Allium hookeri Root Extract Inhibits Adipogenesis by Promoting Lipolysis in High Fat Diet-Induced Obese Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Regents
2.2. Cell Culture and Differentiation
2.3. MTT Assay
2.4. Real-Time Polymerase Chain Reaction (RT-PCR) Analysis
2.5. Oil Red O Staining and Triglyceride Assay
2.6. Animal Studies
2.7. Serum Biochemical Analyses
2.8. Gene Expression Analysis
2.9. Histological Analysis
2.10. Statistical Analysis
3. Results
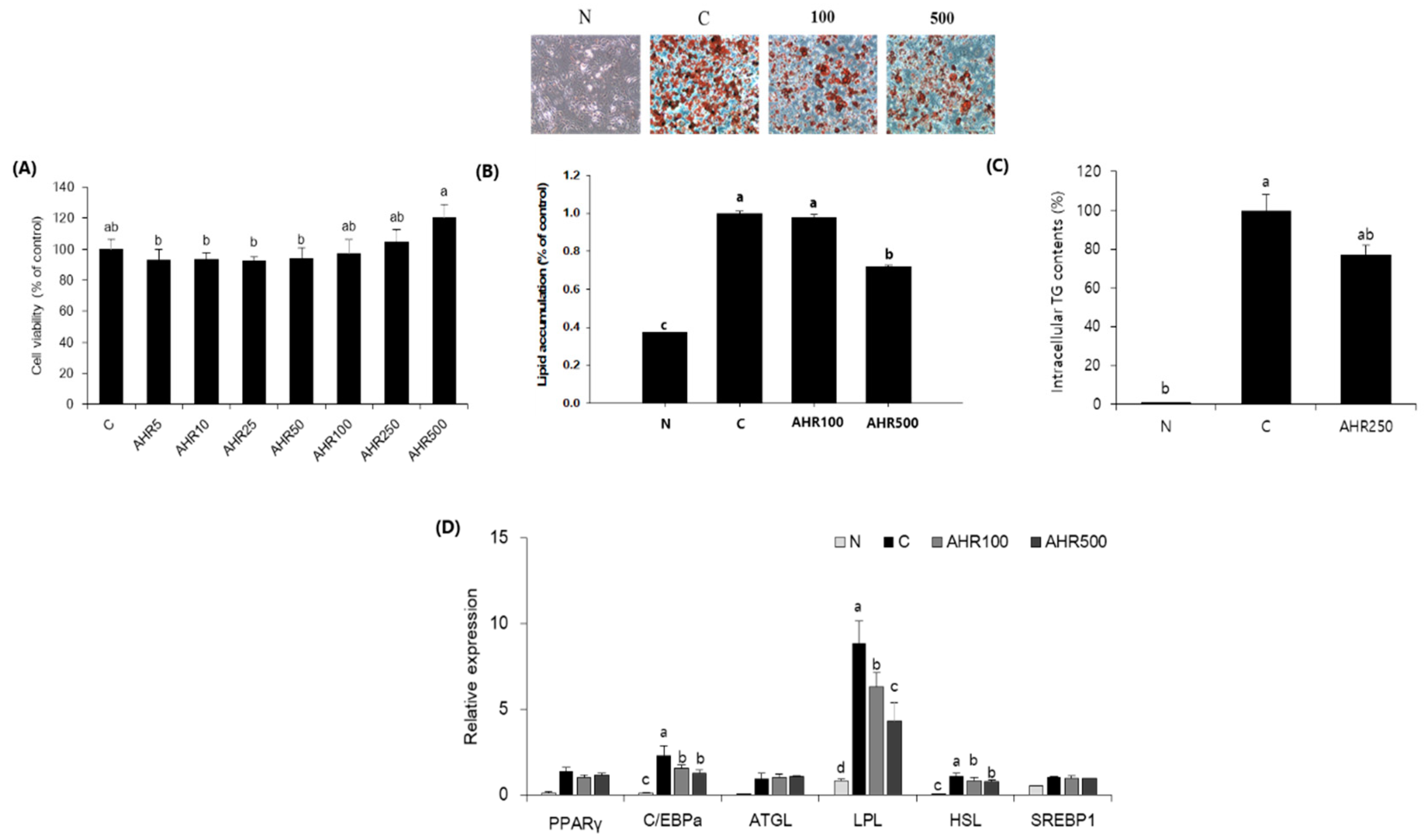
3.1. Fat Accumulation and mRNA Expression in 3T3-L1 Adipocytes
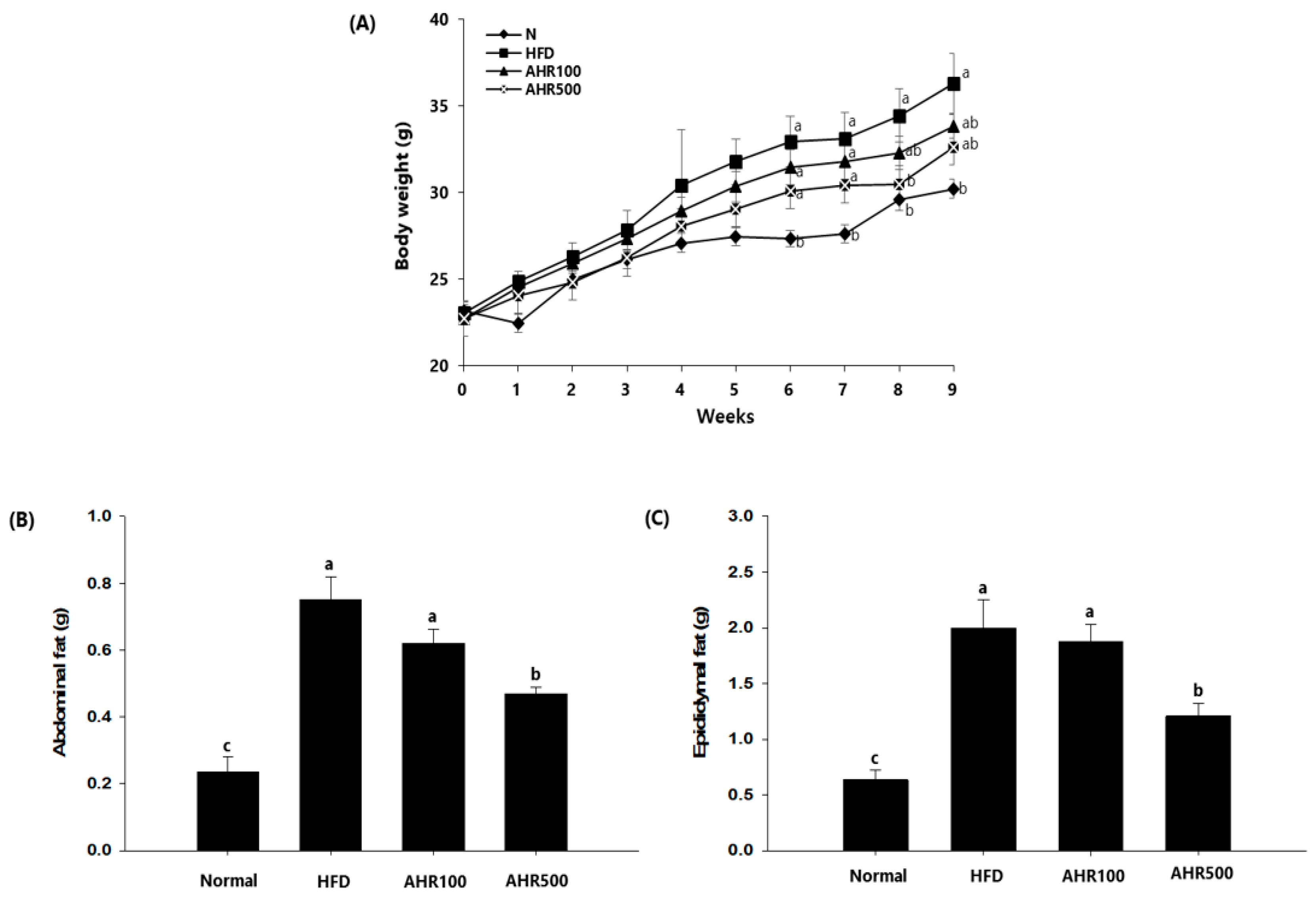
3.2. Body and Tissue Weights
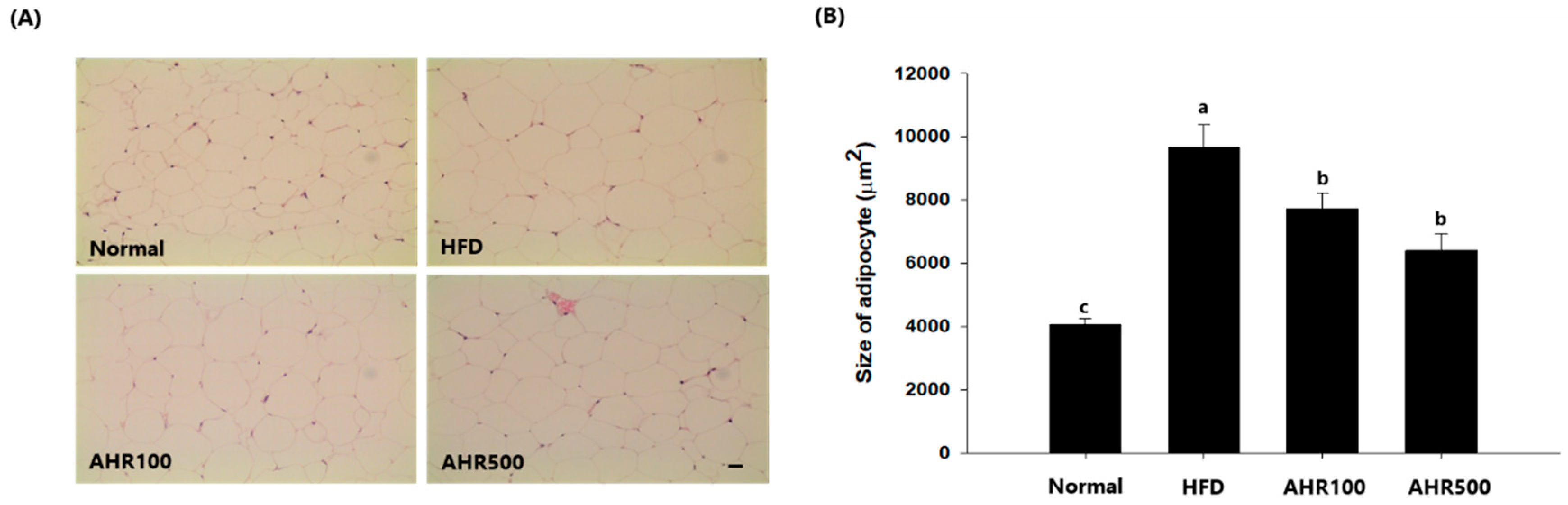
3.3. Cell Size of Epididymal Adipose Tissue
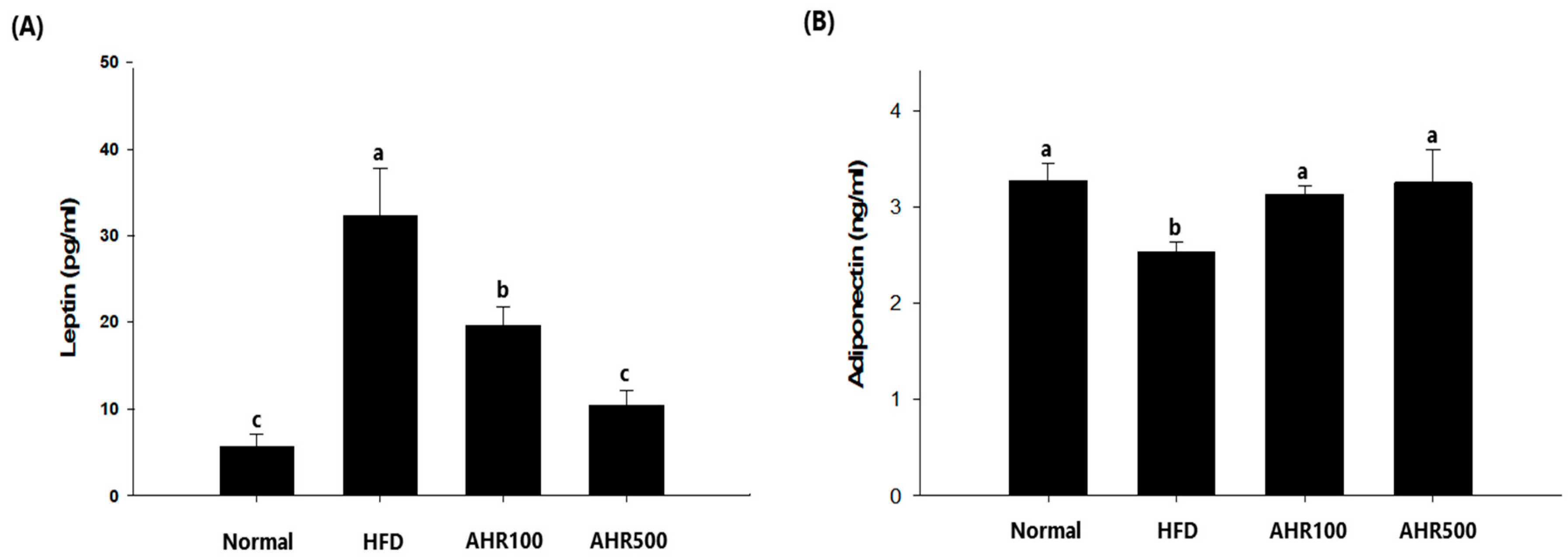
3.4. Serum Measurements
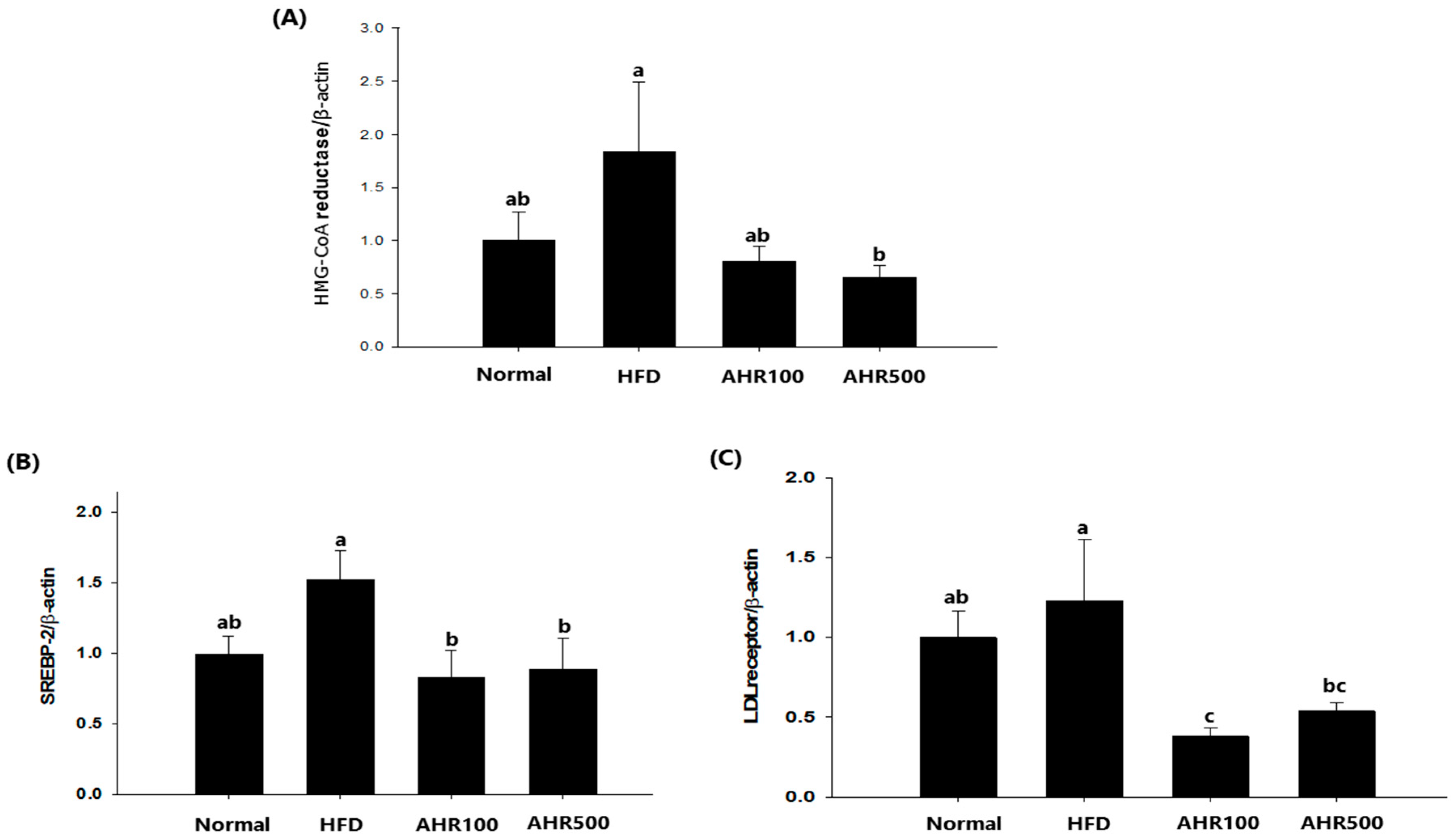
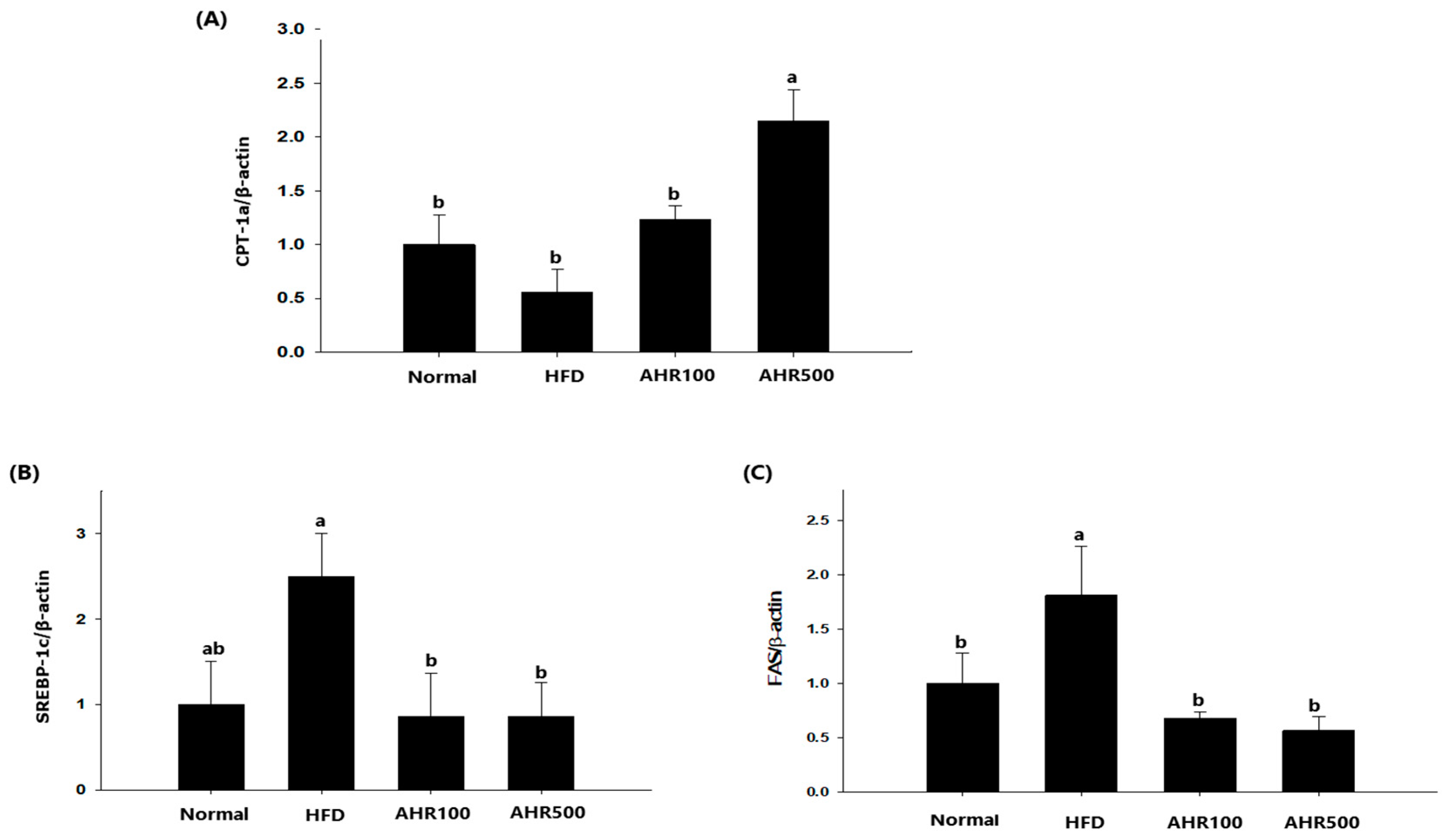
3.5. mRNA Expression Level in the Liver
3.6. mRNA Expression Level in Epididymal Adipose Tissue
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Heymsfield, S.B.; Wadden, T.A. Mechanism, pathophysiology, and management of obesity. N. Engl. J. Med. 2017, 376, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.O.; Peters, J.C. Environmental contributions to the obesity epidemic. Science 1998, 280, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Gavrilova, O.; Pack, S.; Jou, W.; Mullen, S.; Sumner, A.E.; Cushman, S.W.; Periwal, V. Hypertrophy and/or hyperplasia: Dynamics of adipose tissue growth. PLoS Comput. Biol. 2009, 5, e1000324. [Google Scholar] [CrossRef] [PubMed]
- Scherer, P.E. Adipose tissue: From lipid storage compartment to endocrine organ. Diabetes 2006, 55, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Lass, A.; Haemmerle, G.; Zechner, R. Fate of fat: The role of adipose triglyceride lipase in lipolysis. Biochim. Biophys. Acta 2009, 1791, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Zechner, R.; Kienesberger, P.C.; Haemmerle, G.; Zimmermann, R.; Lass, A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 2009, 50, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Yajima, H.; Kobayashi, Y.; Kanaya, T.; Horino, Y. Identification of peroxisome-proliferator responsive element in the mouse HSL gene. Biochem. Biophys. Res. Commun. 2007, 352, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Ko, E.H.; Kim, J.E.; Kim, E.; Lee, H.; Choi, H.; Yu, J.H.; Kim, H.J.; Seong, J.K.; Kim, K.S.; et al. Nuclear receptor PPARγ–regulated monoacylglycerol O-acyltransferase 1 expression is responsible for the lipid accumulation in diet-induced hepatic steatosis. Proc. Natl. Acad. Sci. USA 2012, 109, 13656–13661. [Google Scholar] [CrossRef]
- Den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J.; et al. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ–dependent switch from lipogenesis to fat oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Petrovska, B.B. Historical review of medicinal plants’ usage. Pharmacogn. Rev. 2012, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Gohil, R.N.; Kaul, V. Cytological status of Allium hookeri Thwaites (2n = 22). Genet. Resour. Crop Evol. 2011, 58, 1041–1050. [Google Scholar] [CrossRef]
- Rhyu, D.Y.; Park, S.H. Characterization of alkyl thiosulfinate in Allium hookeri root using HPLC-ESI-MS. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 457–459. [Google Scholar] [CrossRef]
- Ayam, V.S. Allium hookeri, Thw.Enum. A lesser known terrestrial perennial herb used as food and its ethnobotanical relevance in Manipur. Afr. J. Food Agric. Nutr. Dev. 2011, 11, 5389–5412. [Google Scholar]
- Hwang, J.S.; Lee, B.H.; An, X.; Jeong, H.R.; Kim, Y.E.; Lee, I.; Lee, H.; Kim, D.O. Total phenolics, total flavonoids, and antioxidant capacity in the leaves, bulbs, and roots of Allium hookeri. Korean J. Food Sci. Technol. 2015, 47, 261–266. [Google Scholar] [CrossRef]
- Lee, E.B.; Kim, J.H.; Yang, J.H.; Kim, Y.S.; Jun, H.I.; Kim, B.H.; Lee, S.H.; Kim, Y.S.; Han, S.C.; Kim, D.K. Antioxidant and longevity properties of the root of Allium hookeri in Caenorhabditis elegans. Korean J. Pharmacogn. 2015, 46, 234–242. [Google Scholar]
- Jun, H.I.; Jang, H.; Ahn, D.; Kim, D.K.; Yang, J.H.; Yun, B.S.; Kim, Y.S. Isolation and characterization of phenolic compound from Allium hookeri root for potential use as antioxidant in foods. Food Sci. Biotechnol. 2015, 24, 2031–2034. [Google Scholar] [CrossRef]
- Li, R.; Wang, Y.F.; Sun, Q.; Hu, H.B. Chemical composition and antimicrobial activity of the essential oil from Allium hookeri consumed in Xishuangbanna, southwest China. Nat. Prod. Commun. 2014, 9, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.S.; Choi, B.K.; Lee, S.H.; Jang, H.H.; Kim, J.B.; Kim, H.R.; Kim, D.K.; Kim, Y.S.; Yang, J.H.; Kim, H.J.; et al. Effects of Allium hookeri on glucose metabolism in type II diabetic mice. Korean J. Pharmacogn. 2015, 46, 78–83. [Google Scholar]
- Singh, K.D.; Chetia, D.; Mazumdar, S. Anti-diabetic activity of methanolic extract of Allium hookeri leaves. Indo Am. J. Pharm. Sci. 2013, 3, 4098–4104. [Google Scholar]
- Roh, S.S.; Kwon, O.J.; Yang, J.H.; Kim, Y.S.; Lee, S.H.; Jin, J.S.; Jeon, Y.D.; Yokozawa, T.; Kim, H.J. Allium hookeri root protects oxidative stress-induced inflammatory response and β-cell damage in pancreas of streptozotocin-induced diabetic rats. BMC Complement. Altern. Med. 2016, 16, 63. [Google Scholar] [CrossRef]
- Yang, H.S.; Choi, Y.J.; Jin, H.Y.; Lee, S.C.; Huh, C.K. Effects of Allium hookeri root water extracts on inhibition of adipogenesis and GLUT-4 expression in 3T3-L1 adipocytes. Food Sci. Biotechnol. 2016, 25, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; No, K.; Lee, J. Anti-obesity effect of Allium hookeri leaf extract in high-fat diet-fed mice. J. Med. Food 2018, 21, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.C.; Bae, D.Y. The anti-inflammatory effects of ethanol extract of Allium Hookeri cultivated in South Korea. Korea J. Herbol. 2012, 27, 55–61. [Google Scholar] [CrossRef][Green Version]
- Kim, C.H.; Lee, M.A.; Kim, T.W.; Jang, J.Y.; Kim, H.J. Anti-inflammatory effect of Allium hookeri root methanol extract in LPS-induced RAW264.7 cells. J. Korean Soc. Food Sci. Nutr. 2012, 41, 1645–1648. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, M.J.; You, B.R.; Jin, J.S.; Lee, S.H.; Yun, Y.R.; Kim, H.J. Allium hookeri root extract exerts anti-inflammatory effects by nuclear factor-κB down-regulation in lipopolysaccharide-induced RAW264.7 cells. BMC Complement. Altern. Med. 2017, 17, 126. [Google Scholar] [CrossRef]
- Park, S.Y.; Je, J.Y.; Ahn, C.B. Phenolic composition and hepatoprotective activities of Allium hookeri against hydrogen-peroxide induced oxidative stress in cultured hepatocytes. J. Food Biochem. 2016, 40, 284–293. [Google Scholar] [CrossRef]
- Park, H.; Jeong, J.; Hyun, H.; Kim, J.; Kim, H.; Oh, H.I.; Choi, J.Y.; Hwang, H.S.; Oh, D.B.; Kim, J.I.; et al. Effects of a hot-water extract of Allium hookeri roots on bone formation in human osteoblast-like MG-63 cells in vitro and in rats in vivo. Planta Med. 2016, 82, 1410–1415. [Google Scholar] [CrossRef]
- Lee, H.A.; Hong, S.; Yoo, J.H.; Chung, Y.; Kim, O. Anti-Helicobacter pylori activity and inhibition of gastritis by Allium hookeri extract. Lab. Anim. Res. 2018, 34, 75–79. [Google Scholar] [CrossRef]
- Lee, Y.S.; Lee, S.H.; Gadde, U.D.; Oh, S.T.; Lee, S.J.; Lillehoj, H.S. Allium hookeri supplementation improves intestinal immune response against necrotic enteritis in young broiler chickens. Poult. Sci. 2018, 97, 1899–1908. [Google Scholar] [CrossRef]
- Yang, M.H.; Kim, N.H.; Heo, J.D.; Rho, J.R.; Ock, K.J.; Shin, E.C.; Jeong, E.J. Comparative evaluation of sulfur compounds contents and antiobesity properties of Allium hookeri prepared by different drying methods. Evid. Based Complement. Altern. Med. 2017, 2017, 2436927–2436936. [Google Scholar] [CrossRef]
- Friedman, J. The long road to leptin. J. Clin. Investig. 2016, 126, 4727–4734. [Google Scholar] [CrossRef] [PubMed]
- Yanagita, T.; Han, S.Y.; Wang, Y.M.; Tsuruta, Y.; Anno, T. Cycloalliin, a cyclic sulfur amino acid, reduces serum triacylglycerol in rats. Nutr. J. 2003, 19, 140–143. [Google Scholar] [CrossRef]
- Jang, G.J.; Sung, M.J.; Hur, H.J.; Yoo, M.; Choi, J.H.; Hwang, I.K.; Lee, S. Metabolomics analysis of the lipid-regulating effect of Allium hookeri in a hamster model of high-fat diet-induced hyperlipidemia by UPLC/ESI-Q-TOF mass spectrometry. Evid. Based Complement. Altern. Med. 2018, 2018, 5659174–5659181. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.E.; Ahmadian, M.; Jaworski, K.; Sarkadi-Nagy, E.; Sul, H.S. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 2007, 27, 79–101. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004, 306, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Shen, W.J.; Patel, S.; Natu, V.; Wang, J.; Osuga, J.; Ishibashi, S.; Kraemer, F.B. Resistance to high fat diet-induced obesity and altered expression of adipose-specific genes in HSL-deficient mice. Am. J. Physiol. Endocrinol. Metab. 2003, 285, E1182–E1195. [Google Scholar] [CrossRef] [PubMed]
- Dludla, P.V.; Nkambule, B.B.; Jack, B.; Mkandla, Z.; Mutize, T.; Silvestri, S.; Orlando, P.; Tiano, L.; Louw, J.; Mazibuko-Mbeje, S.E. Inflammation and oxidative stress in an obese state and the protective effects of gallic acid. Nutrients 2019, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Wakil, S.J.; Abu-Elheiga, L.A. Fatty acid metabolism: Target for metabolic syndrome. J. Lipid Res. 2009, 50, S138–S143. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Ontko, J.A. Interactions between fatty acid synthesis, oxidation, and esterification in the production of triglyceride-rich lipoproteins by the liver. J. Lipid Res. 1984, 25, 831–842. [Google Scholar] [PubMed]
- Large, V.; Reynisdottir, S.; Langin, D.; Fredby, K.; Klannemark, M.; Holm, C.; Arner, P. Decreased expression and function of adipocyte hormone-sensitive lipase in subcutaneous fat cell of obese subjects. J. Lipid Res. 1999, 40, 2059–2066. [Google Scholar] [PubMed]
- Reynisdottir, S.; Langin, D.; Carlstrom, K.; Holm, C.; Rossner, S.; Arner, P. Effects of weight reduction on the regulation of lipolysis in adipocytes of women with upper-body obesity. Clin. Sci. 1995, 89, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Ginsberg, H.N. Insulin resistance and cardiovascular disease. J. Clin. Investig. 2000, 106, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Janani, C.; Kumari, B.R. PPAR gamma gene—A review. Diabetes Metab. Syndr. 2015, 9, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Okuno, A.; Tamemoto, H.; Tobe, K.; Ueki, K.; Mori, Y.; Iwamoto, K.; Umesono, K.; Akanuma, Y.; Fujiwara, T.; Horikoshi, H.; et al. Troglitazone increase the number of small adipocytes without change of white adipose tissue mass in obese Zucker rats. J. Clin. Investig. 1998, 101, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Shan, S.; Li, P.P.; Shen, Z.F.; Lu, X.P.; Cheng, J.; Ning, Z.Q. Peroxisome proliferator-activated receptor-γ transcriptionally up-regulates hormone-sensitive lipase via the involvement of specificity protein-1. Endocrinology 2006, 147, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Kamon, J.; Waki, H.; Murakami, K.; Motojima, K.; Komeda, K.; Ide, T.; Kubota, N.; Terauchi, Y.; Tobe, K.; et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J. Biol. Chem. 2001, 276, 41245–41254. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Gogg, S.; Hedjazifar, S.; Jenndahl, L.; Hammarstedt, A.; Smith, U. Inflammation and impaired adipogenesis in hypertrophic obesity in man. Am. J. Endocrinol. Metab. 2009, 279, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Amor, S.; Gonzalez-Hedstrom, D.; Martin-Carro, B.; Inarejos-Garcia, A.M.; Almodovar, P.; Prodanov, M.; Garcia-Villalon, A.L.; Granado, M. Beneficial effects of an aged black garlic extract in the metabolic and vascular alterations induced by a high fat/sucrose diet in male rats. Nutrients 2019, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.W.; Chen, C.S.; Chen, Y.C.; Hsu, Y.A.; Huang, C.C.; Chang, C.Y.; Lin, C.J.; Lin, C.W.; Lin, H.J.; Liu, F.T.; et al. Allyl isothiocyanate ameliorates obesity by inhibiting Galectin-12. Mol. Nutr. Food Res. 2018, 62, e1700616. [Google Scholar] [CrossRef]
- Zhang, H.Q.; Chen, S.Y.; Wang, A.S.; Yao, A.J.; Fu, J.F.; Zhao, J.S.; Chen, F.; Zou, Z.Q.; Zhang, X.H.; Shan, Y.J.; et al. Sulforaphane induces adipocyte browning and promotes glucose and lipid utilization. Mol. Nutr. Food Res. 2016, 60, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Moon, M.H.; Jeong, J.K.; Park, Y.G.; Lee, Y.J.; Seol, J.W.; Park, S.Y. Sulforaphane induced adipolysis via hormone sensitive lipase activation, regulated by AMPK signaling pathway. Biochem. Biophys. Res. Commun. 2012, 426, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Lee, D.H.; Kim, H.J.; Lee, S.J.; Ban, J.O.; Cho, M.C.; Jeong, H.S.; Yang, Y.; Hong, J.T.; Yoon, D.Y. Thiacremonone, a sulfur compound isolated form garlic, attenuates lipid accumulation partially mediated via AMPK activation in 3T3-L1 adipocytes. J. Nutr. Biochem. 2012, 23, 1552–1558. [Google Scholar] [CrossRef] [PubMed]
- Okouchi, R.; Sakanoi, Y.; Tsuduki, T. Miso (fermented soybean paste) suppresses visceral fat accumulation in mice, especially in combination with exercise. Nutrients 2019, 11, 560. [Google Scholar] [CrossRef]
| Gene | Primer Sequence (5′–3′) |
|---|---|
| PPAR-γ | 5′-GCCCACCAACTTCGGAATC-3′ |
| 5′-TGCGAGTGGTCTTCCATCAC-3′ | |
| C/EBPa | 5′-GTGTGCACGTCTATGCTAAACCA-3′ |
| 5′-CTCCACTGCCCACCTGTCA-3′ | |
| ATGL | 5′-AACACCAGCATTCCAGTTCAA-3′ |
| 5′-TTGTGTTGCTTGCCATTCTC-3′ | |
| LPL | 5′-ACTCGCTCTCAGATGCCCTA-3′ |
| 5′-TTGTGTTGCTTGCCATTCTC-3′ | |
| HSL | 5′-ACCGAGACAGGCCTCAGTGTG-3′ |
| 5′-GAATCGGCCACCGGTAAAGAG-3′ | |
| SREBP-1 | 5′-ATGATCATCAGCGTAAATGG-3′ |
| 5′-GCCTTTCATAACACATTCCA-3′ | |
| β-actin | 5′-AGCCTTCCTTCTTGGGTATGG-3′ |
| 5′-CACTTGCGGTGCACGATTGGAG-3′ |
| Gene | Nucleotide Sequences | Annealing (°C) |
|---|---|---|
| HMG-CoA reductase | 5′-GTTCTTTCCGTGCTGTGTTCTGGA-3′ | 60 |
| 5′-CTGATATCTTTAGTGCAGAGTGTGGCAC-3′ | ||
| LDL receptor | 5′-CTGTGGGCTCCATAGGCTATCT-3′ | 60 |
| 5′-GCGGTCCAGGGTCATCTTC-3′ | ||
| SREBP-2 | 5′-GTGGAGCAGTCTCAACGTCA-3′ | 60 |
| 5′-TGGTAGGTCTCACCCAGGAG-3′ | ||
| CPT-1a | 5′-TCCACCCTGAGGCATCTATT-3′ | 62 |
| 5′-ATGACCTCCTGGCATTCTCC-3′ | ||
| FAS | 5′-AGGGGTCGACCTGGTCCTCA-3′ | 60 |
| 5′-GCCATGCCCAGAGGGTGGTT-3′ | ||
| SREBP-1c | 5′-CACTTCTGGAGACATCGCAAAC-3′ | 64 |
| 5′-ATGGTAGACAACAGCCGCATC-3′ | ||
| β-actin | 5′-AGAGAAGCTGTGCTATGTT-3′ | 60 |
| 5′-CACAGGATTCCATACCCAAG-3′ |
| Name | Accession No. | Target Sequence |
|---|---|---|
| ABCA1 | NM_013454.3 | CTCCTTGTCATCTCTAGCCAGGATATTCAGCATCCTCTCCCAGAGCAAAAAGCGACTCCACATAGAAGACTACTCTGTCTCTCAGACAACACTTGACCAA |
| ATGL | NM_025802.2 | ACAGCTCCACCAACATCCACGAGCTTCGCGTCACCAACACCAGCATCCAGTTCAACCTTCGCAATCTCTACCGCCTCTCGAAGGCTCTCTTCCCGCCAGA |
| Adiponectin | NM_009605.4 | GACCACAATGGACTCTATGCAGATAACGTCAACGACTCTACATTTACTGGCTTTCTTCTCTACCATGATACCAACTGACTGCAACTACCCATAGCCCATA |
| CD36 | NM_007643.3 | GGGACCATTGGTGATGAAAAAGCAGAAATGTTCAAAACACAAGTGACTGGGAAAATCAAGCTCCTTGGCATGGTAGAGATGGCCTTACTTGGGATTGGAG |
| CEBP-α | NM_007678.3 | AGGAGGACACGGGGACCATTAGCCTTGTGTGTACTGTATGTCGCCAGCCGCTGTTGCTGAAGGAACTTGAAGCACAATCGATCCATCCCAGAGGGACTGG |
| CPT1-b | NM_009948.2 | ACAAGATGTCTCTGGACGCCATCGAACGTGCTGCTTTCTTTGTGACCCTGGATGAAGATTCTCATTGCTACAACCCTGACGATGAGACCAGTCTTAGCCT |
| DGAT | NM_010046.2 | CTATCACTCCAGTGGGTTCCGTGTTTGCTCTGGCATCATACTCCATCATGTTCCTCAAGCTTTATTCCTACCGGGATGTCAACCTGTGGTGCCGCCAGCG |
| FABP4 | NM_024406.2 | TCGAAGGTTTACAAAATGTGTGATGCCTTTGTGGGAACCTGGAAGCTTGTCTCCAGTGAAAACTTCGATGATTACATGAAAGAAGTGGGAGTGGGCTTTG |
| FASN | NM_007988.3 | TTCTCCTCTGTAAGCTGCGGGCGTGGTAATGCTGGCCAAACTAACTACGGCTTCGCCAACTCTACCATGGAGCGTATATGTGAACAGCGCAGGCACGATG |
| GLUT4 | NM_009204.2 | CTGATGTGTCTGACGCACTAGCTGAGCTGAAGGATGAGAAACGGAAGTTGGAGAGAGAGCGTCCAATGTCCTTGCTCCAGCTCCTGGGCAGCCGCACCCA |
| HSL | NM_001039507.2 | CAGGAGTGCTCTTCTTCGAGGGTGATGAAGGACTCACCGCTGACTTCCTGCAAGAGTATGTCACGCTACACAAAGGCTGCTTCTACGGCCGCTGCCTGGG |
| LPL | NM_008509.1 | CCATGCTGTAACCAAGTCTGGCCTAGAACTAAACTATGTATTTCAGGCTGGCCTTGAACTCTCAACCATCCTGCCTTAGCTTCCTGTGTCCTGGGAGCTT |
| Leptin | NM_008493.3 | CCTATTGATGGGTCTGCCCAAGGCAAACCTAATTTTTGAGTGACTGGAAGGAAGGTTGGGATCTTCCAAACAAGAGTCTATGCAGGTAGCGCTCAAGCTT |
| PDH | NM_001098231.1 | AGTCTGCCACTGTTCTCTGATGCCATGCCAGCACCAACTCAACTGTTTTTTCCTCTCGTCCGTAACTGTGAACTGAGCAGAATCTATGGCACTGCATGTT |
| PPAR-α | NM_011144.2 | GGACTTGAACGACCAAGTCACCTTGCTAAAGTACGGTGTGTATGAAGCCATCTTCACGATGCTGTCCTCCTTGATGAACAAAGACGGGATGCTGATCGCG |
| PPAR-γ | NM_011146.1 | ACCAAGTGACTCTGCTCAAGTATGGTGTCCATGAGATCATCTACACGATGCTGGCCTCCCTGATGAATAAAGATGGAGTCCTCATCTCAGAGGGCCAAGG |
| Perilipin | NM_001113471.1 | TACCAAAGGGAGGGCCATGTCCCTATCCGATGCCCTGAAGGGTGTTACGGATAACGTGGTAGACACTGTGGTACACTATGTGCCGCTTCCCAGGCTGTCC |
| SCD1 | NM_009127.3 | GTGTTGCCTGGGTTGCCAGTTTCTTTCGTGGCTGGGCAGGAACTAGTGAGGTTGAGGGGCAGTGTCTGTAAGTAGCTGCTAAGAGGTGCATTTCCAGATG |
| SREBP-1c | NM_011480.1 | GACTACATCCGCTTCTTGCAGCACAGCAACCAGAAGCTCAAGCAGGAGAACCTGACCCTACGAAGTGCACACAAAAGCAAATCACTGAAGGACCTGGTGT |
| β-actin | NM_007393.1 | CAGGTCATCACTATTGGCAACGAGCGGTTCCGATGCCCTGAGGCTCTTTTCCAGCCTTCCTTCTTGGGTATGGAATCCTGTGGCATCCATGAAACTACAT |
| Group | Body Weight Gain (g) | Food Intake (g) | FER * |
|---|---|---|---|
| Normal | 7.06 ± 0.56 b | 2.76 ± 0.35 a | 2.66 ± 0.33 c |
| HFD | 12.99 ± 3.98 a | 2.32 ± 0.16 b | 5.71 ± 1.79 a |
| AHR100 | 11.31 ± 1.58 a | 2.33 ± 0.17 b | 4.96 ± 0.76 ab |
| AHR500 | 10.20 ± 0.87 a | 2.41 ± 0.15 b | 4.23 ± 0.43 b |
| Group | Liver (g) | Heart (g) | Kidney (g) |
|---|---|---|---|
| Normal | 0.97 ± 0.02 a | 0.13 ± 0.01 | 0.33 ± 0.01 |
| HFD | 0.99 ± 0.04 a | 0.12 ± 0.00 | 0.33 ± 0.01 |
| AHR100 | 0.91 ± 0.02 ab | 0.12 ± 0.00 | 0.32 ± 0.01 |
| AHR500 | 0.82 ± 0.04 b | 0.13 ± 0.01 | 0.034 ± 0.01 |
| Group | T-Chol (mg/dL) | TG (mg/dL) |
|---|---|---|
| Normal | 95.78 ± 4.22 a | 97.64 ± 7.33 b |
| HFD | 117.64 ± 8.02 a | 116.80 ± 9.87 a |
| AHR100 | 119.21 ± 1.99 a | 117.98 ± 2.84 a |
| AHR500 | 109.61 ± 4.62 a | 105.52 ± 2.52 ab |
| Gene Function | Name | Fold Change | |||
|---|---|---|---|---|---|
| AHR100 * vs. HFD | p-Value | AHR500 ** vs. HFD | p-Value | ||
| Cholesterol efflux | ABCA1 | −1.13 | 0.152 | −1.00 | 0.978 |
| Regulates the metabolism of lipid and glucose | Adiponectin | −1.02 | 0.870 | 1.21 | 0.217 |
| Imports fatty acids | CD36 | 1.08 | 0.664 | 1.14 | 0.468 |
| Induction of adipogenesis | CEBP-α | −1.10 | 0.515 | 1.23 | 0.178 |
| Adipocyte differentiation | PPAR-γ | 1.19 | 0.415 | 1.79 | 0.023 |
| Fatty acid β-oxidation | CPT1-β | −1.06 | 0.666 | 1.02 | 0.914 |
| FABP4 | −1.08 | 0.500 | 1.13 | 0.312 | |
| PPAR-α | −1.02 | 0.912 | 1.55 | 0.073 | |
| Glucose transporter | GLUT4 | −1.01 | 0.934 | 1.31 | 0.161 |
| TG lipolysis | HSL | 1.03 | 0.894 | 1.62 | 0.031 |
| LPL | 1.16 | 0.291 | 1.44 | 0.028 | |
| ATGL | 1.08 | 0.813 | 1.78 | 0.094 | |
| Regulate appetite | Leptin | 1.31 | 0.259 | −1.38 | 0.176 |
| Adipocyte lipolysis | Perilipin | 1.05 | 0.814 | 1.53 | 0.056 |
| Lipogenesis | SCD1 | 1.04 | 0.873 | 1.70 | 0.067 |
| SREBP-1c | 1.02 | 0.911 | 1.18 | 0.325 | |
| PDH | −1.05 | 0.774 | 1.27 | 0.085 | |
| DGAT | 1.05 | 0.774 | 1.61 | 0.045 | |
| FASN | 1.44 | 0.084 | 1.54 | 0.145 | |
| House keeping | β-actin | 1 | 1 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.J.; Lee, M.-J.; Jang, J.-Y.; Lee, S.-H. Allium hookeri Root Extract Inhibits Adipogenesis by Promoting Lipolysis in High Fat Diet-Induced Obese Mice. Nutrients 2019, 11, 2262. https://doi.org/10.3390/nu11102262
Kim HJ, Lee M-J, Jang J-Y, Lee S-H. Allium hookeri Root Extract Inhibits Adipogenesis by Promoting Lipolysis in High Fat Diet-Induced Obese Mice. Nutrients. 2019; 11(10):2262. https://doi.org/10.3390/nu11102262
Chicago/Turabian StyleKim, Hyun Ju, Min-Jung Lee, Ja-Young Jang, and Sung-Hyen Lee. 2019. "Allium hookeri Root Extract Inhibits Adipogenesis by Promoting Lipolysis in High Fat Diet-Induced Obese Mice" Nutrients 11, no. 10: 2262. https://doi.org/10.3390/nu11102262
APA StyleKim, H. J., Lee, M.-J., Jang, J.-Y., & Lee, S.-H. (2019). Allium hookeri Root Extract Inhibits Adipogenesis by Promoting Lipolysis in High Fat Diet-Induced Obese Mice. Nutrients, 11(10), 2262. https://doi.org/10.3390/nu11102262




