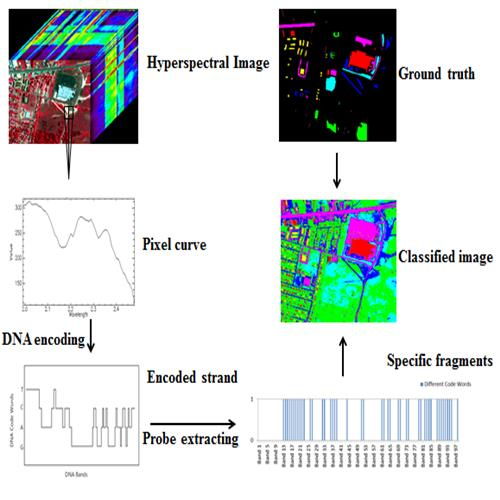
Multi-Probe Based Artificial DNA Encoding and Matching Classifier for Hyperspectral Remote Sensing Imagery
Abstract
:1. Introduction
2. DNA Encoding
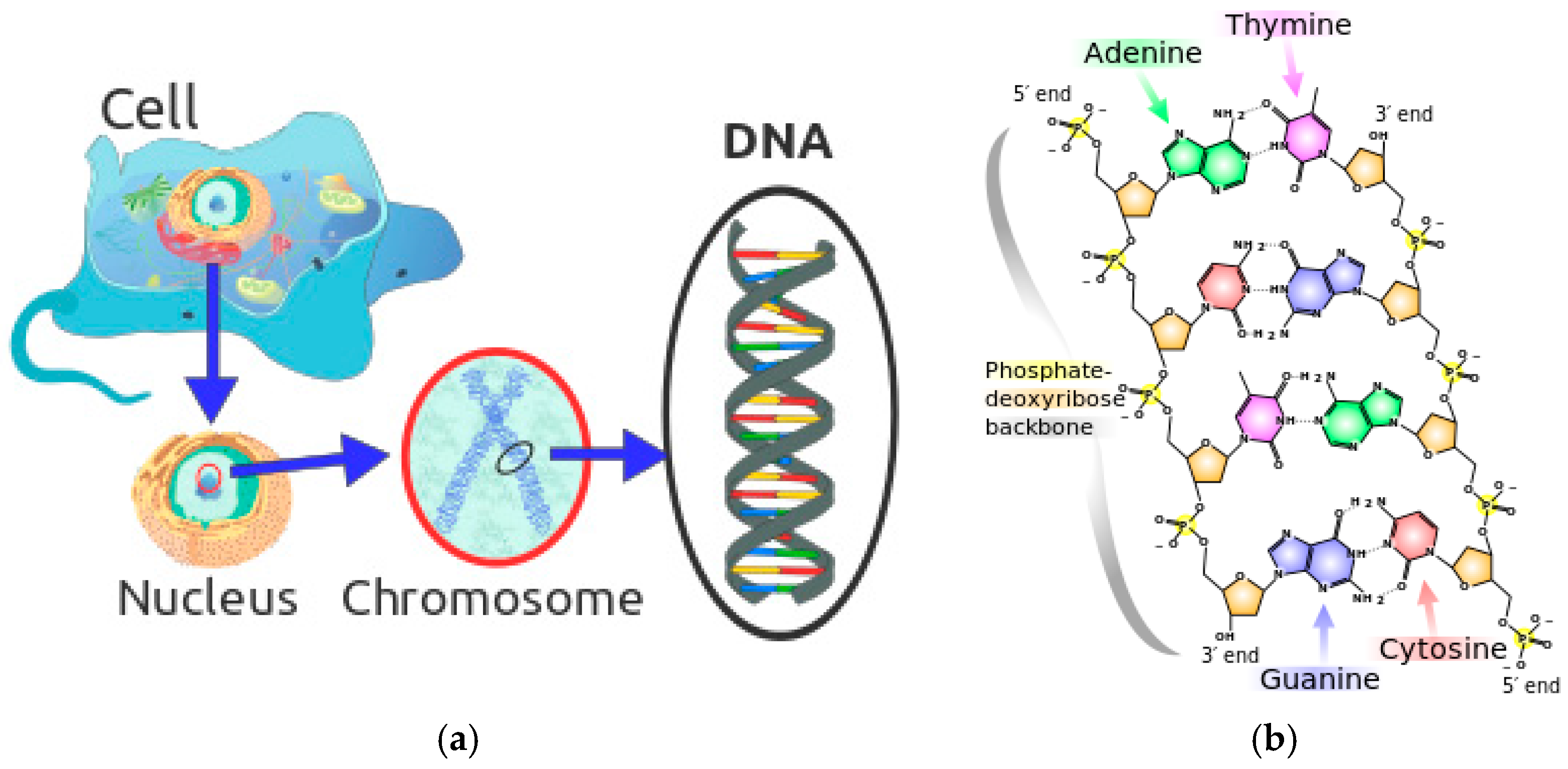
2.1. The Basic Theory of DNA
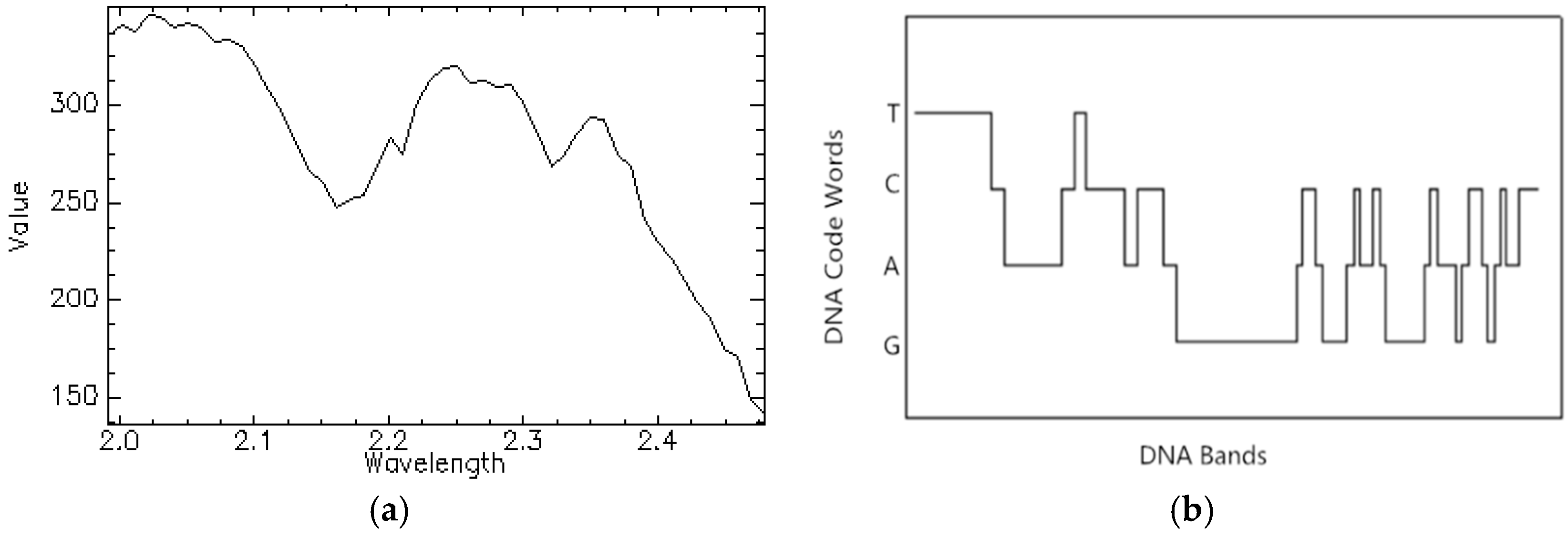
2.2. The DNA Encoding Method
2.2.1. DNA Encoding for Spectral Brightness Information
2.2.2. DNA Encoding for Spectral Shape Information
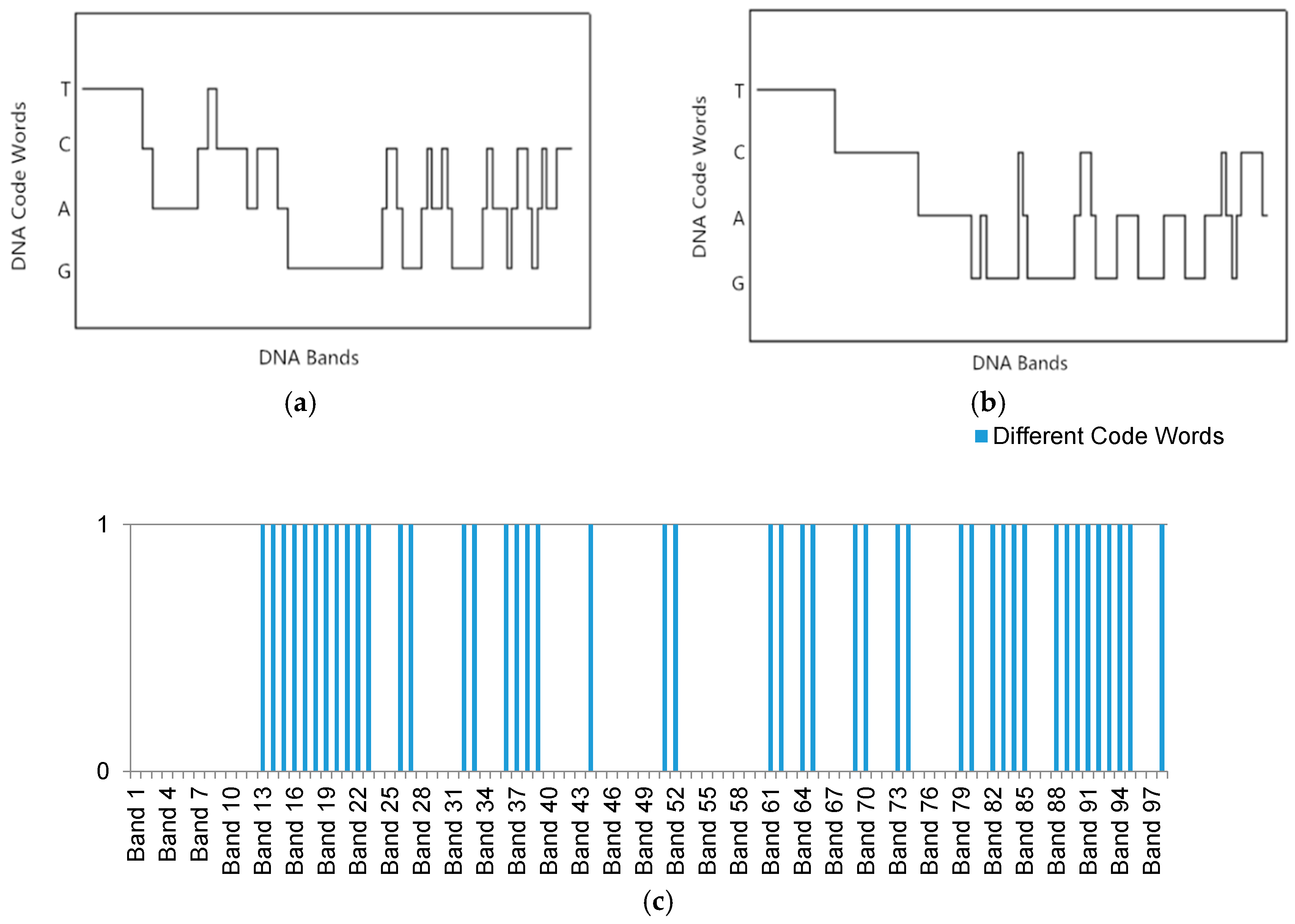
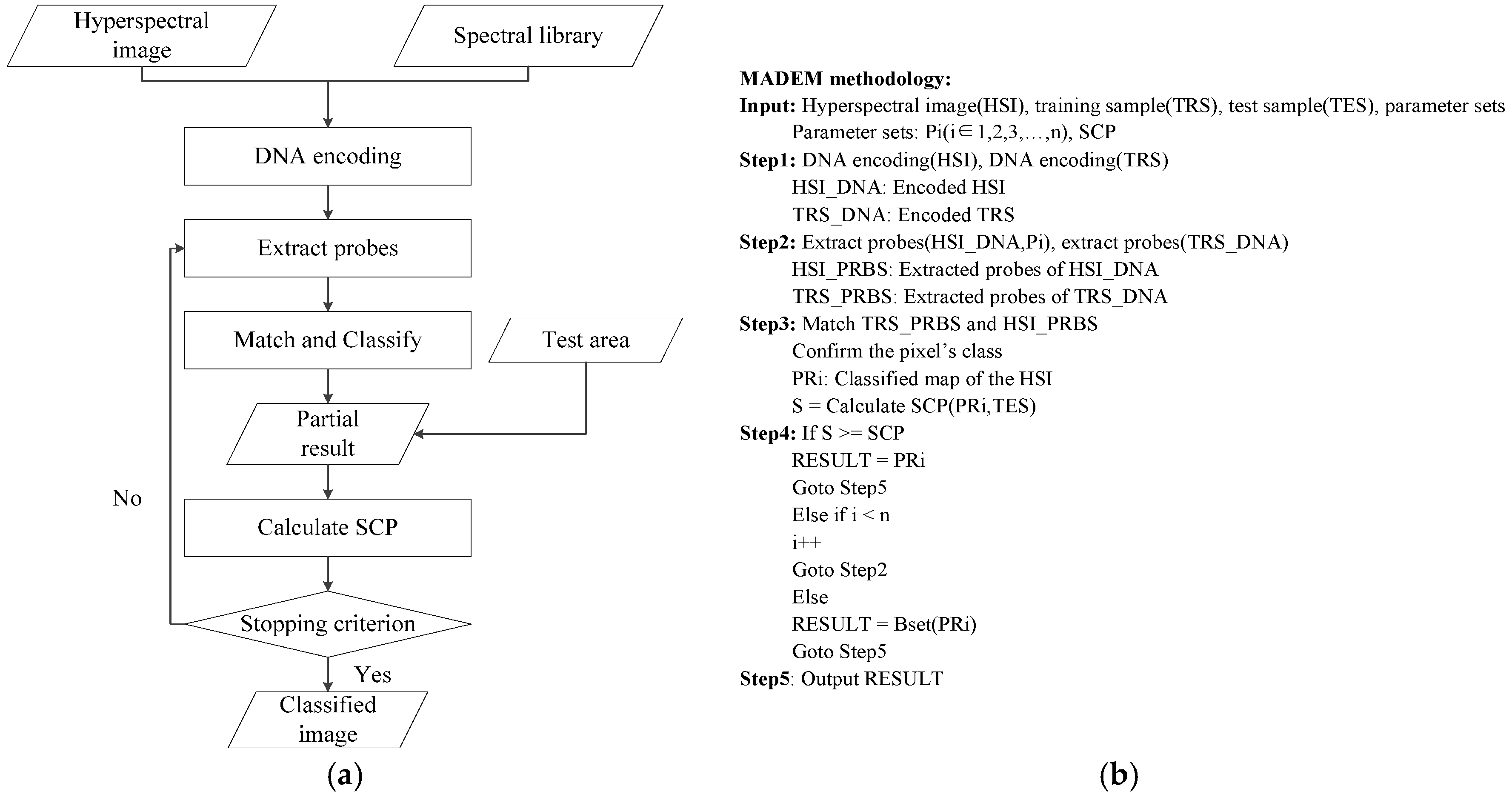
3. The Multi-Probe Based Artificial DNA Encoding and Matching Method
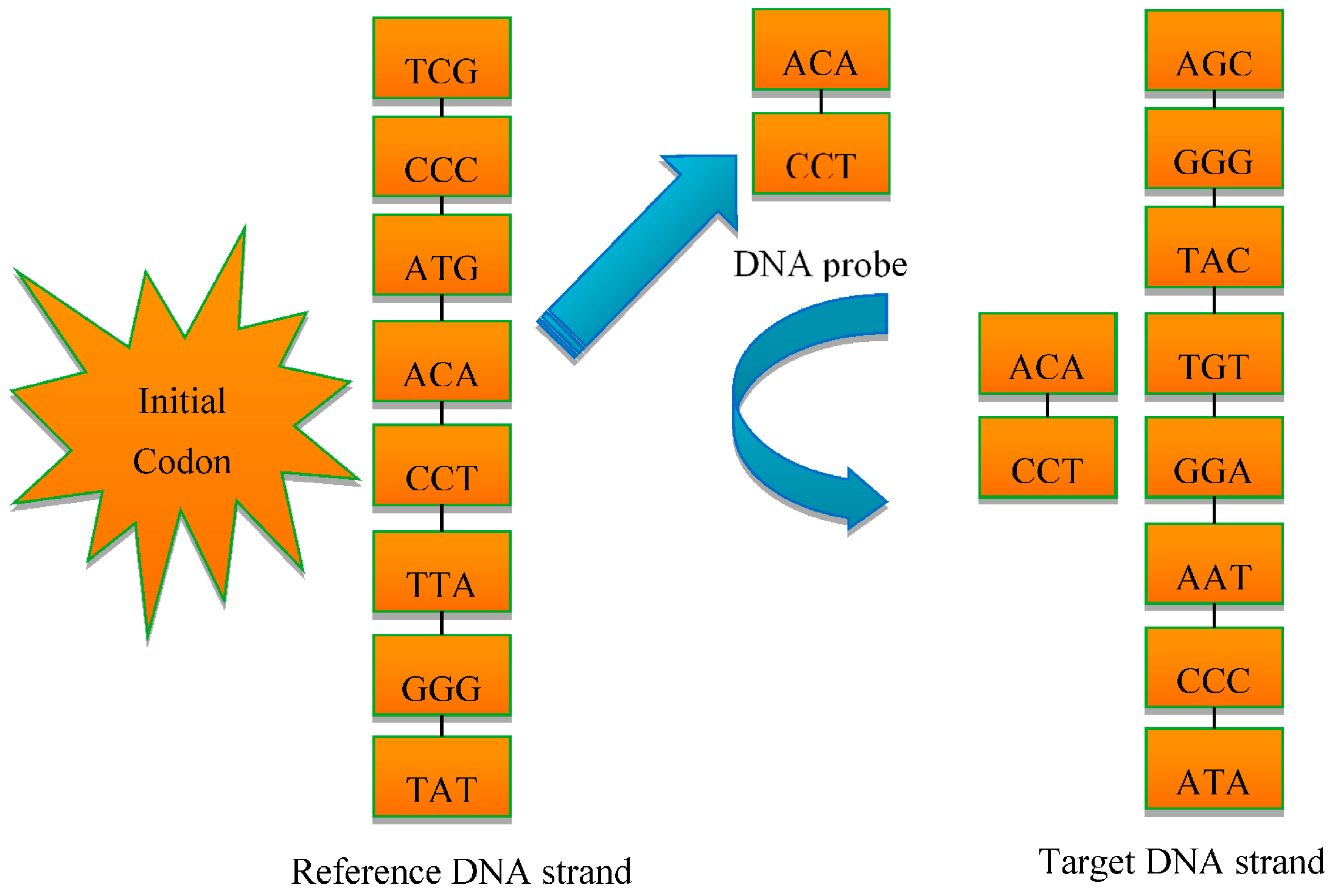
3.1. The DNA Probe Technology
3.2. The DNA Multi-Probe Extracting Strategy Based on Hyperspectral Remote Sensed Image
- (1)
- The starting positions of the probe, which means the initial codon in Figure 2, should be determined in the range of strands;
- (2)
- The length of the strands should not be beyond the strand length; and
- (3)
- Each probe should be selected from the strands, which is the probe selection rule.
4. Experiment
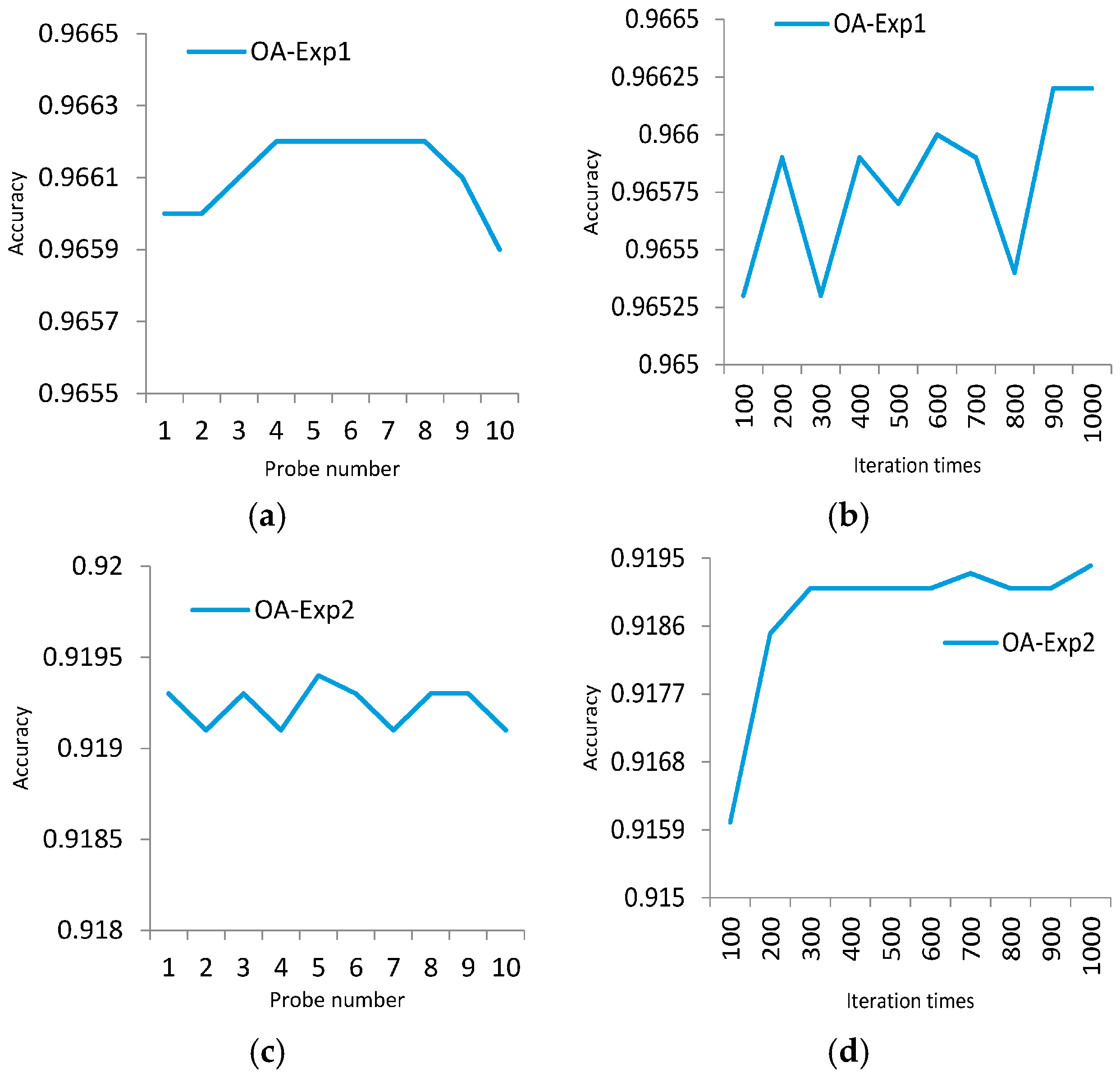
4.1. Experiment 1
4.2. Experiment 2
5. Computational Complexity Analysis
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lillesand, T.; Kiefer, R.W.; Chipman, J. Remote Sensing and Image Interpretation; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Brown, A.J. Spectral curve fitting for automatic hyperspectral data analysis. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1601–1608. [Google Scholar]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, J.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, S110–S122. [Google Scholar]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar]
- Kriegel, H.P.; Kröger, P.; Zimek, A. Clustering high-dimensional data: A survey on subspace clustering, pattern-based clustering, correlation clustering. ACM Trans. Knowl. Discov. Data (TKDD) 2009, 3. [Google Scholar] [CrossRef]
- Lark, R.M. A reappraisal of unsupervised classification, I: Correspondence between spectral and conceptual classes. Int. J. Remote Sens. 1995, 16, 1425–1443. [Google Scholar] [CrossRef]
- Jia, X.; Richards, J.A. Binary coding of imaging spectrometer data for fast spectral matching and classification. Remote Sens. Environ. 1993, 43, 47–53. [Google Scholar] [CrossRef]
- Sweet, J.; Granahan, J.; Sharp, M. An objective standard for hyperspectral image quality. In Proceedings of the AVIRIS Workshop, Jet Propulsion Laboratory, Pasadena, CA, USA, 23–27 February 2000.
- Sweet, J.N. The spectral similarity scale and its application to the classification of hyperspectral remote sensing data. In Proceedings of the 2003 IEEE Workshop on Advances in Techniques for Analysis of Remotely Sensed Data, Greenbelt, MD, USA, 27–28 October 2003; pp. 92–99.
- De Carvalho, O.A.; Menese, P.R. Spectral Correlation Mapper (SCM): An improvement on the Spectral Angle Mapper (SAM). In Proceedings of the Summaries of the 9th JPL Airborne Earth Science Workshop; JPL Publication 00-18; JPL Publication: Pasadena, CA, USA, 2000; Volume 9. [Google Scholar]
- Granahan, J.C.; Sweet, J.N. An evaluation of atmospheric correction techniques using the spectral similarity scale. In Proceedings of the Geoscience and Remote Sensing Symposium, Sydney, Australia, 9–13 July 2001; pp. 2022–2024.
- Van der Meero, F.; Bakker, W. Cross correlogram spectral matching application to surface mineralogical mapping by using AVIRIS data from Cuprite. Remote Sens. Environ. 1997, 61, 371–382. [Google Scholar] [CrossRef]
- Lazebnik, S.; Schmid, C.; Ponce, J. Beyond bags of features: Spatial pyramid matching for recognizing natural scene categories. In Proceedings of the 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, New York, NY, USA, 17–22 June 2006; pp. 2169–2178.
- Yang, J.; Yu, K.; Gong, Y.; Huang, T. Linear spatial pyramid matching using sparse coding for image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2009), Miami, FL, USA, 20–25 June 2009; pp. 1794–1801.
- Peng, X.; Yan, R.; Zhao, B.; Tang, H.; Yi, Z. Fast low rank representation based spatial pyramid matching for image classification. Knowl. Based Syst. 2015, 90, 14–22. [Google Scholar] [CrossRef]
- Peng, X.; Lu, J.; Yan, R.; Zhang, Y.; Rui, Y. Automatic Subspace Learning via Principal Coefficients Embedding. IEEE Trans. Cybern. 2016, 99, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lipton, R.J. DNA solution of hard computational problems. Science 1995, 268, 542. [Google Scholar] [CrossRef] [PubMed]
- Maley, C.C. DNA computation: Theory, practice, and prospects. Evol. Comput. 1998, 6, 201–229. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Zhong, Y.; Zhang, L. Artificial DNA computing-based spectral encoding and matching algorithm for hyperspectral remote sensing data. IEEE Trans. Geosci. Remote Sens. 2012, 50, 4085–4104. [Google Scholar] [CrossRef]
- Jiao, H.; Zhong, Y.; Zhang, L. An unsupervised spectral matching classifier based on artificial DNA computing for hyperspectral remote sensing imagery. IEEE Trans. Geosci. Remote Sens. 2014, 52, 4524–4538. [Google Scholar] [CrossRef]
- Adleman, L.M. Computing with DNA. Sci. Am. 1998, 279, 34–41. [Google Scholar] [CrossRef]
- Zhang, L.; Zhong, Y.; Huang, B.; Gong, J.; Li, P. Dimensionality reduction based on clonal selection for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2007, 45, 4172–4186. [Google Scholar] [CrossRef]
- Knight, R.D.; Freeland, S.J.; Landweber, L.F. Selection, history and chemistry: The three faces of the genetic code. Trends Biochem. Sci. 1999, 24, 241–247. [Google Scholar] [CrossRef]
- Alivisatos, A.P.; Johnsson, K.P.; Peng, X.; Wilson, T.E.; Loweth, C.J.; Bruchez, M.P., Jr.; Schultz, P.G. Organization of ‘nanocrystal molecules’ using DNA. Nature 1996, 382, 609–611. [Google Scholar] [CrossRef] [PubMed]
- Jonoska, N.; Mahalingam, K. Languages of DNA based code words. In DNA Computing; Springer: Berlin, Germany, 2003; pp. 61–73. [Google Scholar]
- Sponk, Eukaryote DNA.svg. Available online: https://commons.wikimedia.org/wiki/File:Eukaryote_DNA-en.svg (accessed on 5 August 2014).
- Madprime, DNA Chemical Structure.svg. Available online: https://commons.wikimedia.org/wiki/File:DNA_chemical_structure.svg (accessed on 5 August 2014).
- Sobell, H.M. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA 1985, 82, 5328–5331. [Google Scholar] [CrossRef] [PubMed]
- Garzon, M.H.; Deaton, R.J. Biomolecular computing and programming. IEEE Trans. Evol. Comput. 1999, 3, 236–250. [Google Scholar] [CrossRef]
- Watada, J. DNA computing and its applications. In Computational Intelligence: A Compendium; Springer: Berlin, Germany, 2008; pp. 1065–1089. [Google Scholar]
- Cao, Y.W.C.; Mirkin, C.A. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 2002, 297, 1536–1540. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, G.A.; Grossberg, S. Adaptive resonance theory. In The Handbook of Brain Theory and Neural Networks; MIT Press: Cambridge, MA, USA, 2003; pp. 87–90. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2. [Google Scholar] [CrossRef]
- Hypercube. Available online: http://www.tec.army.mil/hypercube (accessed on 1 September 2014).
- Fu, Z.; Robles-Kelly, A. Discriminate absorption-feature learning for material classification. IEEE Trans. Geosci. Remote Sens. 2011, 49, 1536–1556. [Google Scholar] [CrossRef]
- Bue, B.D.; Merényi, E.; Csathó, B. Automated labeling of materials in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4059–4070. [Google Scholar] [CrossRef]
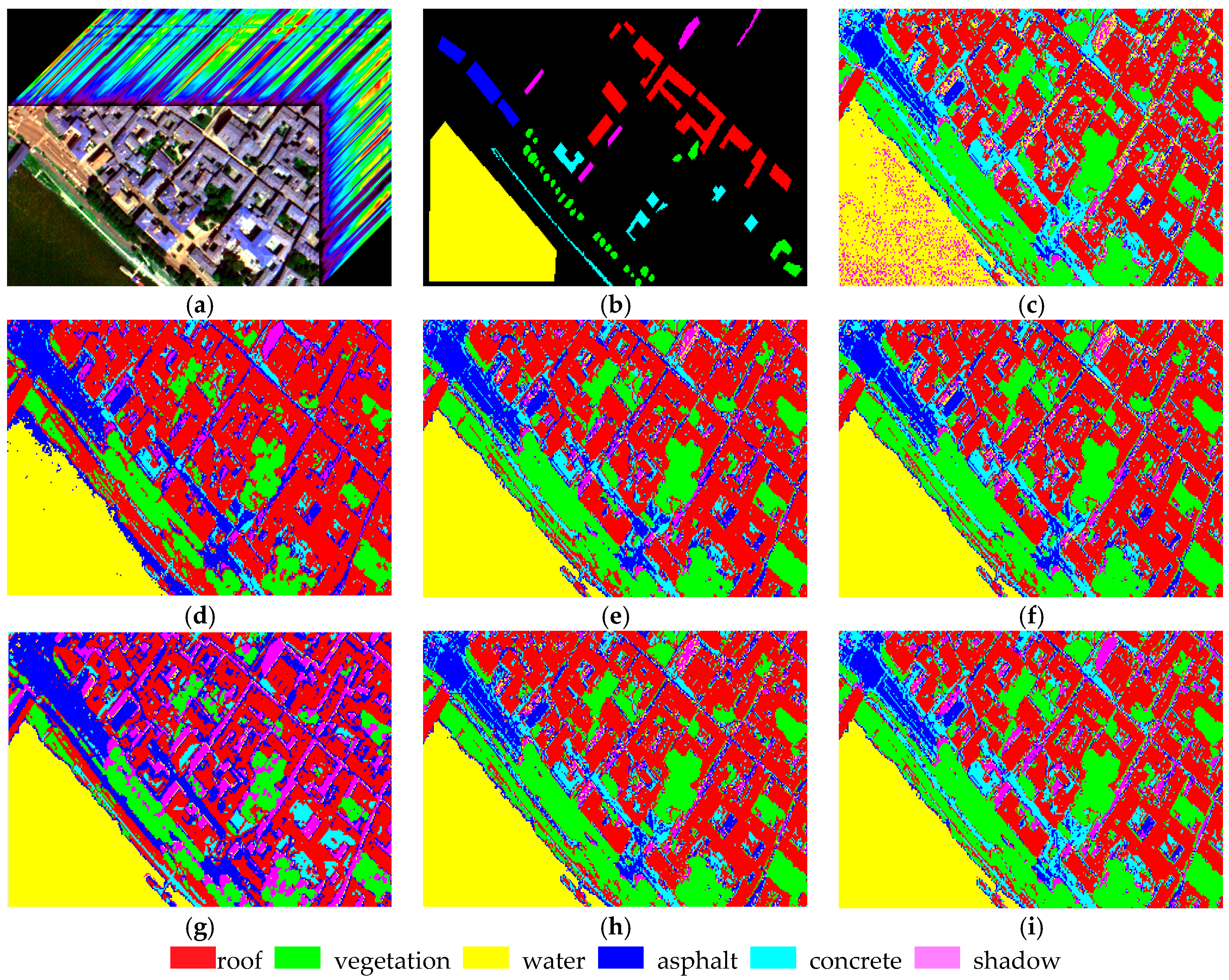
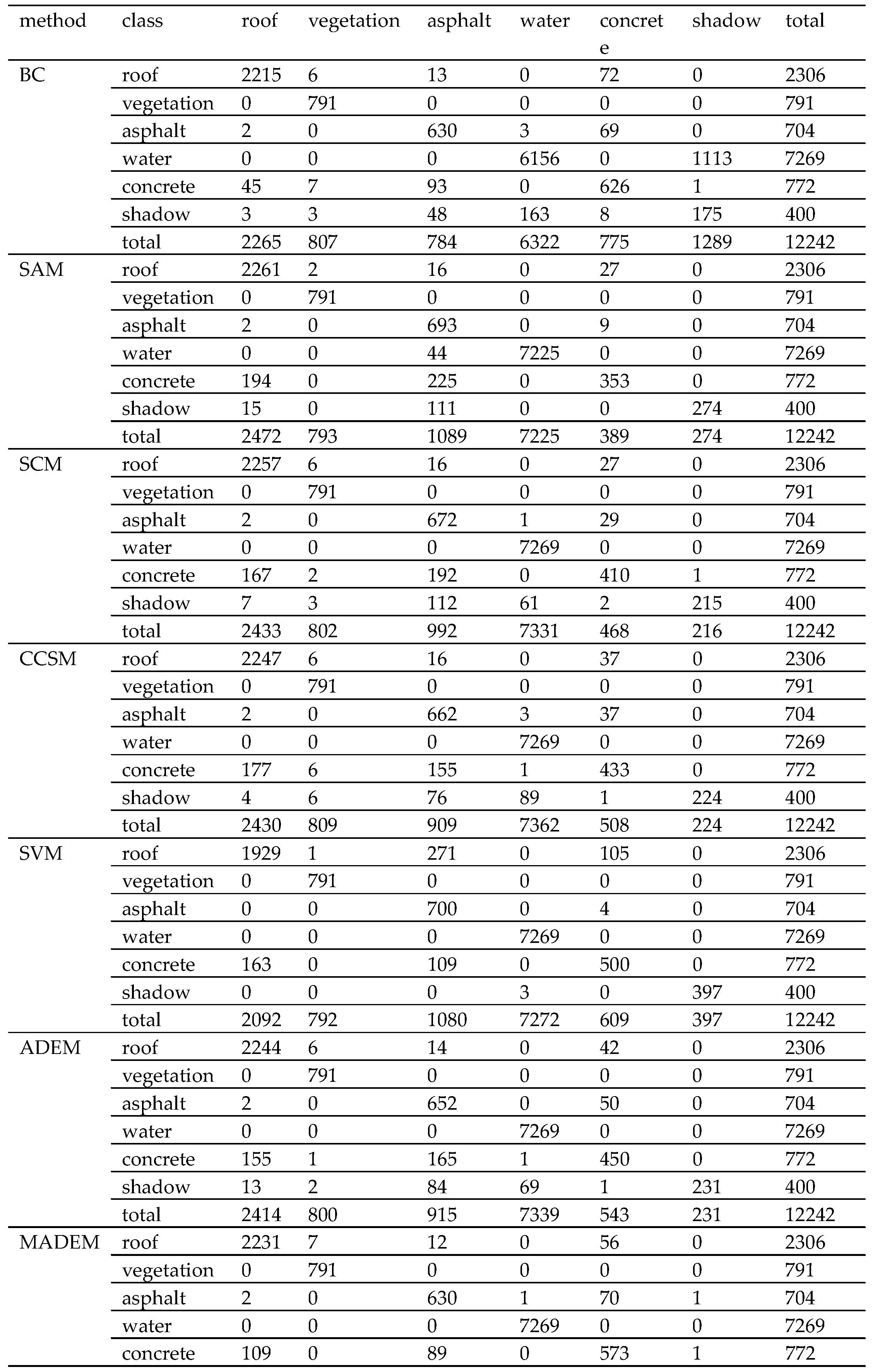
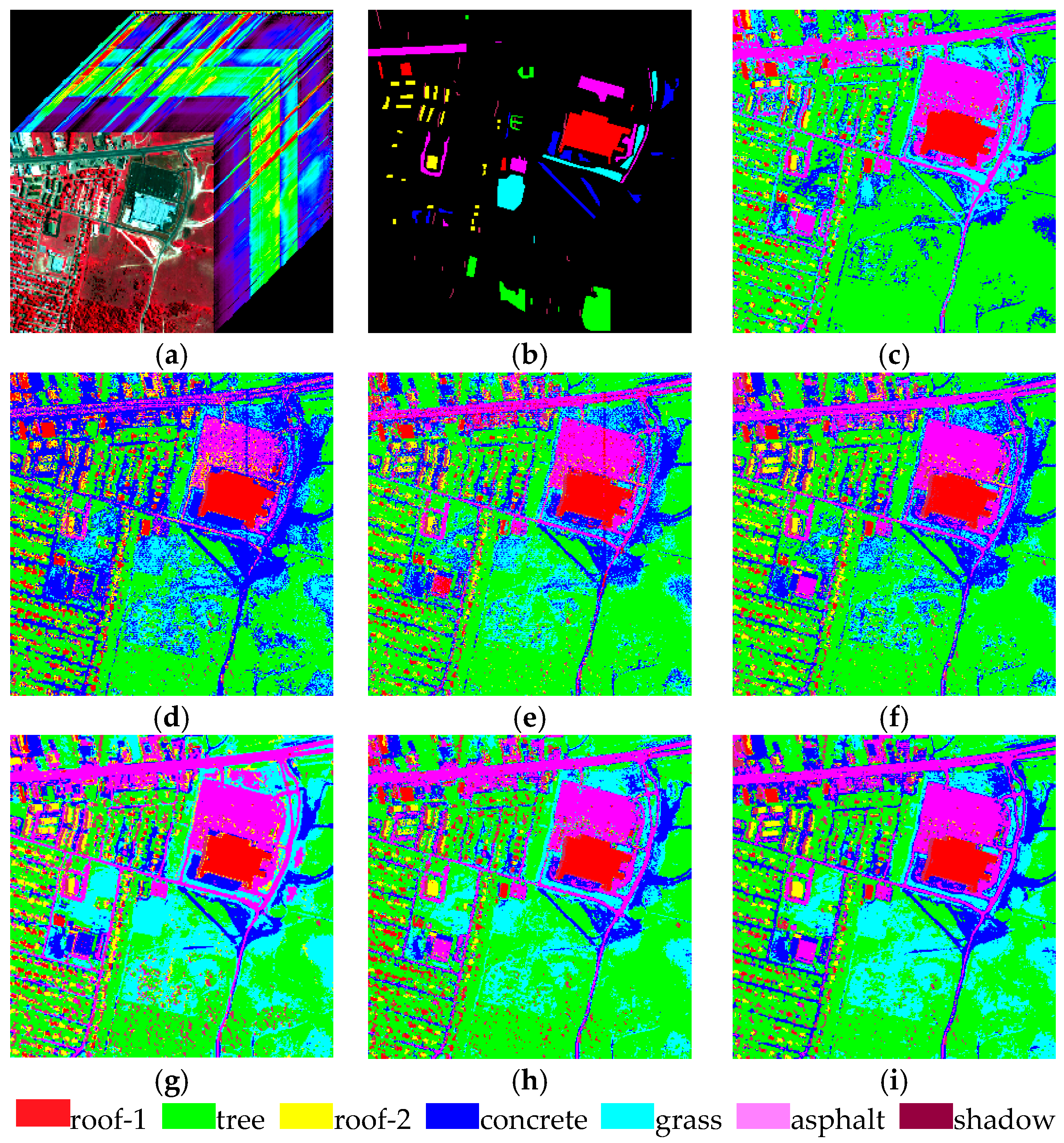
| Experimental Samples | Roof | Vegetation | Asphalt | Water | Concrete | Shadow | Total |
|---|---|---|---|---|---|---|---|
| training samples | 20 | 27 | 13 | 46 | 7 | 6 | 119 |
| testing samples | 2306 | 791 | 704 | 7269 | 772 | 400 | 12,242 |
| Method | BC | SAM | SCM | CCSM | SVM | ADEM | MADEM |
|---|---|---|---|---|---|---|---|
| overall accuracy (%) | 86.53 | 94.73 | 94.87 | 94.97 | 94.63 | 95.06 | 96.62 |
| kappa coefficient (%) | 79.06 | 91.21 | 91.38 | 91.53 | 91.10 | 91.69 | 94.35 |
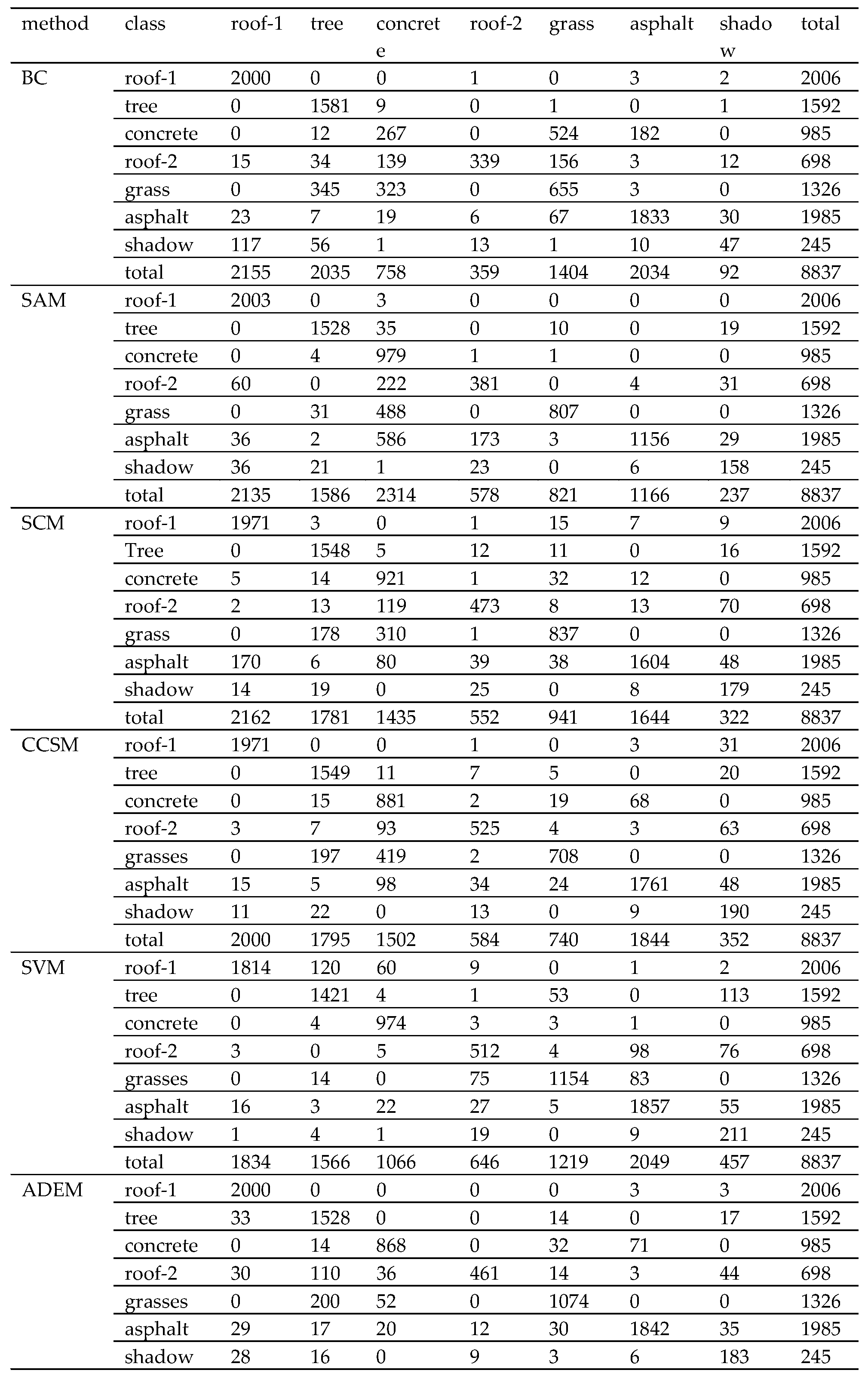
| Experimental samples | Roof-1 | Tree | Concrete | Roof-2 | Grass | Asphalt | Shadow | Total |
|---|---|---|---|---|---|---|---|---|
| training samples | 8 | 8 | 10 | 10 | 8 | 14 | 10 | 68 |
| testing samples | 2006 | 1592 | 985 | 698 | 1326 | 1985 | 245 | 8837 |
| Method | BC | SAM | SCM | CCSM | SVM | ADEM | MADEM |
|---|---|---|---|---|---|---|---|
| overall accuracy (%) | 76.07 | 79.35 | 85.24 | 85.83 | 89.88 | 90.03 | 91.94 |
| kappa coefficient (%) | 70.62 | 75.24 | 82.14 | 82.87 | 87.78 | 87.86 | 90.21 |
| Method | Calculation Times | Space Cost (Byte) |
|---|---|---|
| BC | 2LSB + 2NB + NSLB + SLN | LBS + NB + 4NLS + SL |
| SAM | NSL + SLN | 4(LBS + NB + 2B + NLS) + SL |
| SCM | NSL + SLN | 4(LBS + NB + 2B + NLS) + SL |
| CCSM | 21NSL + 2SLN | 4(LBS + NB + 2B + 42 + NLS) + SL |
| SVM | 8SLB + 6P + 10N + + 9SL + I( + 2N + 2P) | I, P, B, S, L, N |
| ADEM | 6NB + 6SLB + 2SLNB + SLN | 2(LBS + NB + 2NLS) + SL |
| MADEM | 6NB + 6SLB + I(NPB + PBSL + NBSL + 2SLN) | 2(LBS + NB + 2NLS) + SL + 4I |
| Time (ms) | BC | SAM | SCM | CCSM | SVM | ADEM | MADEM |
|---|---|---|---|---|---|---|---|
| Experiment 1 | 2574 | 3572 | 3932 | 42,932 | 38,392 | 12,520 | 17,145 |
| Experiment 2 | 3962 | 4961 | 4821 | 74,241 | 44,397 | 9953 | 12,543 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, K.; Zhao, D.; Zhong, Y.; Du, Q. Multi-Probe Based Artificial DNA Encoding and Matching Classifier for Hyperspectral Remote Sensing Imagery. Remote Sens. 2016, 8, 645. https://doi.org/10.3390/rs8080645
Wu K, Zhao D, Zhong Y, Du Q. Multi-Probe Based Artificial DNA Encoding and Matching Classifier for Hyperspectral Remote Sensing Imagery. Remote Sensing. 2016; 8(8):645. https://doi.org/10.3390/rs8080645
Chicago/Turabian StyleWu, Ke, Dong Zhao, Yanfei Zhong, and Qian Du. 2016. "Multi-Probe Based Artificial DNA Encoding and Matching Classifier for Hyperspectral Remote Sensing Imagery" Remote Sensing 8, no. 8: 645. https://doi.org/10.3390/rs8080645
APA StyleWu, K., Zhao, D., Zhong, Y., & Du, Q. (2016). Multi-Probe Based Artificial DNA Encoding and Matching Classifier for Hyperspectral Remote Sensing Imagery. Remote Sensing, 8(8), 645. https://doi.org/10.3390/rs8080645