1. Introduction
The accomplishment of many reclamation projects has changed tidal flats into agricultural areas on the West Sea coast of the Korean Peninsula. The Saemangeum is one of the largest reclamation projects in the Republic of Korea. The reclaimed areas have typical ecological characteristics. The soil salinity of the land is usually high, which leads to harsh conditions for plants, restricting the colonization of new plants and the development of vegetation. Until the soil salinity reaches a normal level, it is a strong driving force in regulating the distribution of vegetation communities within the area. The soil salinity in the area is continuously changing and heterogeneous [
1,
2], and as a result, it shapes the unique and dynamic characteristics of the ecosystems.
To understand the development and change of these vegetation communities, it is critical to determine the distribution of vegetation types that have different resistances to salinity. Studies of vegetation in reclaimed areas are relatively rare [
1,
2,
3,
4,
5] in Korea compared to studies in salt marshes [
6,
7,
8,
9,
10]. The studies have primarily focused on the relationship between the soil conditions and physiological traits of plants in reclaimed areas. Lee et al. [
4] surveyed four tidal reclamation project areas and concluded that the distribution of plants was strongly restricted by the soil salinity. The clear relationship between the soil salinity and the distribution of vegetation was also found in the Saemangeum area [
1,
3]. Kim et al. [
3] classified the vegetation types of the Saemangeum area as: (1) halophyte vegetation located in a high saline area (mean electrical conductivity (EC): 14 dSm-1), dominated by
Salicornia europaea,
Suaeda asparagoides, and
S. japonica; (2) mixed halophyte vegetation located in mid saline areas (mean EC: 6.7 dSm-1), dominated by
Phragmites communis,
Puccinellia nipponica, and
Carex pumila; and (3) low-saline vegetation located in low saline areas (mean EC: 3.0 dSm-1), dominated by diverse plants, including
Aster subulatus,
A. tripolium, and
Echinochloa spp. Despite the physiological relationships, there is a limited understanding of the ecosystem conditions in a spatial context because these studies were usually conducted at experimental sites [
1,
2,
3,
5].
Remote sensing is an effective method for estimating or mapping target quantities in a large area. It is cost-effective, rapid, and able to cover a large area. Remote sensing has also been extensively used to estimate vegetation in salt marshes [
6,
7,
11,
12,
13,
14,
15,
16,
17], but there is little research on using remotely sensed data for reclaimed areas [
18]. In salt marsh studies, many researchers have tackled classifying halophyte plant and/or vegetation types that share many common characteristics of vegetation in an area reclaimed from estuarine tidal flats. Some studies have focused on (multi-temporal) spectral differences [
6,
7,
11,
16,
17], whilst others have concentrated on classification methods such as Neural Networks Classification [
12] or the random forest method [
19].
For the application of remote sensing techniques to be effective, remotely sensed data should have a strong connection to the target quantity or characteristics, in addition to the proper resolution, in order to take into account spatial variability. Vegetation in the Saemangeum is dominated by grass/herbaceous plants, and the biomass is low. The vegetation types show similar spectral responses, which makes it difficult to classify the types using spectral characteristics alone. Additionally, the vegetation patches are diverse in terms of size and shape. The patch sizes vary from several m2 to thousands of m2, and the shapes are irregular. To take into account spatial heterogeneity, it is important to select the proper resolution of the image.
It is well known that the phenological traits of vegetation can be used to improve the accuracy of vegetation or plant species mapping [
11,
20,
21,
22]. The plants in the study area primarily appear after the rainy season (mid-June to mid-July), due to the dry soil conditions in the spring. The biomass reaches a maximum in late-August to mid-September. Gilmore et al. [
11] found that information on phenological variability in the growing season was useful for distinguishing dominant marsh plant species. However, it is difficult to obtain multi-temporal images with a high resolution during the short growing season. If images with short revisit frequencies and coarse resolutions can be downscaled (i.e., increased in spatial resolution), the utility of the images can be effectively increased.
The downscaling of imagery has gained attention in recent years, and many approaches have been developed (see Atkinson [
23]). Among them, the use of cokriging for image sharpening provides several advantages. Cokriging has a well-established theoretical model and uses semivariograms and cross-variograms of two or more images of different resolutions [
24,
25,
26]. Because cokriging is a generic tool, it is possible to include diverse types of data, such as topographic maps, thematic maps, or experimental data. Additionally, the downscaling cokriging method has demonstrated a lower error (mean error and mean squared error) than the method of using a high pass filter [
24].
The objective of this study was to propose a modified approach to map vegetation in a reclaimed area using multi-temporal downscaled images. This study used cokriging methods to downscale Landsat imagery to the resolution of a RapidEye image. This study provides an effective method for creating an accurate vegetation map, which is essential for monitoring and managing the ecosystem of the reclaimed Saemangeum area.
2. Materials and Methods
2.1. Study Area
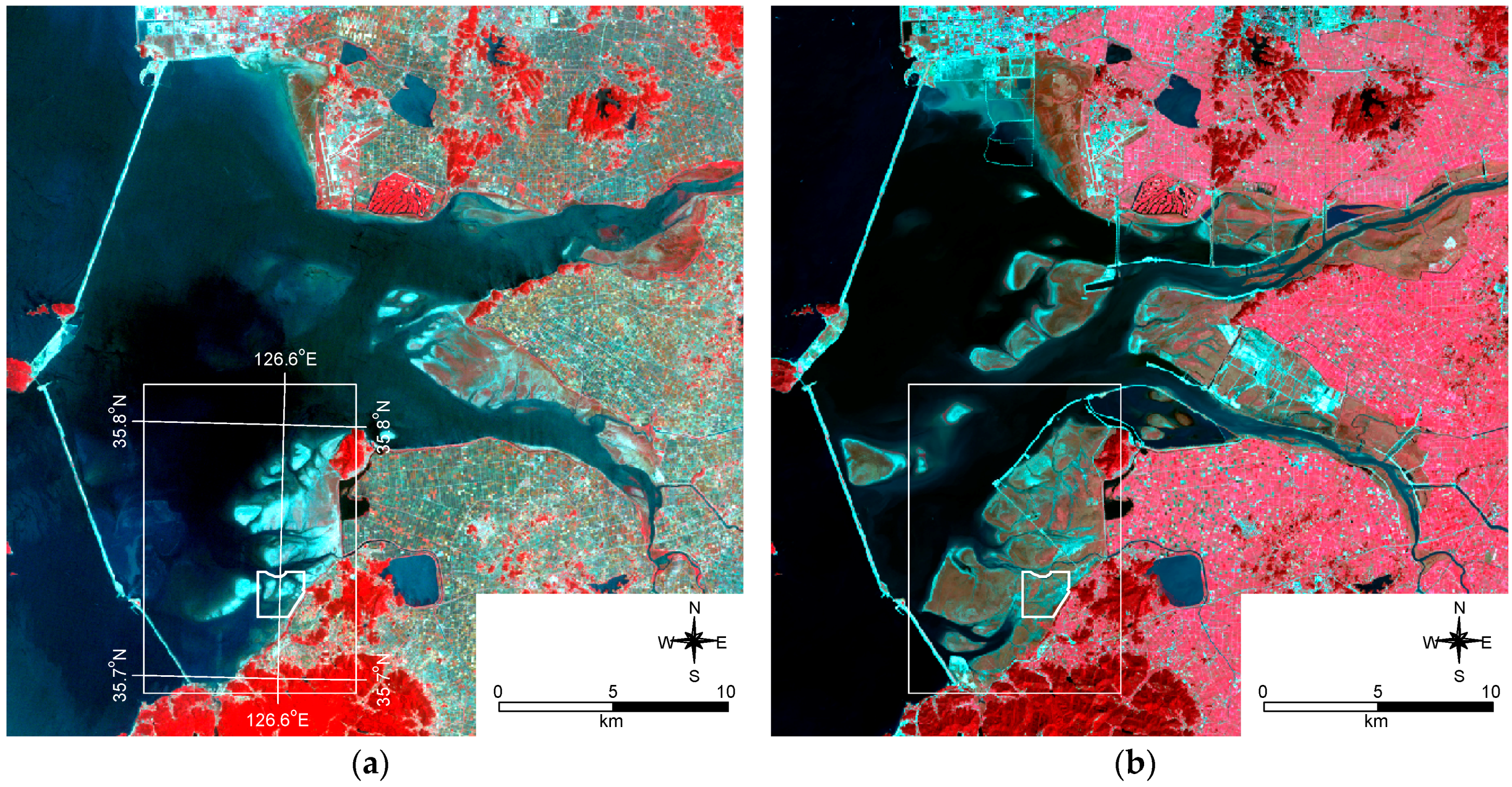
The Saemangeum is a reclaimed area that was created by the construction of a sea dike (33 km) across an estuarine tidal flat on the West Sea coast of South Korea (completed in April 2010). The dike encloses a total area of 401
(281
of land and 120
of fresh water reservoir). Although there are many reclaimed areas in Korea, the Saemangeum is unique because of its large scale and relatively long development period. These conditions allow large areas to remain under natural conditions, with very limited anthropogenic disturbance to the vegetation. It is worth noting that the soil is not imported from outside of the reclamation area, due to its large scale. The reclaimed land was created by lowering the water elevation level (EL). Since the end of 2010, the EL has been maintained at about −1.5 m (below sea level,
Figure 1). At the EL, a large area of estuarine tidal flat (ca. 180
) became land [
27], while some portions of the area remained natural. The study area was selected, based on the criteria of: (1) minimal anthropogenic disturbance; and (2) heterogeneity and typical vegetation type of the reclamation area (
Figure 1).
2.2. Collecting Field Data
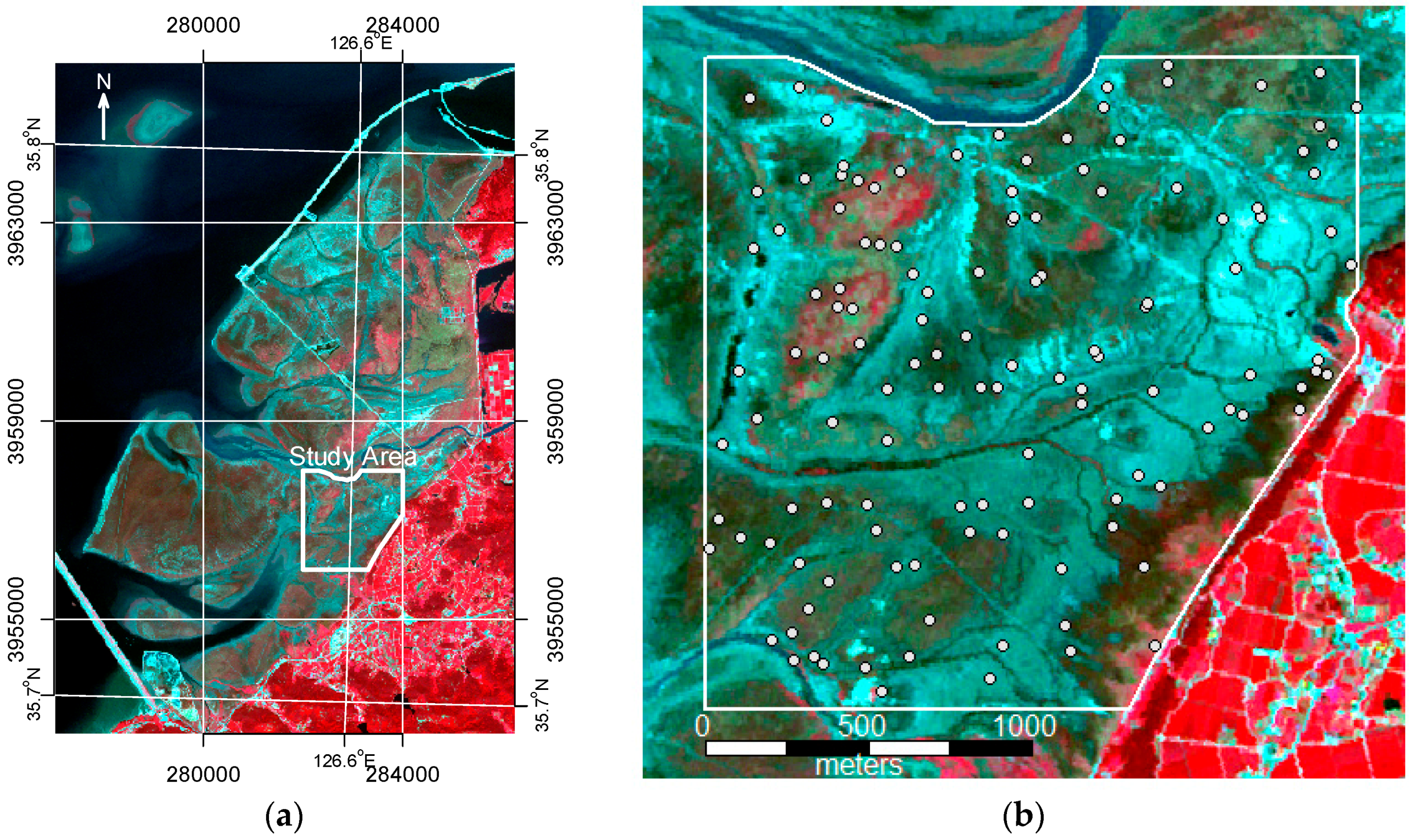
The study area was approximately 3.5 km
2. A total of 127 sample plots (each plot: 5
) were selected by simple random sampling (
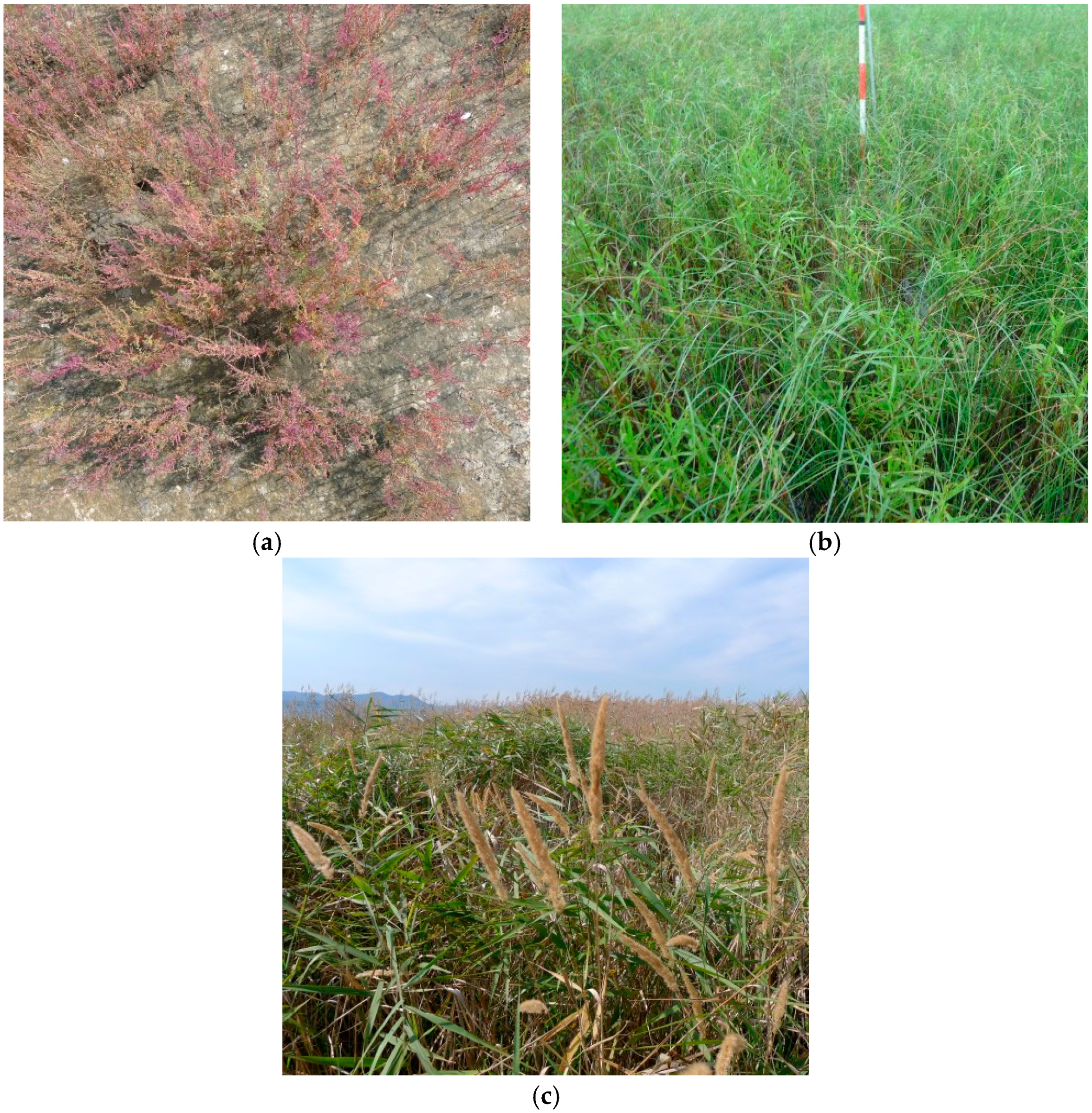
Figure 2) within the study area. A field survey was conducted in September 2014. We recorded the coverage of each species and dominant vegetation type within 25 m
2 of the sample points and classified the typical vegetation types. There were three typical vegetation types: halophyte vegetation (HV), mid- to low saline vegetation (MLV), and reed/small-reed vegetation (Reed,
Figure 3). A total of three randomly selected soil core samples (0–5 cm depth) were collected from each sample point. The soil samples, which were air dried, crushed, and uniformly mixed, were sifted through a 2 mm sieve for EC and pH analysis and a 0.5 mm sieve for organic matter content analysis. The soil EC and pH were measured in saturated paste extract and saturated paste, respectively, according to the methods described by the NAAS [
28]. The organic matter content of the soils was measured by Tyurin’s method.
2.3. Satellite Data Acquisition and Processing
RapidEye and Landsat OLI (Operational Land Imager) satellite images were used. A RapidEye image was acquired on 1 September 2014. The RapidEye imagery is multispectral, with a red edge (690–730 nm) and blue (440–510 nm), green (520–590 nm), red (630–685 nm), and near-infrared (NIR, 760–785 nm) bands. A level 2B RapidEye image was used, and the image was georeferenced to the Universal Transverse Mercator (UTM) coordinate system using the WGS 1984 datum with an accuracy of less than the root mean square of 0.01 pixels. The image was resampled to a 6 spatial resolution. Image processing was conducted with ERDAS Image software (version 9.1).
Landsat 8 OLI images were acquired on 1 July and 5 October 2014. The Landsat 8 OLI blue (450–515 nm), green (525–600 nm), red (630–680 nm), and NIR (845–885 nm) bands were used for downscaling. The digital number (DN) values of the image were converted to reflectance with a multiplicative rescaling factor and an additive rescaling factor. Topographic correction was not necessary because the study area was very flat. The atmospheric correction was carried out with the Second Simulation of Satellite Signal in the Solar Spectrum (6S) model [
29], using GRASS software [
30].
2.4. Downscaling Cokriging
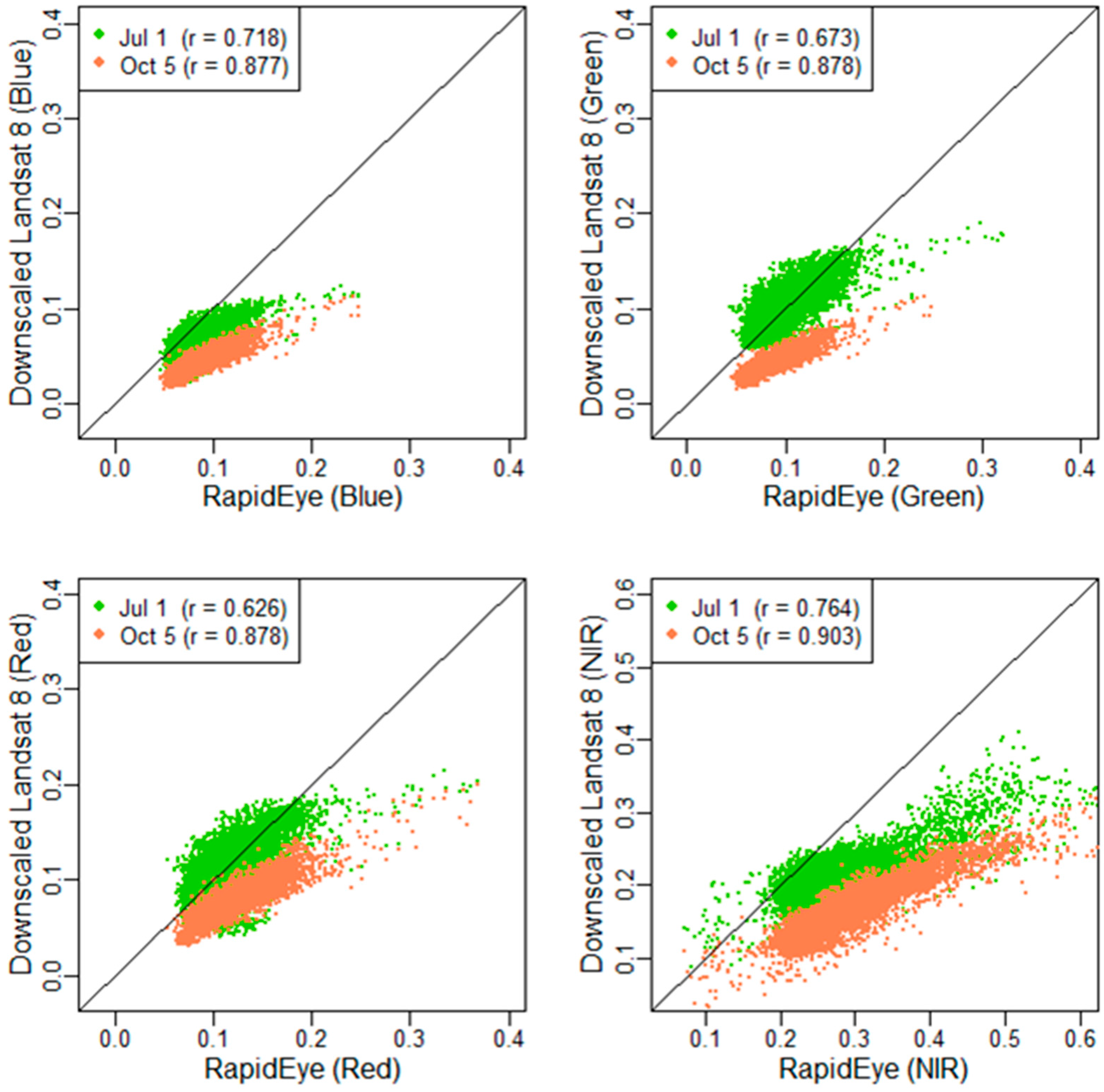
The following description of downscaling cokriging is mainly based on the work of Pardo-Iguzquiz et al. [
24]. All of the formulas and notations followed Pardo-Iguzquiz et al. [
24]. The cokriged finer-spatial-resolution image (downscaled Landsat) of band
k, calculated from a Landsat (band
k) image and RapidEye (band
l), is given by:
where:
is a random variable (RV) of a pixel of areal size u (RapidEye), with the spatial location and spectral band k estimated by cokriging.
is a RV of the pixel of the coarse spatial resolution image with areal size V (Landsat OLI) and spectral band k. N of these pixels are used. The weight assigned to the random variable of the i-th pixel is . The number of window pixels for Landsat (N) was nine (=3 3).
is a RV of the pixel of the fine spatial resolution image with areal size u (RapidEye) and spectral band l. M of these pixels are used. The weight assigned to the random variable of the j-th pixel is . The number of window pixels for RapidEye (M) was 25 (=5 5).
The two sets of weights {; i = 1,…,N} and {; j = 1,…,M} are obtained through cokriging. To be optimal in the sense of giving a minimum variance unbiased estimator, the sum weights of the variable must be 1 (), and the sum of the weights of the variable must be 0 (). The weights are calculated by formulizing the spatial structure of the image data and minimizing the unbiased estimation variance.
There are two ways to address the local mean in the downscaling cokriging process. One is to consider non-stationarity (taking into account the variation of local mean) [
26], and the other is to use a global model [
24]. This study used a global model due to its ease of application and statistical reliability [
24]. The point spread function (PSF) is important to consider when factoring in area-to-point kriging. A uniform PSF was applied in this study because there was little information available to formulate a specific PSF. The Fortran source code for downscaling cokriging (DISKORI) was obtained via the Computers and Geosciences website [
31].
2.5. Classification of Vegetation Type
The random forest classifier (RFC) proposed by Brieman [
32] was applied to image classification because it is widely used in a range of fields and often yields good results [
19,
33,
34,
35]. The random forest algorithm (RF) is a type of decision tree classification. The RFC only has two parameters: the number of trees in the forest (number of bootstrap samples from the original data) and the number of random variables at each node (the maximum number of this parameter is the number of predictors). The RFC can use diverse types of data with little restriction because it does not make specific assumptions about the data distribution. Additionally, it can easily handle a large number of input variables.
R software [
36] was used for image classification. The RFC was conducted with the ‘randomForest’ package (version 4.6–10), which was implemented by Liaw and Wiener [
37] based on the algorithm proposed by Brieman [
32].
Classification was performed for different combinations of images (
Table 1). In addition to the reflectance of image bands, the normalized difference vegetation index (NDVI) was used. The NDVI was computed with the following equation:
where
and
are the surface reflectance at the red and near infrared band, respectively.
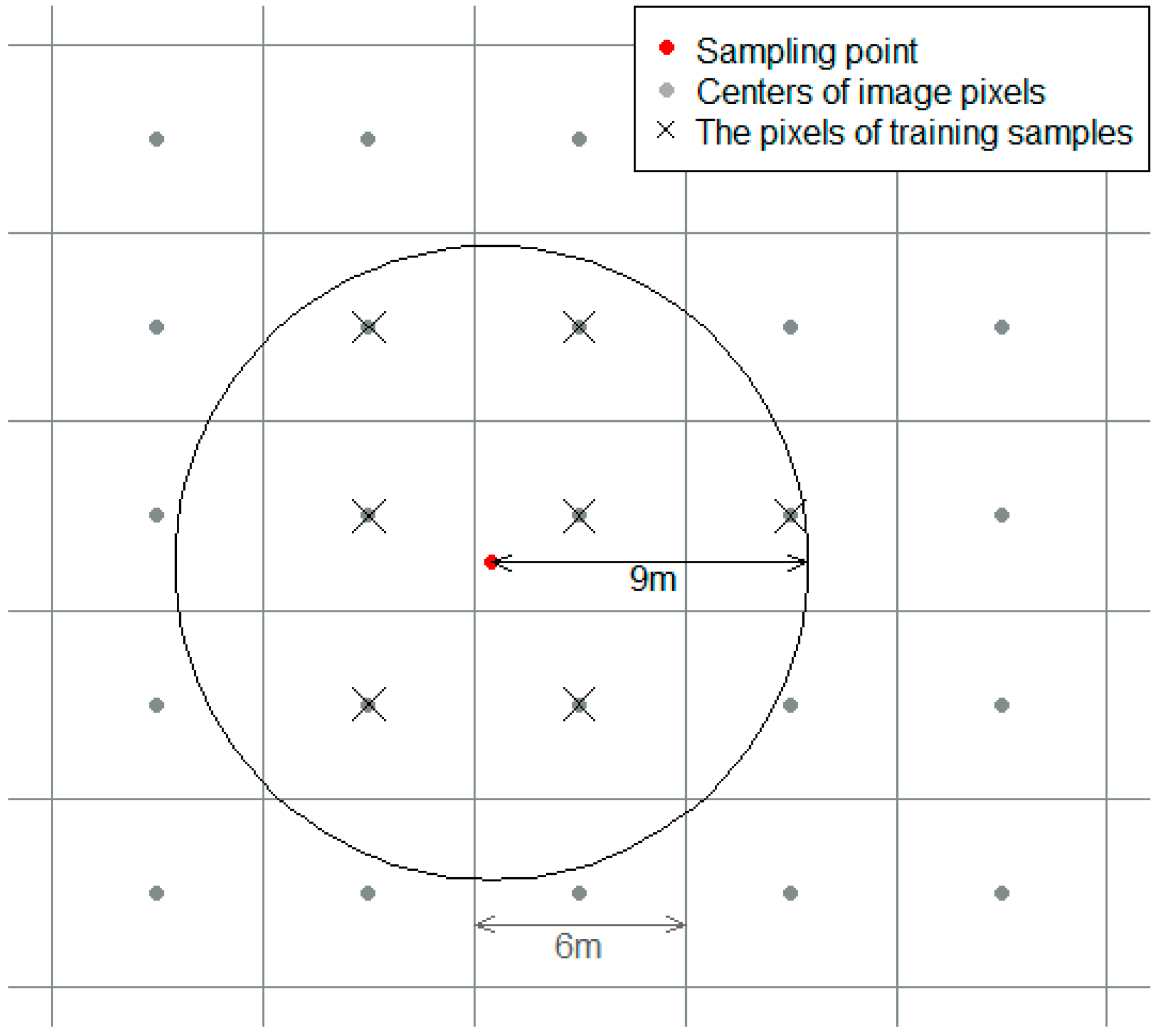
Training samples were selected, based on the 127 randomly selected field survey points. Pixels with centers within a 9 m radius of the training samples were assigned the same vegetation type as the training samples (
Figure 4). The radius criterion was determined by only selecting adjacent pixels (considering the 6 m resolution of the RapidEye and downscaled Landsat imagery) to the sampling points. Diagonal pixels were selectively included, according to the distance from the sampling point. The total sample size was 915. The vegetation classification was trained using randomly selected 200 pixels from each of the three vegetation types (600 training data = 200
three types). After building RF models, the models were used to predict the vegetation type of the remaining 315 pixels. An accuracy assessment of classifications was carried out by comparing the predicted vegetation type and field data. The confusion matrices for each model were created after classification. The overall accuracy, producer’s accuracy, user’s accuracy, and Cohens’ kappa [
38] were calculated, as suggested by Congalton [
39].
2.6. Characterisics of Vegetation Distribution
After creating the vegetation map, the spatial characteristics of the vegetation were analyzed with R software [
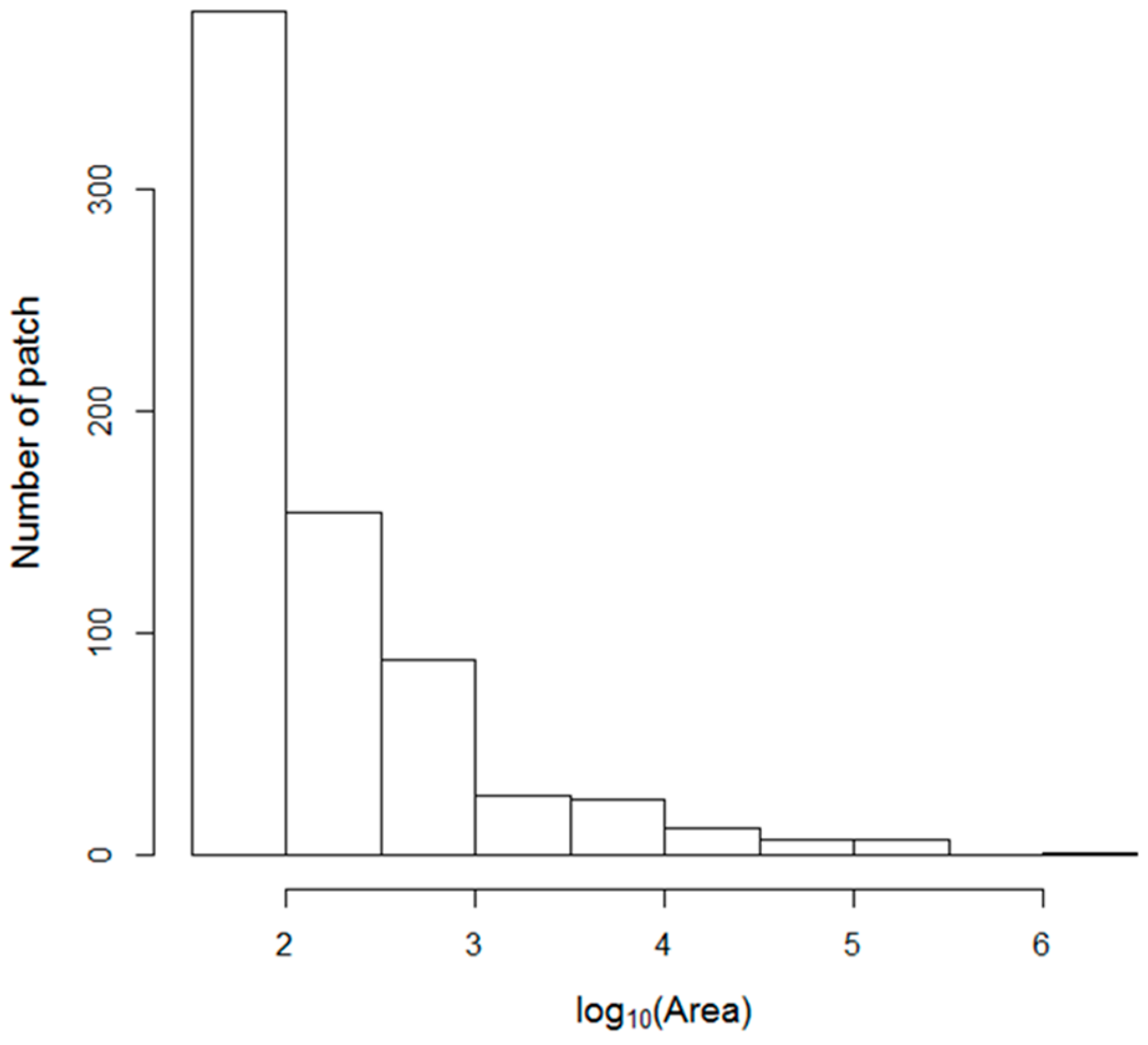
36]. The mean NDVI of each vegetation type was calculated with a ‘raster’ package (version 2.5–8). The patch statistics of the vegetation types were calculated with an ‘SDMTools’ package (version 1.1–221).
4. Discussion
The study area became permanent land after the water level was lowered in 2010, and the soil condition has been changing. Kim et al. [
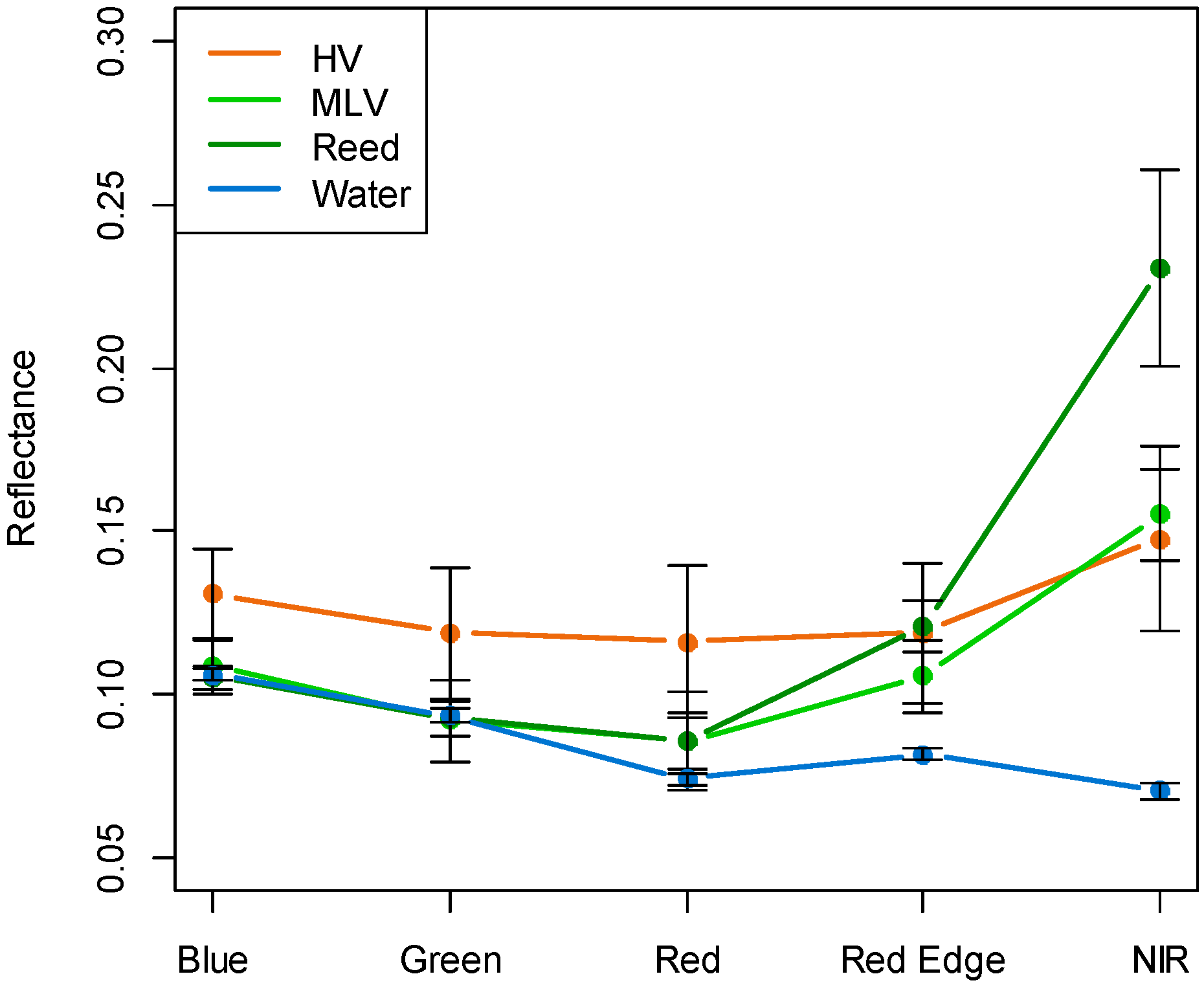
3] reported the mean soil EC of halophyte vegetation as 14 dSm-1, mixed vegetation as 6.7 dSm-1, and reed dominant vegetation as 3.0 dSm-1 in 2010. Compared to an earlier study, the soil EC showed lower levels in 2014. The mean soil EC of HV was 9.5 dSm-1, MLV was 1.0 dSm-1, and Reed was 0.5 dSm-1. The rapid changes in the soil conditions during this relatively short period also influenced the distribution of vegetation.
Although the characteristics and distribution of vegetation are crucial for determining the condition of the ecosystem, it is hard to create accurate vegetation maps of grass/herbaceous plant-dominated areas. There have been a number of studies that have tried to improve the accuracy of vegetation mapping using the phenological characteristics of plants [
11,
20,
21,
22]. Gilmore [
11] suggested that it is possible to discriminate
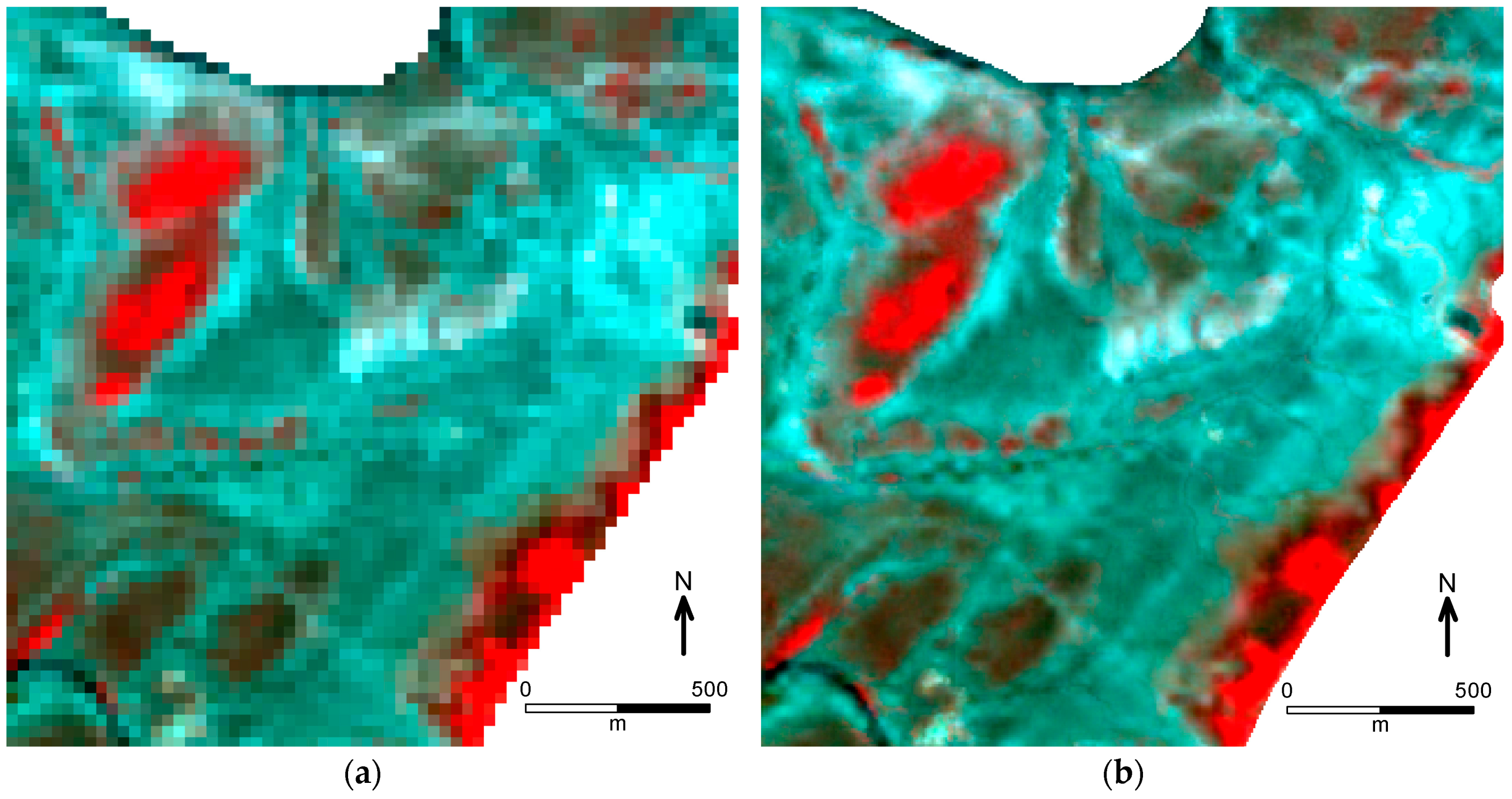
Phragmites spp. using single-date images acquired in the autumn season. However, it was difficult to classify
Phragmites-dominated vegetation using timely acquired RapidEye imagery. More than 20% of the Reed class was misclassified (
Table 4). The accuracy increased when the multi-temporal images were used in classification (
Table 5,
Table 6,
Table 7 and
Table 8).
The goal is to acquire multi-temporal images, especially in the growing season. This research applied cokriging to downscale Landsat imagery, which is easy to obtain, but has limited utility due to its relatively low spatial resolution. Sharpened Landsat imagery taken at different time periods improved the accuracy of the vegetation map. Cokriging is a useful approach because it is based on a well-established geostatistical theory and provides a framework to combine different types of data [
23,
24]. However, the application of the approach has been limited, until recently [
40]. The results show that image downscaling using cokriging can be applied in vegetation studies and is useful for the analysis of multi-temporal data.
In this study, a nested variogram model with two structures was used in cokriging. A nested model is useful for modelling the spatial structure with different scales [
41,
42]. Pardo-Iguzquiz et al. [
24] also used a nested model for downscaling two images with different resolutions. The nested structure model fitted the data well, with a 0 nugget value. The partial sill of the first structure (short range) was much smaller than that of the second structure in a variogram model of a low resolution image. It is consistent with the results of this study. However, in both cases, further research is needed to quantify the uncertainty of using the nested model for downscaling images.
Multi-temporal and multispectral data analysis raises the problem that the number of input variables can rapidly increase. The RFC can easily and effectively handle a large number of input variables [
19,
33,
34,
43]. For that reason, it was used in this study and yielded good results.
5. Conclusions
An area reclaimed from estuarine tidal flats forms a very unique and dynamic ecosystem. The creation of a large reclamation area in itself is a difficult challenge for ecosystem monitoring and management.
This paper presents a novel approach for the classification of vegetation in reclaimed areas, using multi-temporal downscaled Landsat imagery along with RapidEye imagery. The study area has a typical environmental gradient of soil salinity and vegetation types, of which the distribution is mainly controlled by the environmental conditions. This study provides a framework to reduce spatial and temporal resolution by downscaling images with a low spatial and high temporal resolution, with an image that has a high spatial and low temporal resolution. Multi-temporal downscaled images were compared to single high resolution images, to classify the vegetation type. The accuracy of the vegetation map using the multi-temporal downscaled image was approximately 10% higher than the accuracy of the map using a single high resolution image.
These results demonstrate that the downscaling technique is useful when a high resolution image is not available. This technique is also capable of increasing the utility of relatively low spatial resolution images, such as Landsat imagery. The results also confirmed the performance of the RFC. The analysis of multi-temporal multi-band data causes difficulties in handling high dimensional data. The RFC was very versatile and demonstrated a high performance when classifying the vegetation type with a large number of variables.
The results of this study can be utilized in ecosystem monitoring for target objects that are difficult to distinguish with single, high spatial resolution imagery. The method can not only expand the time span of the data, but also increase the accuracy of mapping. It is also possible to apply this approach to the monitoring of ecosystems that witness rapid changes.