Phylogenetic Structure of Foliar Spectral Traits in Tropical Forest Canopies
Abstract
:1. Introduction
2. Methods
2.1. General Approach
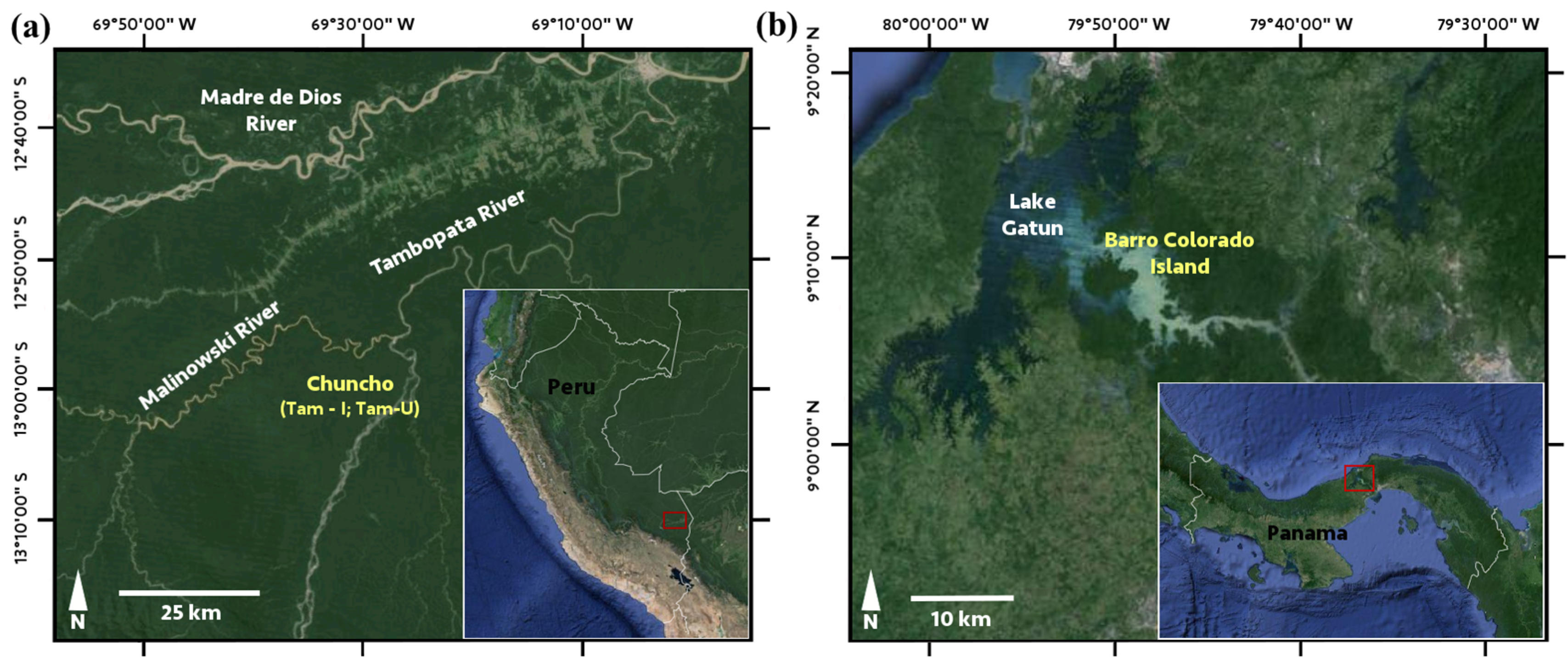
2.2. Site Descriptions
2.3. Species Data for Chemical, Spectral, and Phylogenetic Analyses
2.4. DNA Sequencing, Alignment, and Assembly into a Supermatrix
2.5. Phylogenetic Reconstruction: DNA-Sequence Based Method
2.6. Phylogeny Reconstruction: Supertree Method
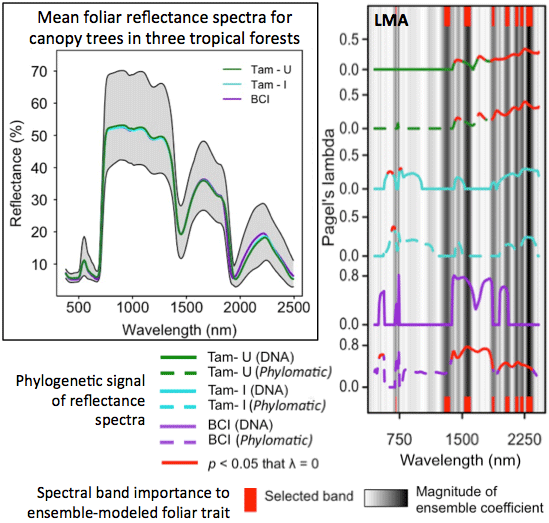
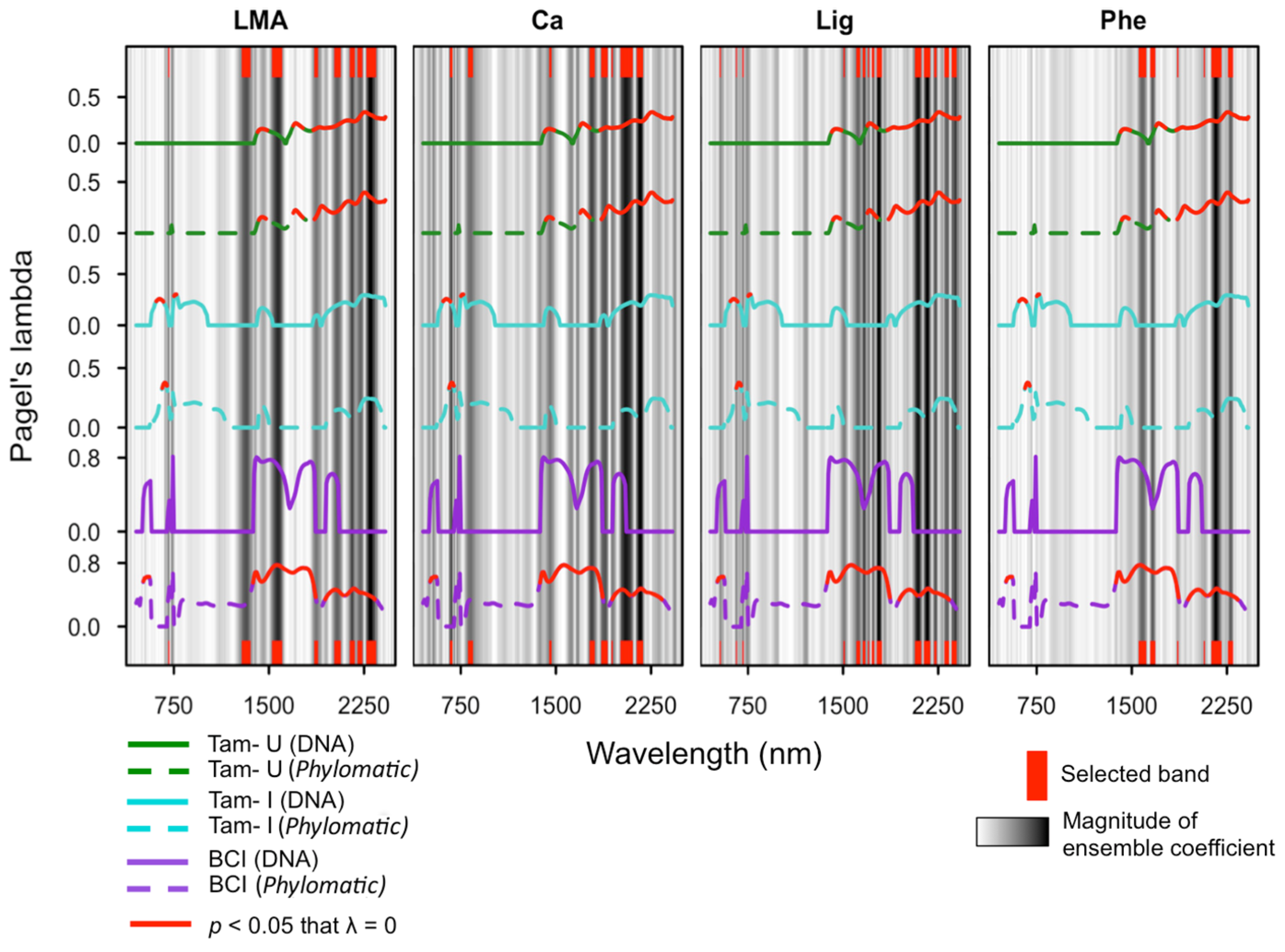
2.7. Quantifying Phylogenetic Signal of Spectra and Traits
2.8. Multi-Method Ensemble Selection of Biochemically-Related Spectral Bands
2.9. Quantification of Trait Correlations and Determination of Phylogenetic Independent Contrasts
3. Results
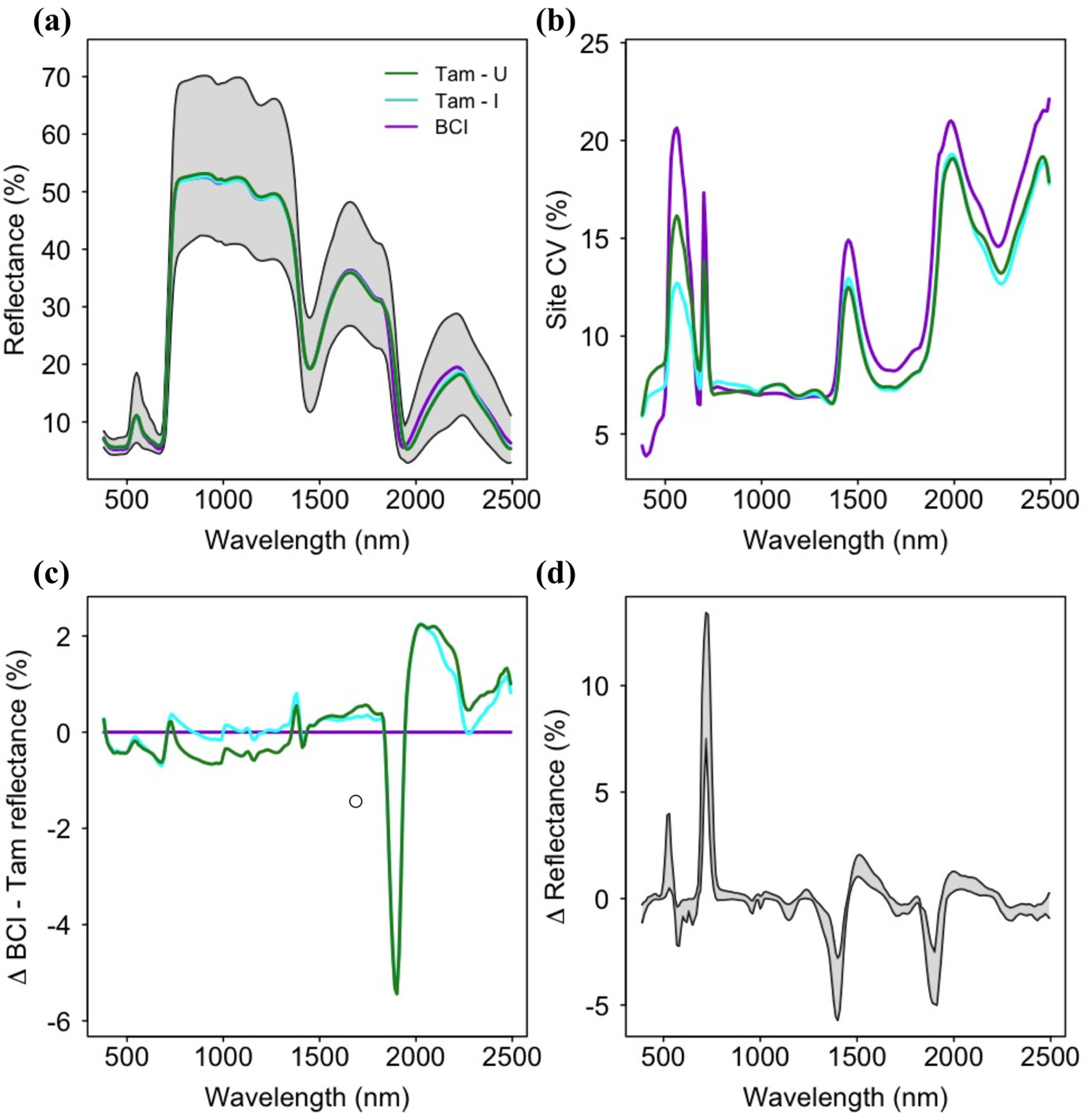
3.1. Spectral Variation across Soil Types
3.2. Patterns of Trait Variation and Co-Variation
3.3. Phylogenetic Structure of Spectra
3.4. Phylogenetic Structure of Traits
3.5. Ensemble Selection of Bands for Biochemical Traits
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Asner, G.P. Organismic remote sensing for tropical forest ecology and conservation. Ann. Mo. Bot. Gard. 2015, 100, 127–140. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gitelson, A.A.; Jacquemoud, S.; Schaepman, M.; Asner, G.P.; Gamon, J.A.; Zarco-Tejada, P. Retrieval of foliar information about plant pigment systems from high resolution spectroscopy. Remote Sens. Environ. 2009, 113, S67–S77. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Hilker, T.; Wulder, M.A.; Carroll, A.L. Detecting mountain pine beetle red attack damage with EO-1 Hyperion moisture indices. Int. J. Remote Sens. 2007, 28, 2111–2121. [Google Scholar] [CrossRef]
- Dahlin, K.M.; Asner, G.P.; Field, C.B. Environmental and community controls on plant canopy chemistry in a Mediterranean-type ecosystem. Proc. Natl. Acad. Sci. USA 2013, 110, 6895–6900. [Google Scholar] [CrossRef] [PubMed]
- Fassnacht, F.E.; Latifi, H.; Ghosh, A.; Joshi, P.K.; Koch, B. Assessing the potential of hyperspectral imagery to map bark beetle-induced tree mortality. Remote Sens. Environ. 2014, 140, 533–548. [Google Scholar] [CrossRef]
- Clark, M.; Roberts, D.; Clark, D. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Kalacska, M.; Bohlman, S.; Sanchez-Azofeifa, G.A.; Castro-Esau, K.; Caelli, T. Hyperspectral discrimination of tropical dry forest lianas and trees: Comparative data reduction approaches at the leaf and canopy levels. Remote Sens. Environ. 2007, 109, 406–415. [Google Scholar] [CrossRef]
- Feret, J.-B.; Asner, G.P. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecol. Appl. 2014, 24, 1289–1296. [Google Scholar] [CrossRef]
- Fyllas, N.M.; Patiño, S.; Baker, T.R.; Bielefeld Nardoto, G.; Martinelli, L.A.; Quesada, C.A.; Paiva, R.; Schwarz, M.; Horna, V.; Mercado, L.M.; et al. Basin-wide variations in foliar properties of Amazonian forest: Phylogeny, soils and climate. Biogeosciences 2009, 6, 2677–2708. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Canopy phylogenetic, chemical and spectral assembly in a lowland Amazonian forest. New Phytol. 2011, 189, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Freckleton, R.P.; Harvey, P.H.; Pagel, M. Phylogenetic analysis and comparative data: A test and review of evidence. Am. Nat. 2002, 160, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, S.P.; Garland, T.; Ives, A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 2003, 57, 717–745. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Perez, R.; Sanjur, O.; Bermingham, E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl. Acad. Sci. USA 2009, 106, 18621–18626. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; Hubbell, S.P. The phylogenetic structure of a neotropical forest tree community. Ecology 2006, 87, S86–S99. [Google Scholar] [CrossRef]
- Kraft, N.J.B.; Cornwell, W.K.; Webb, C.O.; Ackerly, D.D. Trait evolution, community assembly, and the phylogenetic structure of ecological communities. Am. Nat. 2007, 170, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.J.; Kitajima, K.; Kraft, N.J.B.; Reich, P.B.; Wright, I.J.; Bunker, D.E.; Condit, R.; Dalling, J.W.; Davies, S.J.; Díaz, S.; et al. Functional traits and the growth–mortality trade-off in tropical trees. Ecology 2010, 91, 3664–3674. [Google Scholar] [CrossRef] [PubMed]
- Baraloto, C.; Hardy, O.J.; Paine, C.E.T.; Dexter, K.G.; Cruaud, C.; Dunning, L.T.; Gonzalez, M.-A.; Molino, J.-F.; Sabatier, D.; Savolainen, V.; et al. Using functional traits and phylogenetic trees to examine the assembly of tropical tree communities. J. Ecol. 2012, 100, 690–701. [Google Scholar] [CrossRef]
- Kress, W.J.; García-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 2014, 30, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Kraft, N.J.B.; Ackerly, D.D. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 2010, 80, 401–422. [Google Scholar] [CrossRef]
- Townsend, A.R.; Cleveland, C.C.; Asner, G.P.; Bustamante, M.M.C. Controls over foliar N:P ratios in tropical rain forests. Ecology 2007, 88, 107–118. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef]
- Durgante, F.M.; Higuchi, N.; Almeida, A.; Vicentini, A. Species spectral signature: Discriminating closely related plant species in the Amazon with near-infrared leaf-spectroscopy. For. Ecol. Manag. 2013, 291, 240–248. [Google Scholar] [CrossRef]
- Lang, C.; Costa, F.R.C.; Camargo, J.L.C.; Durgante, F.M.; Vicentini, A. Near infrared spectroscopy facilitates rapid identification of both young and mature Amazonian tree species. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M. Inferring the historical patterns of biological evolution. Nature 1999, 401, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Crisp, M.D.; Cook, L.G. Phylogenetic niche conservatism: What are the underlying evolutionary and ecological causes? New Phytol. 2012, 196, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Field, C.; Mooney, H.A. Photosynthesis-nitrogen relationship in wild plants. In Proceedings of the Sixth Maria Moors Cabot Symposium on the Economy of Plant Form and Function, 29 August 1986; Givnish, T.J., Ed.; Cambridge University Press: Cambridge, UK; pp. 25–56.
- Westoby, M. A leaf-height-seed (LHS) plant ecology strategy scheme. Plant Soil 1998, 199, 213–227. [Google Scholar] [CrossRef]
- Poorter, H.; Niinemets, Ü.; Poorter, L.; Wright, I.J.; Villar, R. Causes and consequences of variation in leaf mass per area (LMA): A meta-analysis. New Phytol. 2009, 182, 565–588. [Google Scholar] [CrossRef] [PubMed]
- Vitousek, P.M.; Sanford, R.L. Nutrient cycling in moist tropical forest. Annu. Rev. Ecol. Syst. 1986, 17, 137–167. [Google Scholar] [CrossRef]
- Melillo, J.M.; Aber, J.D.; Muratore, J.F. Nitrogen and lignin control of hardwood leaf litter decomposition dynamics. Ecology 1982, 63, 621–626. [Google Scholar] [CrossRef]
- Feeny, P. Plant apparency and chemical defense. Recent Adv. Phytochem. 1976, 10, 1–40. [Google Scholar]
- Coley, P.D. Costs and benefits of defense by tannins in a neotropical tree. Oecologia 1986, 70, 238–241. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
- Webb, C.O.; Donoghue, M.J. Phylomatic: Tree assembly for applied phylogenetics. Mol. Ecol. Notes 2005, 5, 181–183. [Google Scholar] [CrossRef]
- Woodring, W.P. Geology of Barro Colorado Island, Canal Zone; Smithsonian Institution: Washington, DC, USA, 1958; Volume 4304. [Google Scholar]
- Baillie, I.; Elsenbeer, H.; Barthold, F.; Grimm, R.; Stallard, R.F. Semi-Detailed Soil Survey of Barro Colorado Island; Smithsonian Tropical Research Institute: Panama City, Panama, 2007. [Google Scholar]
- Vitousek, P.M. Litterfall, nutrient cycling, and nutrient limitation in tropical forests. Ecology 1984, 65, 285–298. [Google Scholar] [CrossRef]
- Messmer, T.; Elsenbeer, H.; Wilcke, W. High exchangeable calcium concentrations in soils on Barro Colorado Island, Panama. Geoderma 2014, 217–218, 212–224. [Google Scholar] [CrossRef]
- Leigh, E.G. Tropical Forest Ecology: A View from Barro Colorado Island; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Yavitt, J.B. Nutrient dynamics of soil derived from different parent material on Barro Colorado Island, Panama. Biotropica 2000, 32, 198–207. [Google Scholar] [CrossRef]
- Asner, G.P.; Anderson, C.B.; Martin, R.E.; Tupayachi, R.; Knapp, D.E.; Sinca, F. Landscape biogeochemistry reflected in shifting distributions of chemical traits in the Amazon forest canopy. Nat. Geosci. 2015, 8, 567–573. [Google Scholar] [CrossRef]
- Quesada, C.A.; Lloyd, J.; Schwarz, M.; Baker, T.R.; Phillips, O.L.; Patiño, S.; Czimczik, C.; Hodnett, M.G.; Herrera, R.; Arneth, A.; et al. Regional and large-scale patterns in Amazon forest structure and function are mediated by variations in soil physical and chemical properties. Biogeosci. Discuss. 2009, 6, 3993–4057. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Convergent elevation trends in canopy chemical traits of tropical forests. Glob. Change Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.C. Environmental Genomics; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2008; Volume 410. [Google Scholar]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8.
- Sanderson, M.J. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 1997, 14, 1218–1231. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R Data Import/Export, Version 2.10.0; 2009. [Google Scholar]
- Webb, C.O.; Ackerly, D.D.; Kembel, S.W. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 2008, 24, 2098–2100. [Google Scholar] [CrossRef] [PubMed]
- Gastauer, M.; Meira-Neto, J.A.A. Avoiding inaccuracies in tree calibration and phylogenetic community analysis using Phylocom 4.2. Ecol. Inform. 2013, 15, 85–90. [Google Scholar] [CrossRef]
- Wikström, N.; Savolainen, V.; Chase, M.W. Evolution of the angiosperms: Calibrating the family tree. Proc. Biol. Sci. 2001, 268, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Münkemüller, T.; Lavergne, S.; Bzeznik, B.; Dray, S.; Jombart, T.; Schiffers, K.; Thuiller, W. How to measure and test phylogenetic signal. Methods Ecol. Evol. 2012, 3, 743–756. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Garland, T.; Harvey, P.H.; Ives, A.R. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 1992, 41, 18–32. [Google Scholar] [CrossRef]
- Ollinger, S. V. Sources of variability in canopy reflectance and the convergent properties of plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Mutanga, O.; Skidmore, A.K.; van Wieren, S. Discriminating tropical grass (Cenchrus ciliaris) canopies grown under different nitrogen treatments using spectroradiometry. ISPRS J. Photogramm. Remote Sens. 2003, 57, 263–272. [Google Scholar] [CrossRef]
- Reich, P.B.; Oleksyn, J.; Wright, I.J. Leaf phosphorus influences the photosynthesis-nitrogen relation: A cross-biome analysis of 314 species. Oecologia 2009, 160, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Fine, P.V.A.; Miller, Z.J.; Mesones, I.; Irazuzta, S.; Appel, H.M.; Stevens, M.H.H.; Sääksjärvi, I.; Schultz, J.C.; Coley, P.D. The growth-defense trade-off and habitat specialization by plants in Amazonian forests. Ecology 2006, 87, S150–S162. [Google Scholar] [CrossRef]
- Reich, P.B.; Wright, I.J.; Cavender-Bares, J.; Craine, J.M.; Olesksyn, J.; Westoby, M.; Walters, M.B. The evolution of plant functional variation: Traits, spectra, and strategies. Int. J. Plant Sci. 2003, 164, S143–S164. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Carranza-Jiménez, L.; Martinez, P. Amazonian functional diversity from forest canopy chemical assembly. Proc. Natl. Acad. Sci. USA 2014, 111, 5604–5609. [Google Scholar] [CrossRef] [PubMed]
- Revell, L.J. Size-correction and principal components for interspecific comparative studies. Evolution 2009, 63, 3258–3268. [Google Scholar] [CrossRef] [PubMed]
| BCI | Tam-I | Tam-U | |
|---|---|---|---|
| Demographics | |||
| Individuals | 69 | 196 | 200 |
| Species | 69 (13) | 125 (40) | 150 (34) |
| Genera | 52 (30) | 89 (48) | 90 (41) |
| Families | 27 (25) | 37 (28) | 33 (26) |
| Environment | |||
| Elevation (m) | 189 | 220 | 213 |
| MAP (mm/yr) | 2550 | 2600 | 2600 |
| MAT (°C) | 27.2 | 24 | 24 |
| Primary soil types | Combisols/Ferralsols | Inceptisols | Ultisols |
| Trait | Tam-I | Tam-U | F | ||||||||
| Mean | CV (%) | Range | λDNA | λPhyl | Mean | CV (%) | Range | λDNA | λPhyl | ||
| Nitrogen † (%) | 2.20 a ± 0.64 | 29.0 | 1.04–4.08 | 0.86 ** | 0.66 ** | 2.18 a ± 0.66 | 30.2 | 1.19–4.66 | 0.66 ** | 0.55 ** | 0.0 |
| Phosphorus † (%) | 0.20 b ± 0.08 | 40.0 | 0.09–0.51 | 0.50 ** | 0.58 ** | 0.13 a ± 0.05 | 39.1 | 0.05–0.33 | 0.00 | 0.00 | 72.6 *** |
| Calcium †† (%) | 1.58 a ± 0.82 | 51.8 | 0.25–4.85 | 0.47 ** | 0.52 ** | 0.72 b ± 0.69 | 95.9 | 0.03–3.45 | 0.30 * | 0.23 ** | 77.1 *** |
| LMA †† (g·m−2) | 101 a ± 29 | 28.7 | 47–191 | 0.70 * | 0.45 * | 111 b ± 30 | 27.0 | 38–216 | 0.14 | 0.12 | 12.0 *** |
| Lignin †† (%) | 19.9 b ± 8.4 | 42.0 | 6.3–42.2 | 0.79 ** | 0.84 ** | 23.6 a ± 9.3 | 39.4 | 5.3–50.8 | 0.20 | 0.39 ** | 9.3 ** |
| Cellulose †† (%) | 17.5 a ± 4.6 | 26.3 | 7.3–29.3 | 0.00 | 0.20 | 18.4 a ± 4.9 | 26.7 | 6.4–35.7 | 0.00 | 0.21 | 1.9 |
| Phenols (mg·g−1) | 68 b ± 45 | 66.4 | 1–173 | 0.00 | 0.00 | 90 a ± 50 | 55.6 | 1–225 | 0.42 | 0.52 ** | 12.1 *** |
| Tannins (mg·g−1) | 36 a ± 21 | 57.4 | 1–87 | 0.00 | 0.00 | 43 b ± 22 | 51.1 | 0–116 | 0.39 | 0.54 ** | 4.9 ** |
| Chl † (mg·g−1) | 7.41 b ± 2.29 | 30.9 | 2.28–15.11 | 0.30 | 0.10 | 6.69 c ± 2.18 | 32.5 | 2.16–15.39 | 0.13 | 0.12 | 22.2 *** |
| Trait | BCI | All sites | |||||||||
| Mean | CV (%) | Range | λDNA | λPhyl | Mean | CV (%) | Range | λDNA | λPhyl | ||
| Nitrogen † (%) | 2.18 a ± 0.78 | 35.8 | 1.01–5.04 | 0.00 | 0.37 | 2.19 ± 0.67 | 30.5 | 1.01–5.04 | 0.82 *** | 0.65 *** | |
| Phosphorus † (%) | 0.12 a ± 0.06 | 49.6 | 0.05–0.43 | 0.00 | 0.00 | 0.16 ± 0.07 | 47.6 | 0.05–0.51 | 0.22 | 0.23 * | |
| Calcium †† (%) | 1.40 a ± 0.77 | 55.0 | 0.30–3.72 | 0.00 | 0.47 * | 1.18 ± 0.86 | 72.7 | 0.03–4.85 | 0.51 *** | 0.38 *** | |
| LMA †† (g·m−2) | 93 a ± 27 | 29.0 | 30–155 | 0.00 | 0.00 | 104 ± 30 | 28.8 | 30–216 | 0.39 ** | 0.22 *** | |
| Lignin †† (%) | 23.0 a ± 9.4 | 51.0 | 7.0–43.1 | 0.61 | 0.51 * | 22.0 ± 9.1 | 41.4 | 5.3–50.8 | 0.79 *** | 0.70 *** | |
| Cellulose †† (%) | 18.1 a ± 4.9 | 27.1 | 9.9–31.0 | 0.17 | 0.17 | 18.0 ± 4.8 | 26.5 | 6.4–35.7 | 0.20 * | 0.54 *** | |
| Phenols (mg·g−1) | 92 a ± 56 | 60.7 | 5–256 | 0.00 | 0.00 | 81 ± 50 | 61.9 | 1–256 | 0.44 | 0.41 *** | |
| Tannins (mg·g−1) | 34 a ± 21 | 61.7 | 0–100 | 0.00 | 0.10 | 39 ± 22 | 55.6 | 0–116 | 0.49 * | 0.40 *** | |
| Chl † (mg·g−1) | 7.71 a ± 2.77 | 35.9 | 3.81–19.68 | 0.00 | 0.00 | 7.15 ± 2.35 | 32.9 | 2.16–19.68 | 0.18 * | 0.15 * | |
| PLS R2 | RF R2 | SVM R2 | Ensemble R2 | Bands (n) | |
|---|---|---|---|---|---|
| Nitrogen | 0.70 | 0.18 | 0.66 | 0.51 | 31 |
| Phosphorus | 0.40 | 0.20 | 0.38 | 0.33 | 34 |
| Calcium | 0.48 | 0.16 | 0.48 | 0.38 | 33 |
| LMA | 0.76 | 0.59 | 0.77 | 0.71 | 40 |
| Lignin | 0.39 | 0.07 | 0.41 | 0.29 | 36 |
| Cellulose | 0.52 | 0.08 | 0.47 | 0.36 | 39 |
| Phenols | 0.69 | 0.33 | 0.73 | 0.58 | 24 |
| Tannins | 0.59 | 0.25 | 0.59 | 0.49 | 30 |
| Chlorophyll | 0.72 | 0.56 | 0.73 | 0.67 | 31 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McManus, K.M.; Asner, G.P.; Martin, R.E.; Dexter, K.G.; Kress, W.J.; Field, C.B. Phylogenetic Structure of Foliar Spectral Traits in Tropical Forest Canopies. Remote Sens. 2016, 8, 196. https://doi.org/10.3390/rs8030196
McManus KM, Asner GP, Martin RE, Dexter KG, Kress WJ, Field CB. Phylogenetic Structure of Foliar Spectral Traits in Tropical Forest Canopies. Remote Sensing. 2016; 8(3):196. https://doi.org/10.3390/rs8030196
Chicago/Turabian StyleMcManus, Kelly M., Gregory P. Asner, Roberta E. Martin, Kyle G. Dexter, W. John Kress, and Christopher B. Field. 2016. "Phylogenetic Structure of Foliar Spectral Traits in Tropical Forest Canopies" Remote Sensing 8, no. 3: 196. https://doi.org/10.3390/rs8030196
APA StyleMcManus, K. M., Asner, G. P., Martin, R. E., Dexter, K. G., Kress, W. J., & Field, C. B. (2016). Phylogenetic Structure of Foliar Spectral Traits in Tropical Forest Canopies. Remote Sensing, 8(3), 196. https://doi.org/10.3390/rs8030196