Retrieval of Mangrove Aboveground Biomass at the Individual Species Level with WorldView-2 Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Field Investigation and Estimation of Mangrove Vegetation Biomass


| Species | Tissues | Allometric Equations | Correlation |
|---|---|---|---|
| S. apetala | Foliage | lgWlf = −0.756 + 0.4355 lg (DBH2 × Height) | 0.94 |
| Branch | lgWbr = 0.1590 + 0.3879 lg (DBH2 × Height) | 0.95 | |
| Trunk | lgWst = 0.3067 + 0.3302 lg (DBH2 × Height) | 0.95 | |
| Bark | lgWba = −0.3790 + 0.3559 lg (DBH2 × Height) | 0.91 | |
| Flowers and fruit | lgWfr = −2.3456 + 0.3791 lg (DBH2 × Height) | 0.97 | |
| K. candel | Foliage | lgWlf = −1.1704 + 0.4855 lg (DBH2 × Height) | 0.87 |
| Branch | lgWbr = −0.9067 + 0.5762 lg (DBH2 × Height) | 0.95 | |
| Trunk and bark | lgWst+ba = −0.3112 + 0.2542 lg (DBH2 × Height) | 0.88 | |
| Flowers and fruit | lgWfr = −3.1582 + 1.061 lg (DBH2 × Height) | 0.85 |
2.3. Remotely Sensed Data and Processing
| Wavebands | Spectral Band Edges | Spatial Resolution | |
|---|---|---|---|
| B1 | coastal band | 400 nm to 450 nm | 2 m |
| B2 | blue band | 450 nm to 510 nm | 2 m |
| B3 | green band | 510 nm to 580 nm | 2 m |
| B4 | yellow band | 585 nm to 625 nm | 2 m |
| B5 | red band | 630 nm to 690 nm | 2 m |
| B6 | red-edge band | 705 nm to 745 nm | 2 m |
| B7 | near-infrared-1 band | 770 nm to 895 nm | 2 m |
| B8 | near-infrared-2 band | 860 nm to 1040 nm | 2 m |
2.4. Mangrove Classification Based on WorldView-2 Images
2.4.1. The Characteristics and Spatial Distribution of Mangrove Species
| Land Cover Types | Characteristics and Spatial Distribution Features | |
|---|---|---|
| Target objects | S. apetala | The tree species are tallest in height, distributed in medium to low tidal zones and occupied the largest area on Qi’ao Island. |
| K. candel | The tree species are mainly located outside the enclosing levee in the high tidal zones. | |
| Other vegetation | Other vegetation types, including shrubs (A. corniculatum and A. ilicifolius) and herbaceous (S. alterniflora and P. australis) vegetation, are mainly distributed in medium and low tidal zones. | |
2.4.2. Image Segmentation
2.4.3. The Classification for Mangrove Species
- (1)
- 8 spectral attributes: mean values of 8 bands of WorldView-2 for each image object;
- (2)
- 5 shape attributes: area, density, length/width, shape index, and width of each image object;
- (3)
- 1 vegetation attribute: NDVI for each image object.
2.5. Calculation of Vegetation Indices
2.6. Biomass Modeling Accuracy Assessment
2.7. Importance of the Variables
3. Results
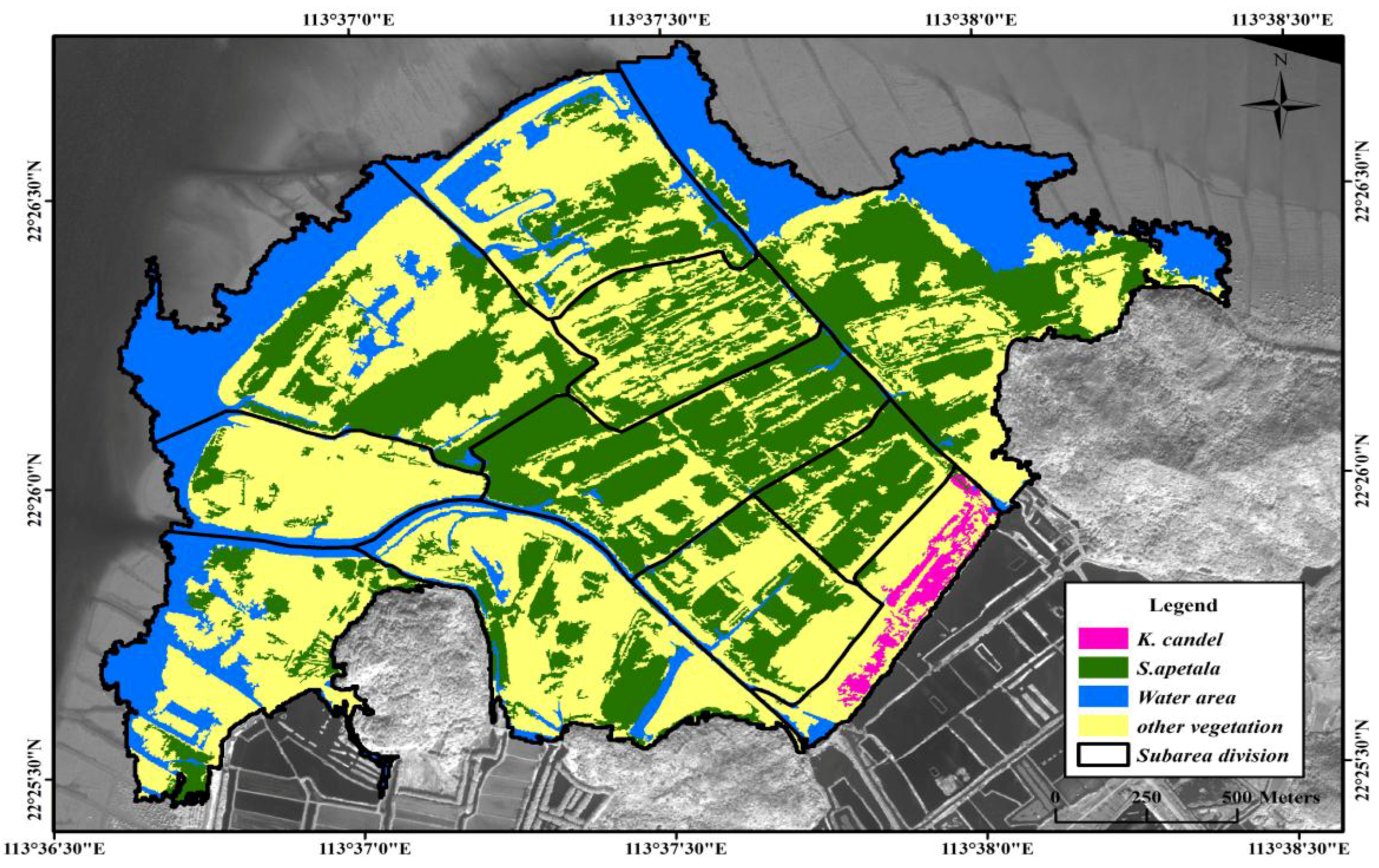
3.1. Mangrove Classification at Species Level
| Classified Data | Reference Data | |||||
|---|---|---|---|---|---|---|
| K. candel | S. apetala | Other Vegetation | Water Area | Total | User’s Accuracy | |
| K. candel | 68 | 0 | 8 | 0 | 76 | 89.47% |
| S. apetala | 0 | 252 | 30 | 0 | 282 | 89.36% |
| Other vegetation | 12 | 48 | 339 | 13 | 412 | 82.28% |
| Water area | 0 | 0 | 12 | 216 | 228 | 94.74% |
| Total | 80 | 300 | 389 | 229 | 998 | |
| Producer’s accuracy | 85.00% | 84.00% | 87.15% | 94.32% | ||
| Overall accuracy: 87.68%;Kappa:0.82 | ||||||
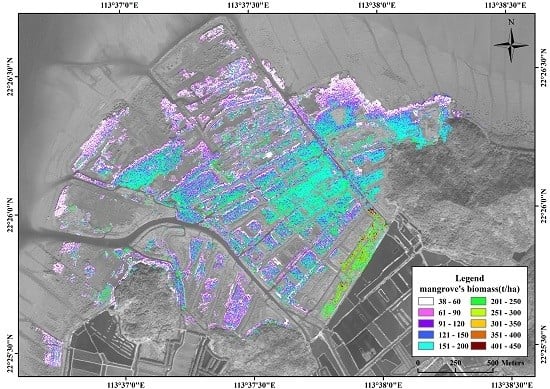
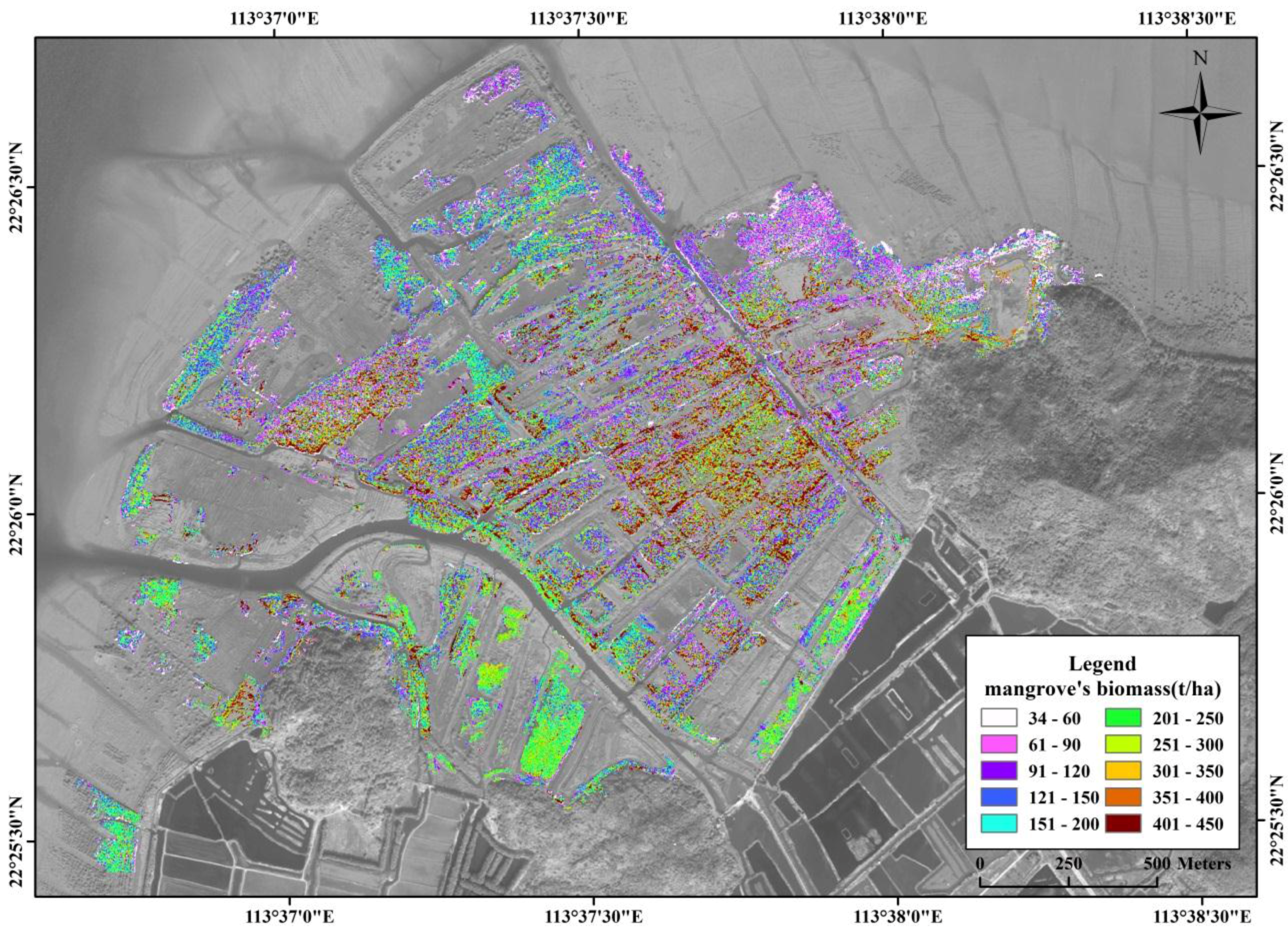
3.2. Estimation of Mangrove Vegetation Biomass
| Species | Average Height(m) | Average DBH (cm) | Number of Trees per ha | Range of Biomass (t/ha) | The Mean of AGB (t/ha) | Standard Deviations (t/ha) |
|---|---|---|---|---|---|---|
| K. candel | 6.62 | 8.32 | 5257 | 170.96~448.19 | 272.97 | 67.27 |
| S. apetala | 11.41 | 13.23 | 1860 | 37.44~258.17 | 125.49 | 55.23 |
3.3. The Relationship between the Biomass and the Selected Variables

3.4. Model Results and Accuracy Test
| BP ANN Model | RMSEs (t/ha) | Average RMSE (t/ha) | Average RMSEr (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Expt.1 | Mixed species | 74.41 | 71.75 | 82.38 | 68.80 | 63.97 | 72.26 | 43.14% |
| Expt.2 | Dummy species | 40.69 | 38.23 | 45.92 | 33.44 | 42.45 | 40.15 | 23.97% |
| Expt.3 | K. candel | 52.12 | 48.37 | 49.48 | 42.23 | 69.70 | 52.38 | 19.52% |
| S. apetala | 31.54 | 26.92 | 22.46 | 18.27 | 22.45 | 24.32 | 18.97% | |
3.5. Spatial Distribution of Mangrove Vegetation Biomass
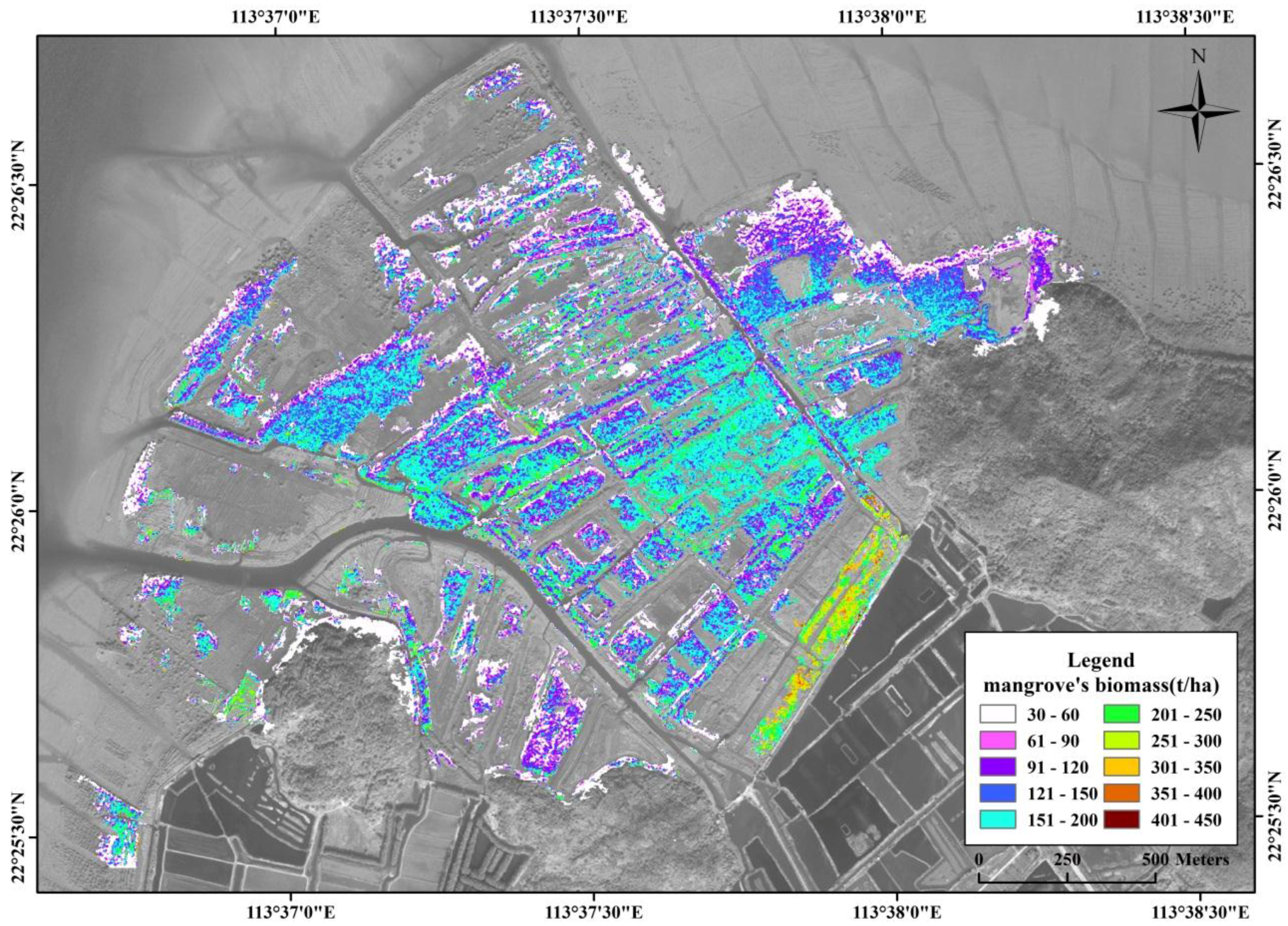

4. Discussion
4.1. Biomass Estimation Accuracy Involving Species Types
4.2. The Remote Sensing-Derived Input Variables Contribution to the Mangrove Biomass
4.3. Spatial Distribution of Mangrove Vegetation Biomass
5. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Myint, S.W.; Giri, C.P.; Wang, L.; Zhu, Z.; Gillete, S.C. Identifying mangrove species and their surrounding land use and land cover classes using object-oriented approach with a lacunarity spatial measure. GISci. Remote Sens. 2008, 45, 188–208. [Google Scholar] [CrossRef]
- Pendleton, L.H.; Sutton-Grier, A.E.; Gordon, D.R.; Murray, B.C.; Victor, B.E.; Griffis, R.B.; Lechuga, J.A.V.; Giri, C. Considering “Coastal Carbon” in existing U.S. Federal Statutes and Policies. Coast. Manag. 2013, 41, 439–456. [Google Scholar] [CrossRef]
- Myint, S.W.; Franklin, J.; Buenemann, M.; Kim, W.K.; Giri, C.P. Examining change detection approaches for tropical mangrove monitoring. Photogramm. Engin. Remote Sens. 2014, 80, 983–993. [Google Scholar] [CrossRef]
- Kankare, V.; Vastaranta, M.; Holopainen, M.; Räty, M.; Yu, X.; Hyyppä, J.; Hyyppä, H.; Alho, P.; Viitala, R. Retrieval of forest aboveground biomass and stem volume with airborne scanning LiDAR. Remote Sens. 2013, 5, 2257–2274. [Google Scholar] [CrossRef]
- Kosoy, N.; Corbera, E. Payments for ecosystem services as commodity fetishism. Ecol. Econ. 2010, 69, 1228–1236. [Google Scholar] [CrossRef]
- Kovacs, J.; King, J.; Flores De Santiago, F.; Flores-Verdugo, F. Evaluating the condition of a mangrove forest of the Mexican Pacific based on an estimated leaf area index mapping approach. Environ. Monit. Assess. 2009, 157, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Lovelock, C. Soil respiration and belowground carbon allocation in mangrove forests. Ecosystems 2008, 11, 342–354. [Google Scholar] [CrossRef]
- Thapa, R.B.; Watanabe, M.; Motohka, T.; Shimada, M. Potential of high-resolution ALOS-PALSAR mosaic texture for aboveground forest carbon tracking in tropical region. Remote Sens. Environ. 2015, 160, 122–133. [Google Scholar] [CrossRef]
- Chen, L.Z.; Lin, P.; Wang, W.Q. The Eco-Engineering Problems of Plantable Areas Selection in Mangroves Forestation. Available online: www.paper.edu.cn/download/downPaper/200503-48 (accessed on 14 September 2015). (In Chinese)
- Peng, Y.; Zhou, Y.; Chen, G. The restoration of mangrove wetland: A review. Acta Ecol. Sin. 2008, 28, 786–797. (In Chinese) [Google Scholar]
- Jia, M.; Wang, Z.; Zhang, Y.; Ren, C.; Song, K. Landsat-Based estimation of mangrove forest loss and restoration in Guangxi Province, China, influenced by human and natural factors. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 311–323. [Google Scholar] [CrossRef]
- Feng, J.; Guo, J.; Huang, Q.; Jiang, J.; Huang, G.; Yang, Z.; Lin, G. Changes in the community structure and diet of benthic macrofauna in invasive Spartina alterniflora Wetlands following restoration with native mangroves. Wetlands 2014, 34, 673–683. [Google Scholar] [CrossRef]
- Giri, C.; Ochieng, E.; Tieszen, L.L.; Zhu, Z.; Singh, A.; Loveland, T.; Masek, J.; Duke, N. Status and distribution of mangrove forests of the world using earth observation satellite data. Global Ecol. Biogeogr. 2011, 20, 154–159. [Google Scholar] [CrossRef]
- Heumann, B.W. Satellite remote sensing of mangrove forests: Recent advances and future opportunities. Prog. Phys. Geogr. 2011, 35, 87–108. [Google Scholar] [CrossRef]
- Hirata, Y.; Tabuchi, R.; Patanaponpaiboon, P.; Poungparn, S.; Yoneda, R.; Fujioka, Y. Estimation of aboveground biomass in mangrove forests using high-resolution satellite data. J. For. Res. 2014, 19, 34–41. [Google Scholar] [CrossRef]
- Patil, V.; Singh, A.; Naik, N.; Unnikrishnan, S. Estimation of carbon stocks in Avicennia marina stand using allometry, CHN analysis, and GIS methods. Wetlands 2014, 34, 379–391. [Google Scholar] [CrossRef]
- Ishil, T.; Tateda, Y. Leaf area index and biomass estimation for mangrove plantation in Thailand. In Proceedings of IEEE International Geoscience and Remote Sensing Symposium, Anchorage, AK, USA, 20–24 September 2004; pp. 2323–2326.
- Proisy, C.; Couteron, P.; Fromard, F. Predicting and mapping mangrove biomass from canopy grain analysis using Fourier-based textural ordination of IKONOS images. Remote Sens. Environ. 2007, 109, 379–392. [Google Scholar] [CrossRef]
- Ji, M.; Hu, J.; Feng, J. Measuring mangrove biomass via remote sensing subpixel analysis. Remote Sens. Model. Ecosyst Sustain. 2010, 7809. [Google Scholar] [CrossRef]
- Lucas, R.M.; Mitchell, A.L.; Rosenqvist, A.; Proisy, C.; Melius, A.; Ticehurst, C. The potential of L-band SAR for quantifying mangrove characteristics and change: Case studies from the tropics. Aquat. Conserv. Mar. Freshw. Ecosyst. 2007, 17, 245–264. [Google Scholar] [CrossRef]
- Li, X.; Yeh, A.G.; Wang, S.; Liu, K.; Liu, X.; Qian, J.; Chen, X. Regression and analytical models for estimating mangrove wetland biomass in South China using Radarsat images. Int. J. Remote Sens. 2007, 28, 5567–5582. [Google Scholar] [CrossRef]
- Simard, M.; Rivera-Monroy, V.H.; Mancera-Pineda, J.E.; Castañeda-Moya, E.; Twilley, R.R. A systematic method for 3D mapping of mangrove forests based on Shuttle Radar Topography Mission elevation data, ICEsat/GLAS waveforms and field data: Application to Ciénaga Grande de Santa Marta, Colombia. Remote Sens. Environ. 2008, 112, 2131–2144. [Google Scholar] [CrossRef]
- Chadwick, J. Integrated LiDAR and IKONOS multispectral imagery for mapping mangrove distribution and physical properties. Int. J. Remote Sens. 2011, 32, 6765–6781. [Google Scholar] [CrossRef]
- Koedsin, W.; Vaiphasa, C. Discrimination of tropical mangroves at the species level with EO-1 hyperion data. Remote Sens. 2013, 5, 3562–3582. [Google Scholar] [CrossRef]
- Ni-Meister, W.; Lee, S.; Strahler, A.H.; Woodcock, C.E.; Schaaf, C.; Yao, T.; Ranson, K.J.; Sun, G.; Blair, J.B. Assessing general relationships between aboveground biomass and vegetation structure parameters for improved carbon estimate from LiDAR remote sensing. J. Geophys. Res. 2010, 115. [Google Scholar] [CrossRef]
- Chen, Q.; Vaglio Laurin, G.; Battles, J.J.; Saah, D. Integration of airborne LiDAR and vegetation types derived from aerial photography for mapping aboveground live biomass. Remote Sens. Environ. 2012, 121, 108–117. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using worldview-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinform. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Demuro, M.; Chisholm, L. Assessment of hyperion for characterizing mangrove communities. In Proceedings of the 12th JPL AVIRIS Airborne Earth Science Workshop, Pasadena, CA, USA, 24–28 February 2003.
- Heenkenda, M.; Joyce, K.; Maier, S.; Bartolo, R. Mangrove species identification: Comparing WorldView-2 with aerial photographs. Remote Sens. 2014, 6, 6064–6088. [Google Scholar] [CrossRef]
- Eacute, S.; Rapinel, B.; Eacute, B.C.; Ment; Magnanon, S.; Sellin, V.; Hubert-Moy, L. Identification and mapping of natural vegetation on a coastal site using a Worldview-2 satellite image. J. Environ. Manag. 2014, 144, 236–246. [Google Scholar]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Adam, E.M.I.; Mutanga, O. Estimation of high density wetland biomass: combining regression model with vegetation index developed from Worldview-2 imagery. Proc. SPIE 2012, 8531. [Google Scholar] [CrossRef]
- Eckert, S. Improved forest biomass and carbon estimations using texture measures from worldView-2 satellite data. Remote Sens. 2012, 4, 810–829. [Google Scholar] [CrossRef]
- Jachowski, N.R.A.; Quak, M.S.Y.; Friess, D.A.; Duangnamon, D.; Webb, E.L.; Ziegler, A.D. Mangrove biomass estimation in Southwest Thailand using machine learning. Appl. Geogr. 2013, 45, 311–321. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ciresan, D.; Meier, U.; Schmidhuber, J. Multi-column deep neural networks for image classification. In Proceedings of IEEE Computer Vision and Pattern Recognition (CVPR), Providence, RI, USA, 16–21 June 2012; pp. 3642–3649.
- Mas, J.F.; Flores, J.J. The application of artificial neural networks to the analysis of remotely sensed data. Int. J. Remote Sens. 2008, 29, 617–663. [Google Scholar] [CrossRef]
- Wang, L.; Silván-Cárdenas, J.L.; Sousa, W.P. Neural network classification of mangrove species from multiseasonal IKONOS imagery. Photogramm. Engin. Remote Sens. 2008, 74, 921–927. [Google Scholar] [CrossRef]
- Liu, K.; Li, X.; Wang, S.G.; Liu, W.X. Classification of mangroves by data fusion and neural networks. Remote Sens. Inf. 2006, 3, 32–36. (In Chinese) [Google Scholar]
- Liu, K.; Li, X.; Shi, X.; Wang, S. Monitoring mangrove forest changes using remote sensing and GIS data With decision-tree learning. Wetlands 2008, 28, 336–346. [Google Scholar] [CrossRef]
- Wang, S.G.; Li, X.; Zhou, Y.Z.; Liu, K.; Chen, G.Z. The change of mangrove wetland ecosystem and controlling countermeasures in the Qi’ao Island. Wetl. Sci. 2005, 3, 13–20. (In Chinese) [Google Scholar]
- Liu, K.; Liu, L.; Liu, H.; Li, X.; Wang, S. Exploring the effects of biophysical parameters on the spatial pattern of rare cold damage to mangrove forests. Remote Sens. Environ. 2014, 150, 20–33. [Google Scholar] [CrossRef]
- Liao, B.W.; Guan, W.; Zhang, J.E.; Tang, G.L.; Lei, Z.S. Studies on Dynamic Development of Mangrove Communities on Qi’ao Island, Zhuhai. J. South China Agric. Univ. 2008, 29, 59–64. (In Chinese) [Google Scholar]
- Chen, H.; Liao, B.; Liu, B.E.; Peng, C.; Zhang, Y.; Guan, W.; Zhu, Q.A.; Yang, G. Eradicating invasive Spartina alterniflora with alien Sonneratia apetala and its implications for invasion controls. Ecol. Engin. 2014, 73, 367–372. (In Chinese) [Google Scholar] [CrossRef]
- Zan, Q.J.; Wang, Y.J. Biomass and net productivity of Sonneratia apetala, S.caseolaris mangrove man-made forest. J. Wuhan Bot. Res. 2001, 19, 391–396. (In Chinese) [Google Scholar]
- Aguilar, M.A.; Saldaña, M.M.; Aguilar, F.J. GeoEye-1 and WorldView-2 pan-sharpened imagery for object-based classification in urban environments. Int. J. Remote Sens. 2013, 34, 2583–2606. [Google Scholar] [CrossRef]
- Oumar, Z.; Mutanga, O. Using WorldView-2 bands and indices to predict bronze bug (Thaumastocoris peregrinus) damage in plantation forests. Int. J. Remote Sens. 2013, 34, 2236–2249. [Google Scholar] [CrossRef]
- Sridharan, H.; Qiu, F. Developing an object-based hyperspatial image classifier with a case study using WorldView-2 data. Photogramm. Engin. Remote Sens. 2013, 79, 1027–1036. [Google Scholar] [CrossRef]
- Puetz, A.M.; Lee, K.; Olsen, R.C. WorldView-2 data simulation and analysis results. Proc. SPIE 2009, 73340. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with Random forest using very high spatial resolution 8-band worldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Mitri, G.H.; Gitas, I.Z. A performance evaluation of a burned area object-based classification model when applied to topographically and non-topographically corrected TM imagery. Int. J. Remote Sens. 2004, 25, 2863–2870. [Google Scholar] [CrossRef]
- Laliberte, A.S.; Fredrickson, E.L.; Rango, A. Combining decision trees with hierarchical object-oriented image analysis for mapping arid rangelands. Photogramm. Engin. Remote Sens. 2007, 73, 197–207. [Google Scholar] [CrossRef]
- Laliberte, A.S.; Rango, A.; Havstad, K.M.; Paris, J.F.; Beck, R.F.; McNeely, R.; Gonzalez, A.L. Object-oriented image analysis for mapping shrub encroachment from 1937 to 2003 in southern New Mexico. Remote Sens. Environ. 2004, 93, 198–210. [Google Scholar] [CrossRef]
- Briggs, J.M. An object-oriented approach to urban forest mapping in Phoenix. Photogramm. Engin. Remote Sens. 2007, 73, 577–583. [Google Scholar]
- Wieland, M.; Pittore, M. Performance evaluation of machine learning algorithms for urban pattern recognition from multi-spectral satellite images. Remote Sens. 2014, 6, 2912–2939. [Google Scholar] [CrossRef]
- Meng, Q.; Cieszewski, C.J.; Madden, M.; Borders, B.E. K Nearest neighbor method for forest inventory using remote sensing data. GIScience Remote Sens. 2007, 44, 149–165. [Google Scholar] [CrossRef]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Hall, D.E.; Falkowski, M.J. Nearest neighbor imputation of species level, plot-scale forest structure attributes from LiDAR data. Remote Sens. Environ. 2008, 112, 2232–2245. [Google Scholar] [CrossRef]
- Wang, Q.; Adiku, S.; Tenhunen, J.; Granier, A. On the relationship of NDVI with leaf area index in a deciduous forest site. Remote Sens. Environ. 2005, 94, 244–255. [Google Scholar] [CrossRef]
- Kasawani, I.; Norsaliza, U.; Hasmadi, I.M. Analysis of spectral vegetation indices related to soil-line for mapping mangrove forests using satellite imagery. Appl. Remote Sens. J. 2010, 1, 25–31. [Google Scholar]
- Darvishzadeh, R.; Skidmore, A.; Atzberger, C.; van Wieren, S. Estimation of vegetation LAI from hyperspectral reflectance data: Effects of soil type and plant architecture. Int. J. Appl. Earth Obs. Geoinform. 2008, 10, 358–373. [Google Scholar] [CrossRef]
- Peña-Barragán, J.M.; Ngugi, M.K.; Plant, R.E.; Six, J. Object-based crop identification using multiple vegetation indices, textural features and crop phenology. Remote Sens. Environ. 2011, 115, 1301–1316. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards Jr, T.C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef] [PubMed]
- Muukkonen, P.; Heiskanen, J. Estimating biomass for boreal forests using ASTER satellite data combined with standwise forest inventory data. Remote Sens. Environ. 2005, 99, 434–447. [Google Scholar] [CrossRef]
- Demuth, H.; Beale, M.; Hagan, M. Neural Network Toolbox™ 6. User’s Guide. 2008. Available Online: http://kashanu.ac.ir/Files/Content/neural_network_toolbox_6.pdf (accessed on 15 September 2015).
- Garson, G.D. Interpreting neural-network connection weights. Ai Expert 1991, 6, 46–51. [Google Scholar]
- Gevrey, M.; Dimopoulos, I.; Lek, S. Review and comparison of methods to study the contribution of variables in artificial neural network models. Ecol. Model. 2003, 160, 249–264. [Google Scholar] [CrossRef]
- Chave, J.; Muller-Landau, H.C.; Baker, T.R.; Easdale, T.A.; Hans Steege, T.E.R.; Webb, C.O. Regional and phylogenetic variation of wood density across 2456 neotropical tree species. Ecol. Appl. 2006, 16, 2356–2367. [Google Scholar] [CrossRef]
- Niklas, K.J. Size-dependent allometry of tree height, diameter and trunk-taper. Ann. Bot. 1995, 75, 217–227. [Google Scholar] [CrossRef]
- Komiyama, A.; Ong, J.E.; Poungparn, S. Allometry, biomass, and productivity of mangrove forests: A review. Aquat. Bot. 2008, 89, 128–137. [Google Scholar] [CrossRef]
- Nelson, R.; Krabill, W.; Tonelli, J. Estimating forest biomass and volume using airborne laser data. Remote Sens. Environ. 1988, 24, 247–267. [Google Scholar] [CrossRef]
- Adelabu, S.; Mutanga, O.; Adam, E.; Sebego, R. Spectral discrimination of insect defoliation levels in mopane woodland using hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 177–186. [Google Scholar] [CrossRef]
- Tang, G.L.; Chen, L.H.; Weng, W.H.; Zhang, J.E.; Liao, B.W. Effects of using sonneratia apetala to control the growth of spartina alterniflora loisel. J. South China Agric. Univ. 2007, 28, 10–13. (In Chinese) [Google Scholar]
- Liao, B.W.; Tian, G.H.; Yang, X.B.; Tang, G.L.; Zhang, J.E. The analysis of natural regeneration and diffusion of the seedling of Sonneratia apetala in the Qi’ao Island, Zhuhai. Ecol. Sci. 2006, 25, 485–488. (In Chinese) [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Liu, K.; Liu, L.; Wang, S.; Liu, H. Retrieval of Mangrove Aboveground Biomass at the Individual Species Level with WorldView-2 Images. Remote Sens. 2015, 7, 12192-12214. https://doi.org/10.3390/rs70912192
Zhu Y, Liu K, Liu L, Wang S, Liu H. Retrieval of Mangrove Aboveground Biomass at the Individual Species Level with WorldView-2 Images. Remote Sensing. 2015; 7(9):12192-12214. https://doi.org/10.3390/rs70912192
Chicago/Turabian StyleZhu, Yuanhui, Kai Liu, Lin Liu, Shugong Wang, and Hongxing Liu. 2015. "Retrieval of Mangrove Aboveground Biomass at the Individual Species Level with WorldView-2 Images" Remote Sensing 7, no. 9: 12192-12214. https://doi.org/10.3390/rs70912192
APA StyleZhu, Y., Liu, K., Liu, L., Wang, S., & Liu, H. (2015). Retrieval of Mangrove Aboveground Biomass at the Individual Species Level with WorldView-2 Images. Remote Sensing, 7(9), 12192-12214. https://doi.org/10.3390/rs70912192