Effects of Flaxseed and Its Components on Mammary Gland MiRNome: Identification of Potential Biomarkers to Prevent Breast Cancer Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Study and Dietary Treatments
2.2. Serum Lignan Analysis
2.3. NanoString nCounter miRNA Profiling and Data Analysis
2.4. MiRNAs Target Prediction and Pathway Analysis
2.5. Statistical Analysis
3. Results
3.1. Body Weights and Food Intake
3.2. Serum Lignan Concentration
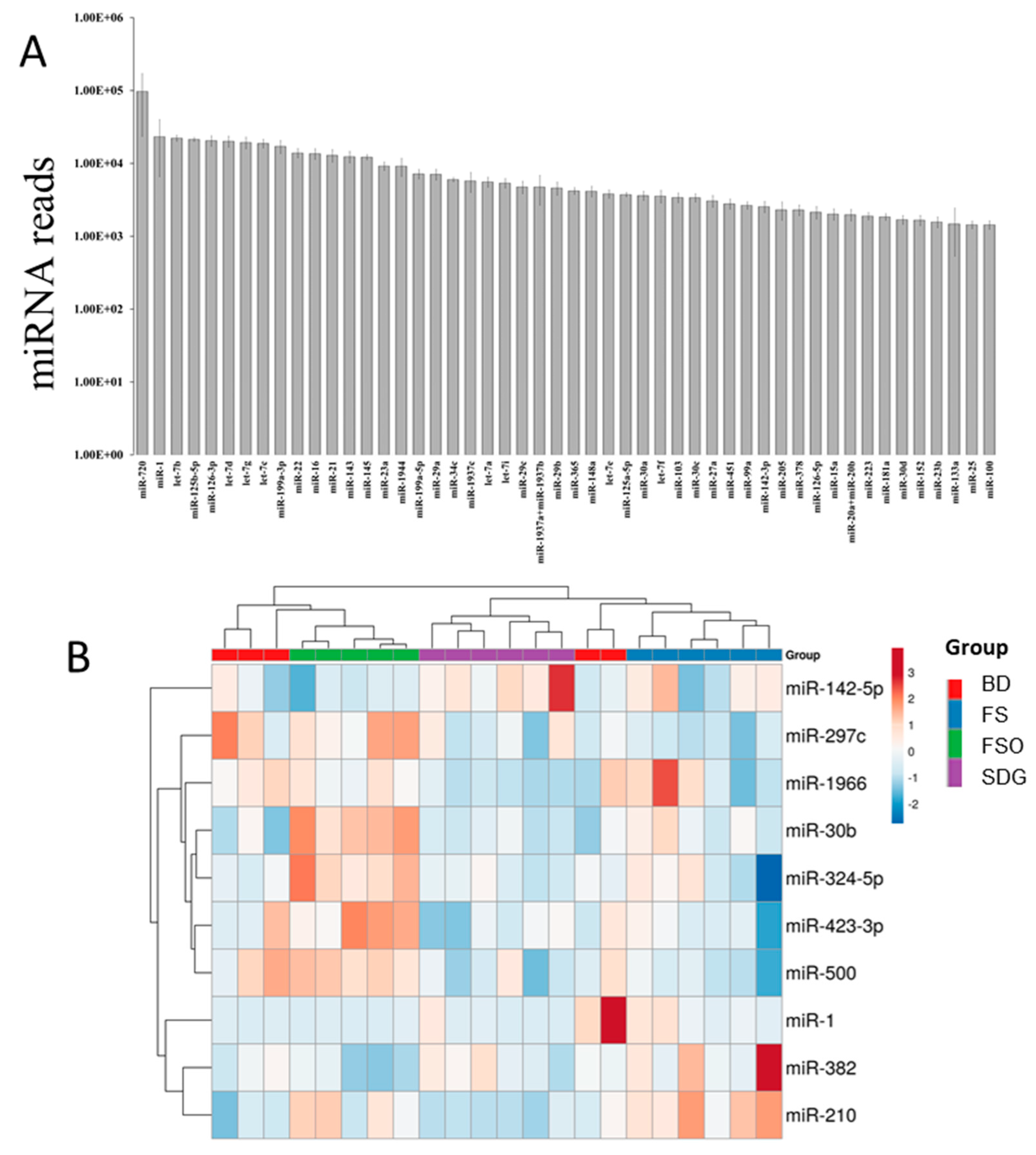
3.3. Mammary Gland miRNome
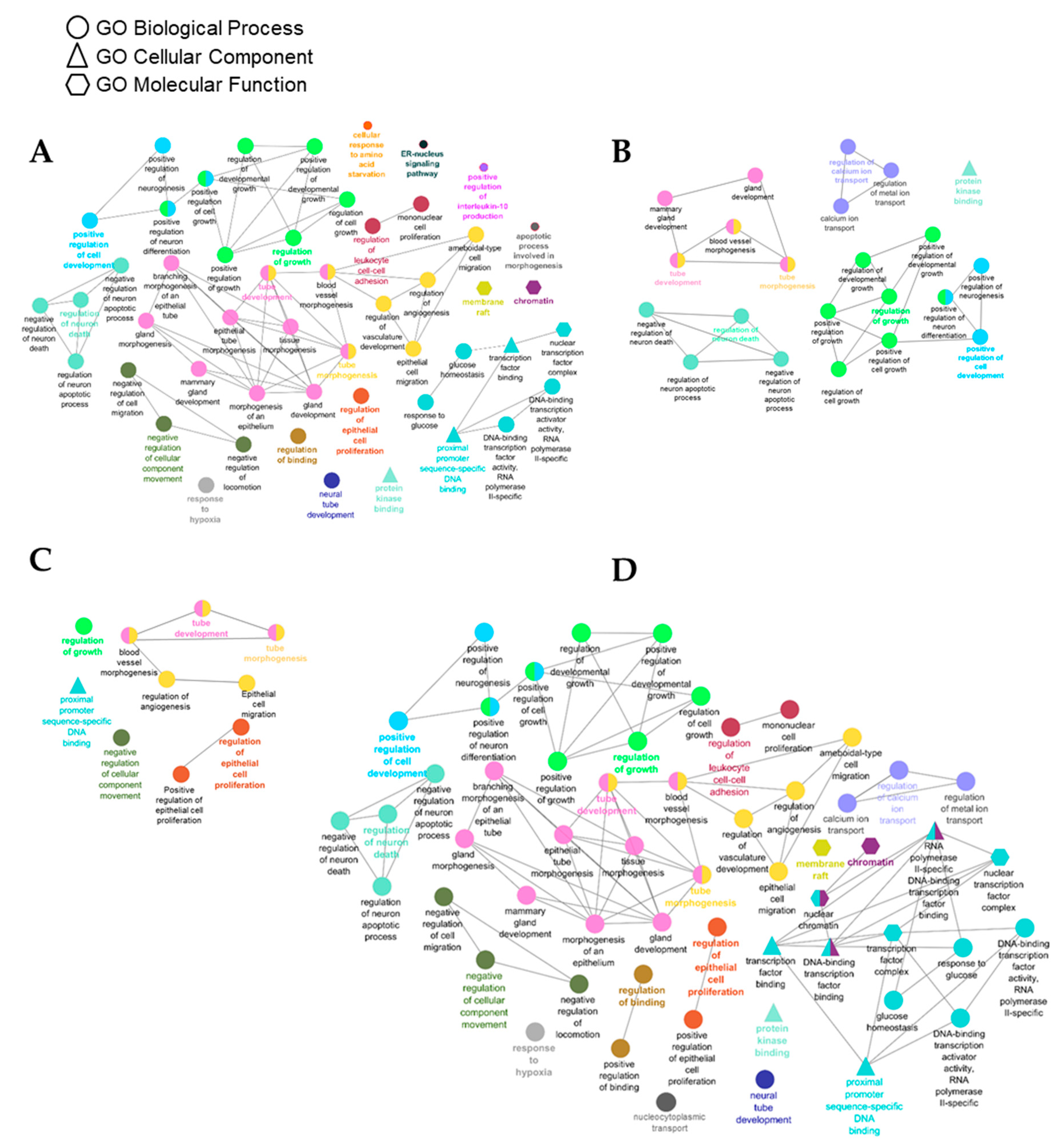
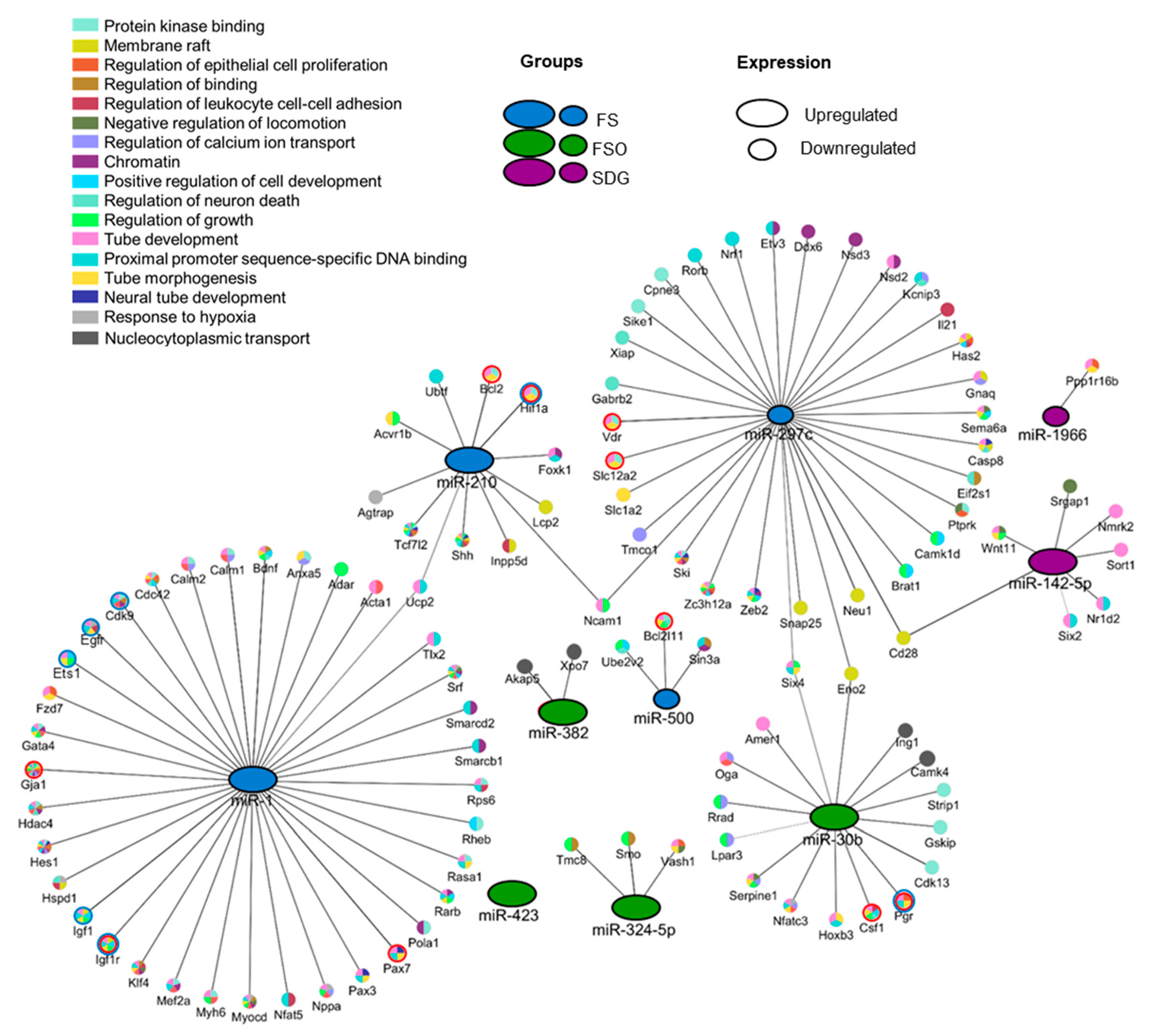
3.4. Identification of Gene Targets of Deregulated miRNAs and Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeSantis, C.; Ma, J.; Bryan, L.; Jemal, A. Breast cancer statistics, 2013. CA A Cancer J. Clin. 2014, 64, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, L. Breast cancer: Challenges, controversies, breakthroughs. Nat. Rev. Clin. Oncol. 2010, 7, 669–670. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.K.; Thompson, L.U. Flaxseed and its lignan and oil components: Can they play a role in reducing the risk of and improving the treatment of breast cancer? Appl. Physiol. Nutr. Metab. 2014, 39, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Hall, C., 3rd; Tulbek, M.C.; Xu, Y. Flaxseed. Adv. Food Nutr. Res. 2006, 51, 1–97. [Google Scholar] [CrossRef]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef]
- Thompson, L.U.; Rickard, S.E.; Orcheson, L.J.; Seidl, M.M. Flaxseed and its lignan and oil components reduce mammary tumor growth at a late stage of carcinogenesis. Carcinogenesis 1996, 17, 1373–1376. [Google Scholar] [CrossRef]
- Chen, J.; Stavro, P.M.; Thompson, L.U. Dietary flaxseed inhibits human breast cancer growth and metastasis and downregulates expression of insulin-like growth factor and epidermal growth factor receptor. Nutr. Cancer 2002, 43, 187–192. [Google Scholar] [CrossRef]
- Tou, J.C.; Thompson, L.U. Exposure to flaxseed or its lignan component during different developmental stages influences rat mammary gland structures. Carcinogenesis 1999, 20, 1831–1835. [Google Scholar] [CrossRef]
- Li, Y.; Kowdley, K.V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012, 431, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Gigli, I.; Maizon, D.O. microRNAs and the mammary gland: A new understanding of gene expression. Genet. Mol. Biol. 2013, 36, 465–474. [Google Scholar] [CrossRef]
- Silveri, L.; Tilly, G.; Vilotte, J.L.; Le Provost, F. MicroRNA involvement in mammary gland development and breast cancer. Reprod. Nutr. Dev. 2006, 46, 549–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. MiRNA dysregulation in breast cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar] [CrossRef] [PubMed]
- Adhami, M.; Haghdoost, A.A.; Sadeghi, B.; Malekpour Afshar, R. Candidate miRNAs in human breast cancer biomarkers: A systematic review. Breast Cancer 2018, 25, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Kapinova, A.; Kubatka, P.; Golubnitschaja, O.; Kello, M.; Zubor, P.; Solar, P.; Pec, M. Dietary phytochemicals in breast cancer research: Anticancer effects and potential utility for effective chemoprevention. Environ. Health Prev. Med. 2018, 23, 36. [Google Scholar] [CrossRef]
- LeMay-Nedjelski, L.; Mason-Ennis, J.K.; Taibi, A.; Comelli, E.M.; Thompson, L.U. Omega-3 polyunsaturated fatty acids time-dependently Reduce cell viability and oncogenic microRNA-21 expression in estrogen receptor-positive breast cancer cells (MCF-7). Int. J. Mol. Sci. 2018, 19, 244. [Google Scholar] [CrossRef]
- Reeves, P.G. Components of the AIN-93 diets as improvements in the AIN-76A diet. J. Nutr. 1997, 127, 838s–841s. [Google Scholar] [CrossRef]
- Mason, J.K.; Fu, M.; Chen, J.; Thompson, L.U. Flaxseed oil enhances the effectiveness of trastuzumab in reducing the growth of HER2-overexpressing human breast tumors (BT-474). J. Nutr. Biochem. 2015, 26, 16–23. [Google Scholar] [CrossRef]
- Rickard, S.E.; Yuan, Y.V.; Thompson, L.U. Plasma insulin-like growth factor I levels in rats are reduced by dietary supplementation of flaxseed or its lignan secoisolariciresinol diglycoside. Cancer Lett. 2000, 161, 47–55. [Google Scholar] [CrossRef]
- Zarepoor, L.; Lu, J.T.; Zhang, C.; Wu, W.; Lepp, D.; Robinson, L.; Wanasundara, J.; Cui, S.; Villeneuve, S.; Fofana, B.; et al. Dietary flaxseed intake exacerbates acute colonic mucosal injury and inflammation induced by dextran sodium sulfate. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G1042–G1055. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tan, K.P.; Ward, W.E.; Thompson, L.U. Exposure to flaxseed or its purified lignan during suckling inhibits chemically induced rat mammary tumorigenesis. Exp. Biol. Med. 2003, 228, 951–958. [Google Scholar] [CrossRef]
- Tan, K.P.; Chen, J.; Ward, W.E.; Thompson, L.U. Mammary gland morphogenesis is enhanced by exposure to flaxseed or its major lignan during suckling in rats. Exp. Biol. Med. (Maywood N.J.) 2004, 229, 147–157. [Google Scholar] [CrossRef]
- Avril-Sassen, S.; Goldstein, L.D.; Stingl, J.; Blenkiron, C.; Le Quesne, J.; Spiteri, I.; Karagavriilidou, K.; Watson, C.J.; Tavare, S.; Miska, E.A.; et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genom. 2009, 10, 548. [Google Scholar] [CrossRef]
- Rudel, R.A.; Fenton, S.E.; Ackerman, J.M.; Euling, S.Y.; Makris, S.L. Environmental exposures and mammary gland development: State of the science, public health implications, and research recommendations. Environ. Health Perspect. 2011, 119, 1053–1061. [Google Scholar] [CrossRef]
- Forman, M.R.; Winn, D.M.; Collman, G.W.; Rizzo, J.; Birnbaum, L.S. Environmental exposures, breast development and cancer risk: Through the looking glass of breast cancer prevention. Reprod. Toxicol. 2015, 54, 6–10. [Google Scholar] [CrossRef]
- Wu, L.; Wang, T.; He, D.; Li, X.; Jiang, Y. miR-1 inhibits the proliferation of breast cancer stem cells by targeting EVI-1. Onco Targets Ther. 2018, 11, 8773–8781. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.; Lai, Y.; Liao, Y.; Liu, R.; Qiu, W. Hsa-miR-1 suppresses breast cancer development by down-regulating K-ras and long non-coding RNA MALAT1. Int. J. Biol. Macromol. 2015, 81, 491–497. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Fukumoto, I.; Kikkawa, N.; Matsushita, R.; Mataki, H.; Mizuno, K.; Okamoto, Y.; Seki, N. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J. Hum. Genet. 2017, 62, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Song, X.; Tian, H.; Miao, Y.; Feng, X.; Li, Y.; Wang, H. MicroRNA-137 inhibits growth of glioblastoma through EGFR suppression. Am. J. Transl. Res. 2017, 9, 1492–1499. [Google Scholar] [PubMed]
- Xu, F.; Li, F.; Zhang, W.; Jia, P. Growth of glioblastoma is inhibited by miR-133-mediated EGFR suppression. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 9553–9558. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Wu, D.; Xu, B.; Qian, W.; Li, P.; Lu, Q.; Yin, C.; Zhang, W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol. Rep. 2012, 27, 1967–1975. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kosaka, N.; Tanaka, M.; Koizumi, F.; Kanai, Y.; Mizutani, T.; Murakami, Y.; Kuroda, M.; Miyajima, A.; Kato, T.; et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009, 14, 529–538. [Google Scholar] [CrossRef]
- Jiang, M.; Zhou, L.Y.; Xu, N.; An, Q. Down-regulation of miR-500 and miR-628 suppress non-small cell lung cancer proliferation, migration and invasion by targeting ING1. Biomed. Pharmacother. 2018, 108, 1628–1639. [Google Scholar] [CrossRef]
- Fishel, R.; Croce Carlo, M.; Kopelovich, L.; Aronow, B.; Martin-Lopez, J. Materials and Methods Related to NSAID Chemoprevention in Colorectal Cancer. US Patent 2015/0292023 A1, 15 October 2015. [Google Scholar]
- Ichikawa, T.; Sato, F.; Terasawa, K.; Tsuchiya, S.; Toi, M.; Tsujimoto, G.; Shimizu, K. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS ONE 2012, 7, e31422. [Google Scholar] [CrossRef]
- Le Guillou, S.; Sdassi, N.; Laubier, J.; Passet, B.; Vilotte, M.; Castille, J.; Laloe, D.; Polyte, J.; Bouet, S.; Jaffrezic, F.; et al. Overexpression of miR-30b in the developing mouse mammary gland causes a lactation defect and delays involution. PLoS ONE 2012, 7, e45727. [Google Scholar] [CrossRef]
- Turashvili, G.; Lightbody, E.D.; Tyryshkin, K.; SenGupta, S.K.; Elliott, B.E.; Madarnas, Y.; Ghaffari, A.; Day, A.; Nicol, C.J.B. Novel prognostic and predictive microRNA targets for triple-negative breast cancer. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018. [Google Scholar] [CrossRef]
- Mar-Aguilar, F.; Mendoza-Ramirez, J.A.; Malagon-Santiago, I.; Espino-Silva, P.K.; Santuario-Facio, S.K.; Ruiz-Flores, P.; Rodriguez-Padilla, C.; Resendez-Perez, D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis. Markers 2013, 34, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.Y.; Hsu, R.J.; Liu, J.M.; Chen, S.C.; Liao, G.S.; Gao, H.W.; Yu, C.P. MicroRNA-382-5p aggravates breast cancer progression by regulating the RERG/Ras/ERK signaling axis. Oncotarget 2017, 8, 22443–22459. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Liu, T.; Huang, W.; Liu, H.; Zhang, H.M.; Li, Q.; Chen, Z.; Guo, A.Y. MicroRNA regulatory pathway analysis identifies miR-142-5p as a negative regulator of TGF-beta pathway via targeting SMAD3. Oncotarget 2016, 7, 71504–71513. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, L.; Yang, R.; Wei, M.; Sheng, Y.; Ji, L. Identification of serum microRNAs as potential toxicological biomarkers for toosendanin-induced liver injury in mice. Phytomed. Int. J. Phytother. Phytopharm. 2019, 58, 152867. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, P.F.; Msaouel, P.; Koutsilieris, M. The role of the insulin-like growth factor-1 system in breast cancer. Mol. Cancer 2015, 14, 43. [Google Scholar] [CrossRef]
- Weroha, S.J.; Haluska, P. The insulin-like growth factor system in cancer. Endocrinol. Metab. Clin. N. Am. 2012, 41, 335–350. [Google Scholar] [CrossRef]
- Mitra, P.; Yang, R.M.; Sutton, J.; Ramsay, R.G.; Gonda, T.J. CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget 2016, 7, 9069–9083. [Google Scholar] [CrossRef] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taibi, A.; Lin, Z.; Tsao, R.; Thompson, L.U.; Comelli, E.M. Effects of Flaxseed and Its Components on Mammary Gland MiRNome: Identification of Potential Biomarkers to Prevent Breast Cancer Development. Nutrients 2019, 11, 2656. https://doi.org/10.3390/nu11112656
Taibi A, Lin Z, Tsao R, Thompson LU, Comelli EM. Effects of Flaxseed and Its Components on Mammary Gland MiRNome: Identification of Potential Biomarkers to Prevent Breast Cancer Development. Nutrients. 2019; 11(11):2656. https://doi.org/10.3390/nu11112656
Chicago/Turabian StyleTaibi, Amel, Zhen Lin, Rong Tsao, Lilian U. Thompson, and Elena M. Comelli. 2019. "Effects of Flaxseed and Its Components on Mammary Gland MiRNome: Identification of Potential Biomarkers to Prevent Breast Cancer Development" Nutrients 11, no. 11: 2656. https://doi.org/10.3390/nu11112656
APA StyleTaibi, A., Lin, Z., Tsao, R., Thompson, L. U., & Comelli, E. M. (2019). Effects of Flaxseed and Its Components on Mammary Gland MiRNome: Identification of Potential Biomarkers to Prevent Breast Cancer Development. Nutrients, 11(11), 2656. https://doi.org/10.3390/nu11112656




