D.A.; Taylor, C.M.; Luo, M.; et al. CD obesity-prone rats, but not obesity-resistant rats, robustly ferment resistant starch without increased weight or fat accretion. Obesity 2018, 26, 570–577. [Google Scholar] [CrossRef]
- Cao, W.; Chin, Y.; Chen, X.; Mi, Y.; Xue, C.; Wang, Y.; Tang, Q. The role of gut microbiota in the resistance to obesity in mice fed a high fat diet. Int. J. Food Sci. Nutr. 2020, 71, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H.; Gholami, A.M.; Berry, D.; Desmarchelier, C.; Hahne, H.; Loh, G.; Mondot, S.; Lepage, P.; Rothballer, M.; Walker, A.; et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014, 8, 295–308. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Loy, A.; Pfann, C.; Steinberger, M.; Hanson, B.; Herp, S.; Brugiroux, S.; Gomes Neto, J.C.; Boekschoten, M.V.; Schwab, C.; Urich, T.; et al. Lifestyle and horizontal gene transfer-mediated evolution of mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems 2017, 2, e00171-16. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Lin, D.; Zhao, Y.; Li, W.; Yang, X. Effects of dietary fiber supplementation on fatty acid metabolism and intestinal microbiota diversity in C57BL/6J mice fed with a high-fat diet. J. Agric. Food Chem. 2018, 66, 12706–12718. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Kuzyk, O.; Rauch, I.; Heider, S.; Schwab, C.; Hainzl, E.; Decker, T.; Müller, M.; Strobl, B.; Schleper, C.; et al. Intestinal microbiota signatures associated with inflammation history in mice experiencing recurring colitis. Front. Microbiol. 2015, 6, 1408. [Google Scholar] [CrossRef] [PubMed]
- Neyrinck, A.M.; Van Hée, V.F.; Piront, N.; De Backer, F.; Toussaint, O.; Cani, P.D.; Delzenne, N.M. Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutr. Diabet. 2012, 2, e28. [Google Scholar] [CrossRef]
- Moya-Pérez, A.; Neef, A.; Sanz, Y. Bifidobacterium pseudocatenulatum CECT 7765 reduces obesity-associated inflammation by restoring the lymphocyte-macrophage balance and gut microbiota structure in high-fat diet-fed mice. PLoS ONE 2015, 10, e0126976. [Google Scholar] [CrossRef]
- Ozato, N.; Saito, S.; Yamaguchi, T.; Katashima, M.; Tokuda, I.; Sawada, K.; Katsuragi, Y.; Kakuta, M.; Imoto, S.; Ihara, K.; et al. Blautia genus associated with visceral fat accumulation in adults 20–76 years of age. NPJ Biofilms Microbiomes 2019, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, C.X.; Katiraei, S.; Kooijman, S.; Zhou, E.; Chung, C.K.; Gao, Y.; van den Heuvel, J.K.; Meijer, O.C.; Berbee, J.F.P.; et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut 2018, 67, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed]




| Ingredient, g/kg | TP 23300 | TP 23302 |
|---|---|---|
| Casein | 259 | 189 |
| Maltodextrin | 166 | 108 |
| Sucrose | 91 | 67 |
| Corn Starch | 0 | 497 |
| Soybean Oil | 33 | 21 |
| Lard | 313 | 21 |
| Cellulose | 67 | 48 |
| Mineral Mix, M1020 | 52 | 36 |
| Vitamin Mix, V1010 | 13 | 9 |
| L-Cystine | 3 | 2 |
| Choline Bitartrate | 3 | 2 |
| Tertiary butylhydroquinone (TBHQ) | 0.067 | 0.067 |
| Total | 1000.067 | 1000.067 |
| Gene | PrimerBank ID | Forward Primer | Reverse Primer |
|---|---|---|---|
| Ucp1 | 6678497a1 | AGGCTTCCAGTACCATTAGGT | CTGAGTGAGGCAAAGCTGATTT |
| Pgc1α | 6679433a1 | TATGGAGTGACATAGAGTGTGCT | CCACTTCAATCCACCCAGAAAG |
| Ppar-α | 31543500a1 | AGAGCCCCATCTGTCCTCTC | ACTGGTAGTCTGCAAAACCAAA |
| Ppar-γ | 6755138a1 | TCGCTGATGCACTGCCTATG | GAGAGGTCCACAGAGCTGATT |
| Fasn | 30911099a1 | GGAGGTGGTGATAGCCGGTAT | TGGGTAATCCATAGAGCCCAG |
| Atgl | 26327465a1 | GGATGGCGGCATTTCAGACA | CAAAGGGTTGGGTTGGTTCAG |
| Mgl | 6754690a1 | CGGACTTCCAAGTTTTTGTCAGA | GCAGCCACTAGGATGGAGATG |
| Hsl | 26325924a1 | CCAGCCTGAGGGCTTACTG | CTCCATTGACTGTGACATCTCG |
| Gapdh | 6679937a1 | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference. Nutrients 2020, 12, 3197. https://doi.org/10.3390/nu12103197
Wang B, Kong Q, Li X, Zhao J, Zhang H, Chen W, Wang G. A High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference. Nutrients. 2020; 12(10):3197. https://doi.org/10.3390/nu12103197
Chicago/Turabian StyleWang, Botao, Qingmin Kong, Xiu Li, Jianxin Zhao, Hao Zhang, Wei Chen, and Gang Wang. 2020. "A High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference" Nutrients 12, no. 10: 3197. https://doi.org/10.3390/nu12103197
APA StyleWang, B., Kong, Q., Li, X., Zhao, J., Zhang, H., Chen, W., & Wang, G. (2020). A High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference. Nutrients, 12(10), 3197. https://doi.org/10.3390/nu12103197





