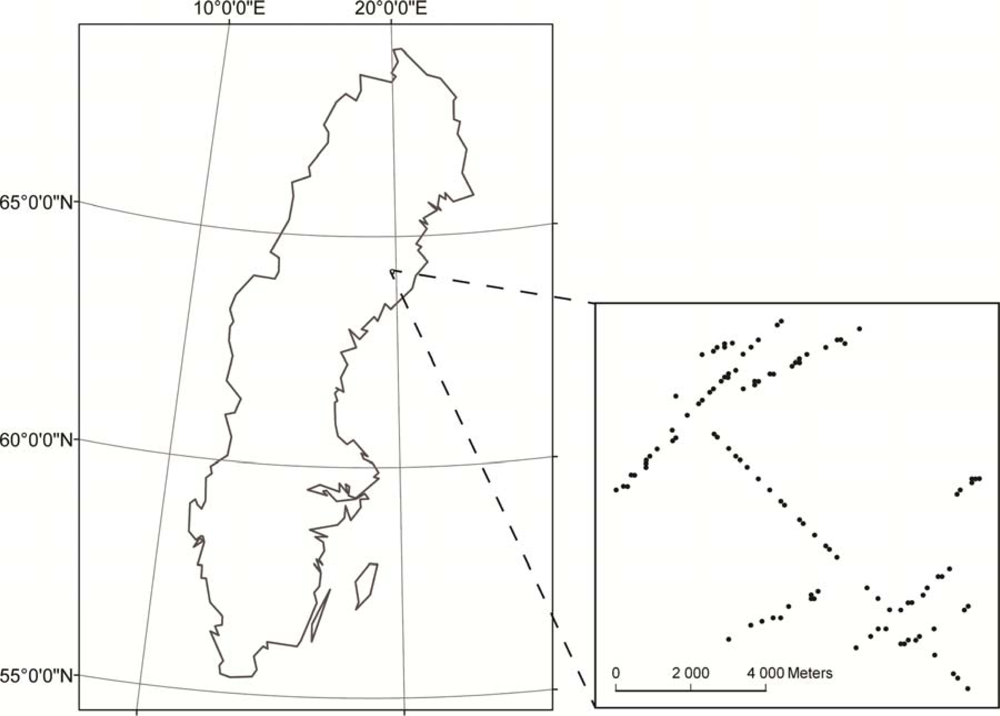
1. Introduction
Many modern systems for forest management planning require information at the individual tree level [
1–
4] or, at least, about the distribution of stem diameters at breast height (DBH) [
5]. For the purpose of forest resource planning, unbiased estimates are also essential.
Data from airborne laser scanning (ALS) are three-dimensional coordinate measurements of light reflections from the ground and other objects. During the last fifteen years, methods have been developed to use ALS data for estimation of forest variables such as tree height and stem volume. The most commonly used method is estimation at an area level when forest variables measured in field plots are modeled from variables derived from ALS data for the same area [
6]. The variables derived from the ALS data are typically measures of the height distribution and the density of the ALS data in different height intervals above the ground. The estimation is usually done with regression models [
6] or with semi-parametric models such as
k-Most Similar Neighbours (
k-MSN), where the similarity is based on canonical correlations [
7].
If the ALS data are dense enough, individual tree crowns (ITC) may also be delineated from the data. This has mostly been done based on surface models [
8–
10], such as a normalized digital surface model (nDSM). Typically, the local maxima in the surface model are defined as tree tops and the area around them is delineated to define tree crowns [
11]. Features extracted from the spatial distribution or the intensity values of the ALS data inside each segment may be used to estimate stem volume and tree species of the individual trees [
12–
14]. This kind of analysis utilizes more details of the ALS data together with the knowledge of the shapes and proportions of tree tops and tree crowns. However, it often fails to detect trees standing close together and trees below the tallest canopy layer [
10,
15].
The failure of ITC segmentation to detect all trees has been addressed with statistical approaches. Maltamo
et al.[
16] used expected tree size distribution functions to predict small trees. Lindberg
et al.[
17] classified the delineated segments to determine the number of trees contained in each segment. The properties of the trees contained in each segment were estimated with regression from the properties of the ALS data in the segment. The resulting tree list in each field plot was then adjusted using the estimated stem volume and distributions of DBH and tree height in the field plot. Holmgren
et al.[
18] used imputation to estimate tree lists based on properties of the ALS data in the segment using harvester data as training data. For estimation of several correlated variables, it is difficult to fit parametric models. Breidenbach
et al.[
19] used a similar imputation approach to determine the properties of the trees contained in each segment, which they named the semi-ITC approach. The results were not significantly biased and more accurate than estimates from regression models at area level. ITC segmentation makes use of the 3D structure of the ALS data and can be based on models of tree crowns, while estimation at an area level usually only considers the vertical distribution and density of the ALS data [
20].
Since the laser pulses can pass through gaps in the canopy, the ALS data include measurements of surfaces below the tallest canopy layer. This makes it possible to derive a digital elevation model of the ground also in dense forests [
21,
22]. Additionally, measurements may originate from small trees below the tallest canopy layer. These 3D properties have been used to delineate tree crowns from ALS data with clustering-based approaches where the initial values for the clustering are derived from local maxima in an nDSM or other means of detecting tree tops from the ALS data [
23–
27]. Other approaches have been to delineate tree crowns based on the mean shift algorithm [
28] and to first determine an approximate number of tree stems by clustering of the ALS data below the tree crowns and then use the estimated stem number to delineate tree crowns with a normalized cut algorithm [
29].
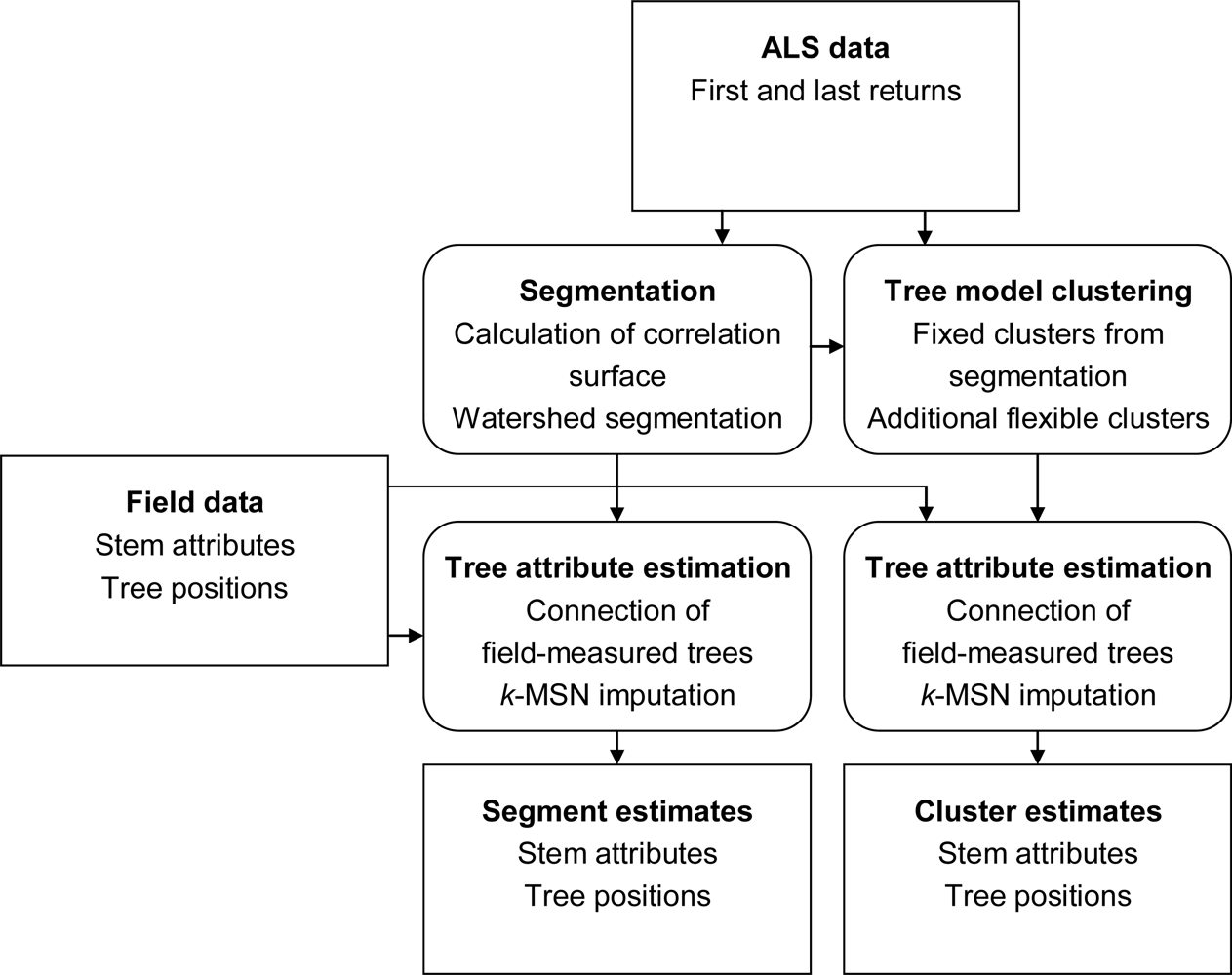
The aim of this study is to extract information from ALS data to estimate lists of individual trees with a higher accuracy when aggregated to area level than what is currently achieved with ITC segmentation of surface models. The idea is to first derive information about the tallest canopy layer from segmentation of a surface model and then use 3D analysis to extract information about trees below the tallest canopy layer with a tree model clustering approach. The information extracted from the ALS data is connected to field data to create models for unbiased plot level estimates of forest variables. The connection and estimation is done both for the tree model clustering approach and, as a comparison, for segmentation of a surface model.
4. Results
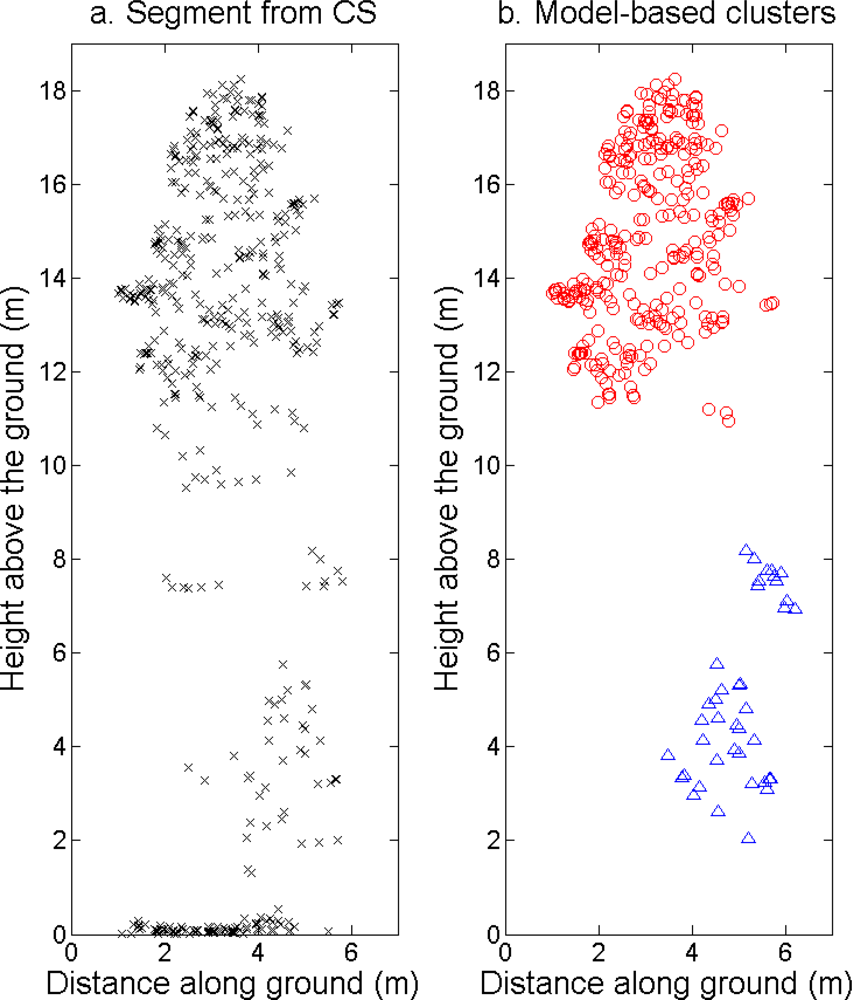
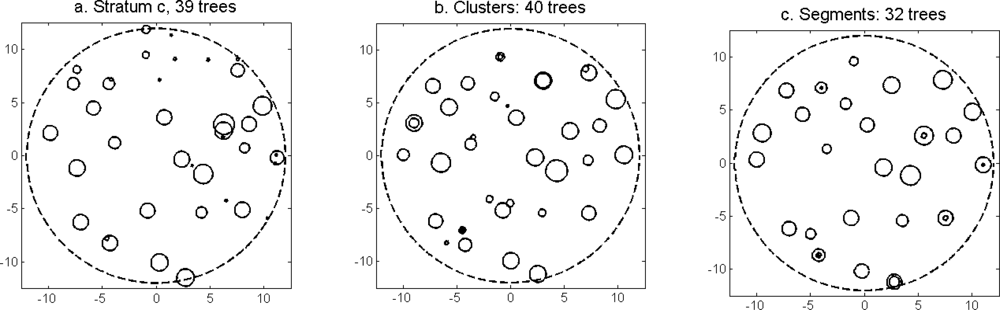
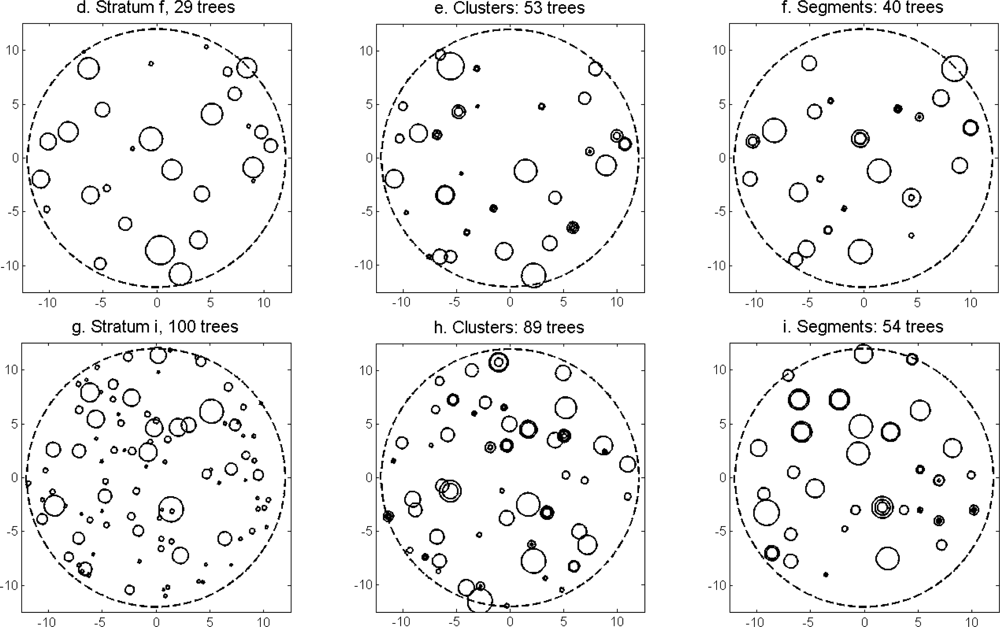
More field-measured trees could be linked to clusters from the tree model clustering than to segments delineated from the CS (
Table 5). However, the tree model clustering also resulted in more clusters that could not be linked to any field-measured tree. The total number of field-measured trees inside the buffer zones was 3,757.
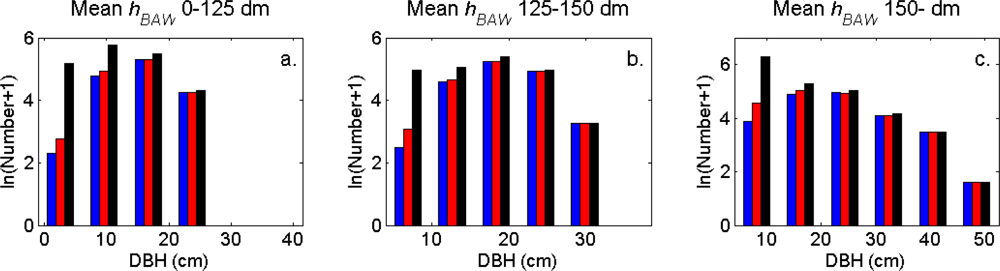
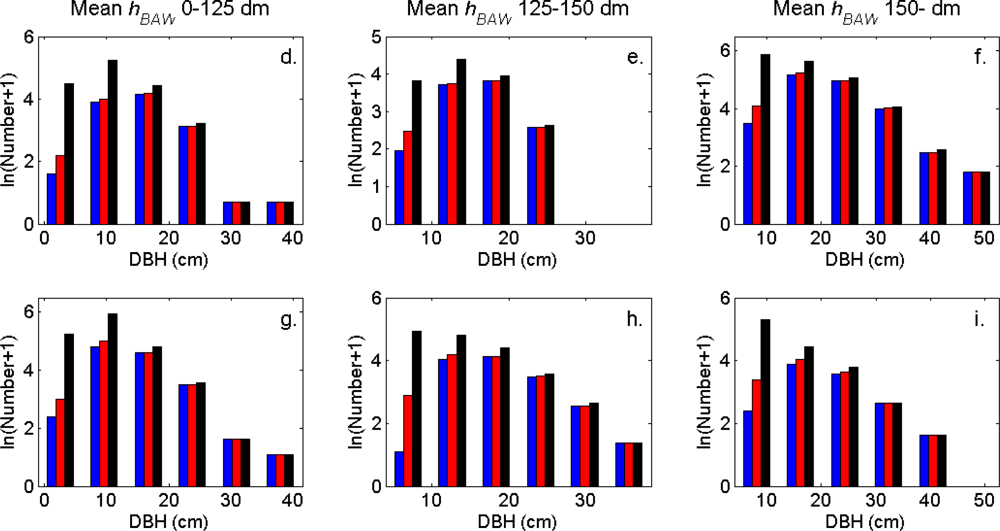
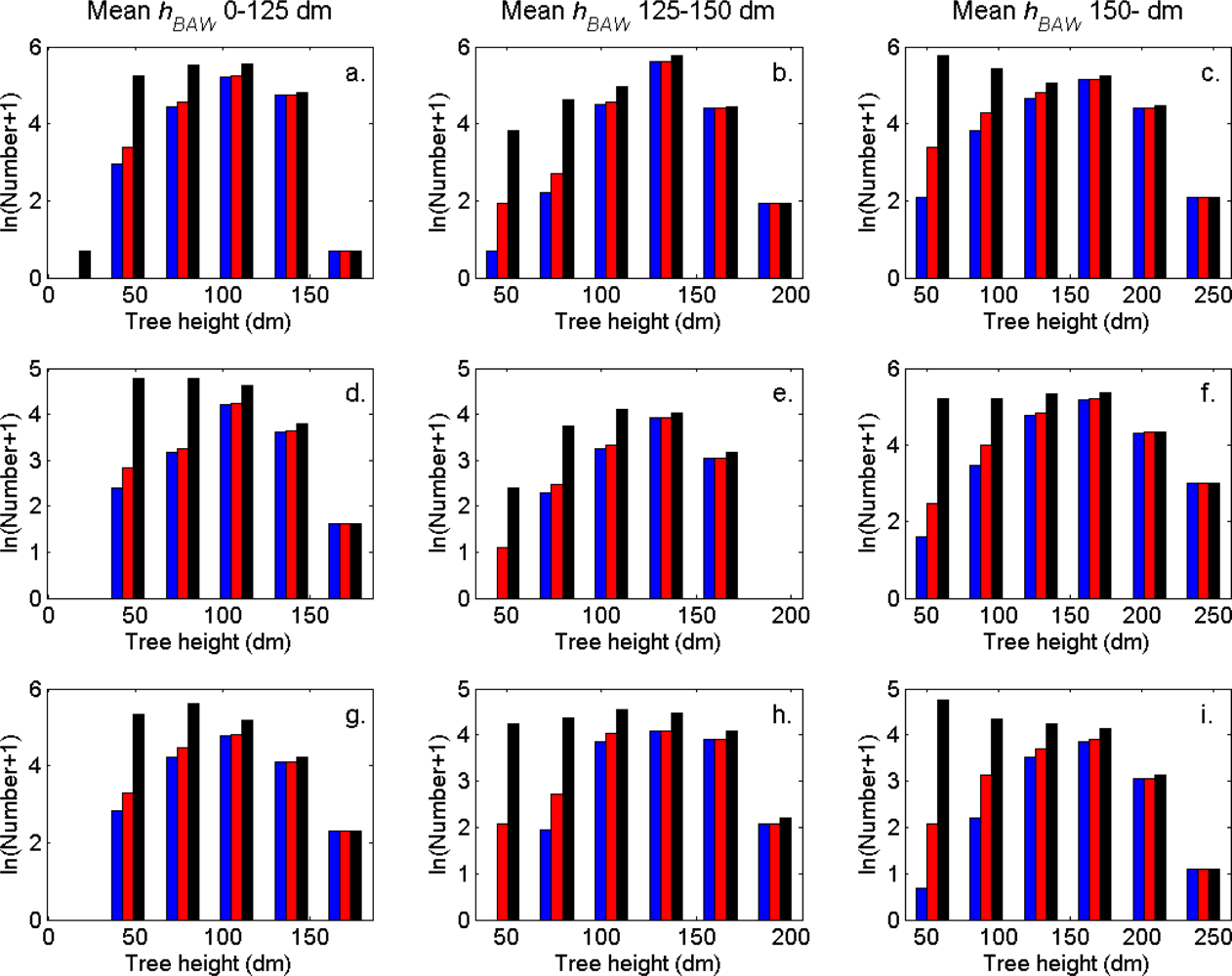
The distributions of individual features of the clusters were similar for clusters linked to zero, one and two or more trees (
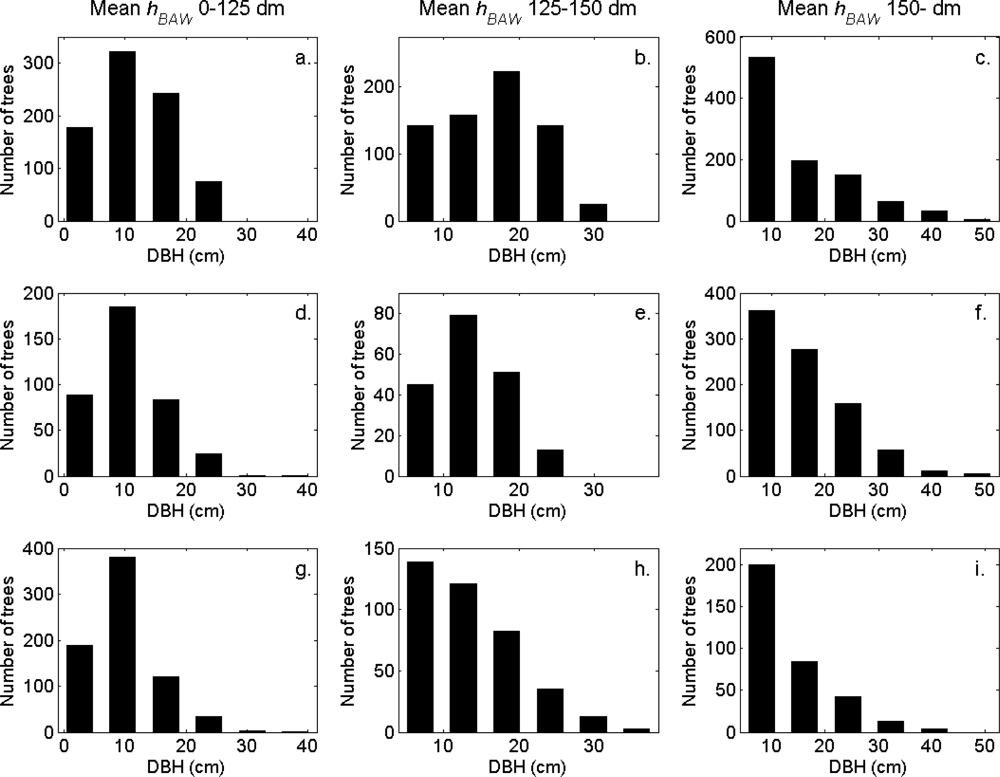
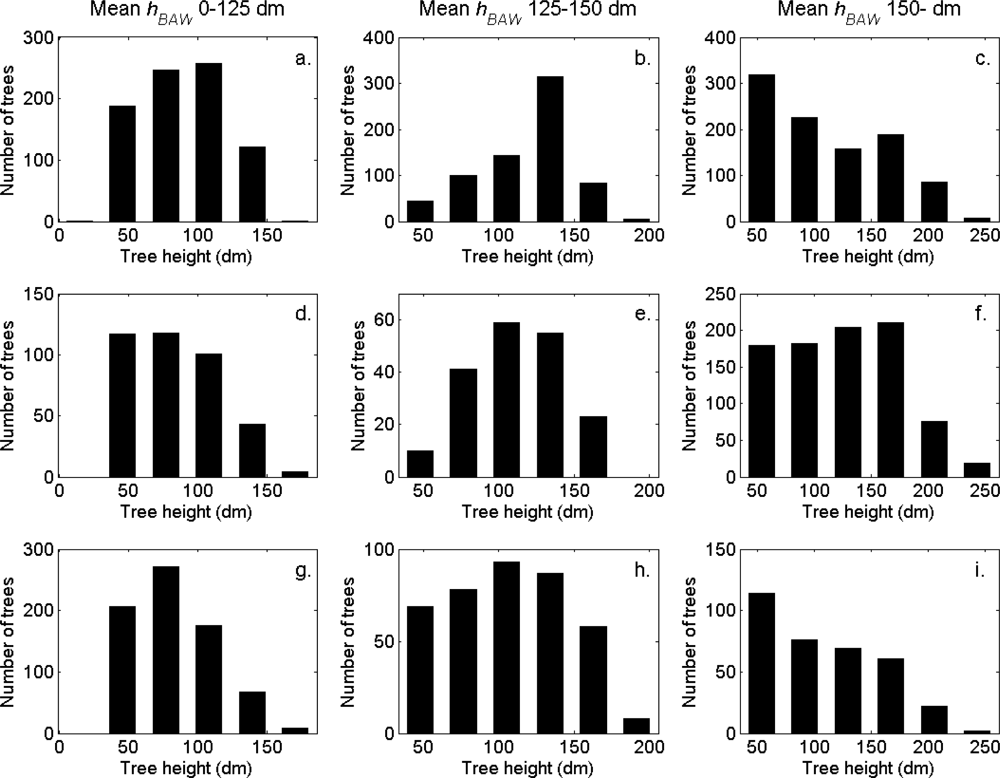
Figure 7). More field-measured trees below the tallest canopy layer and with a DBH < 20 cm could be linked to clusters from the tree model clustering than to segments delineated from the CS, especially in field plots with higher basal area-weighted mean tree height (
Figures 8 and
9).
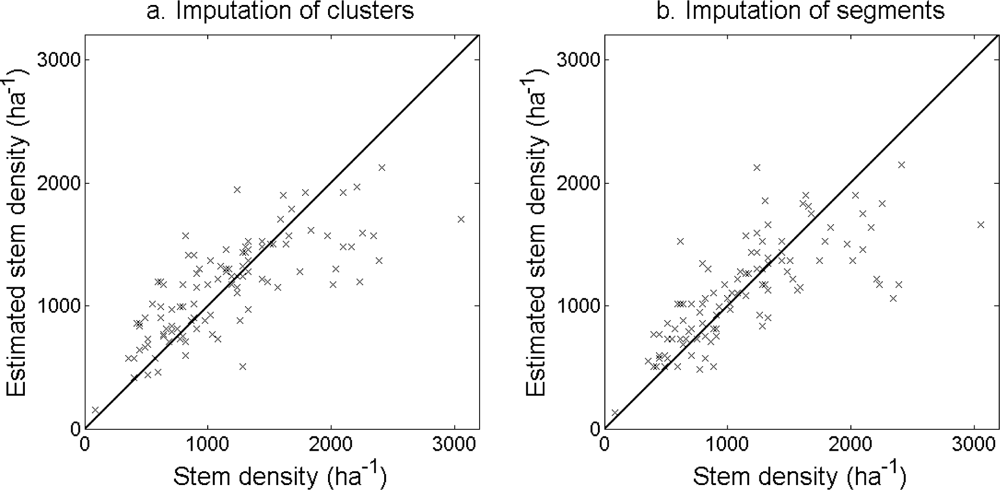
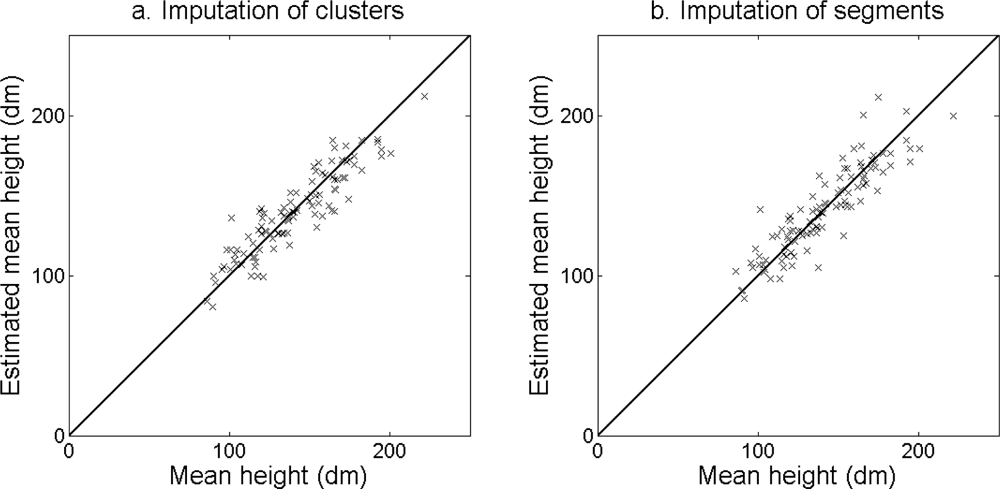
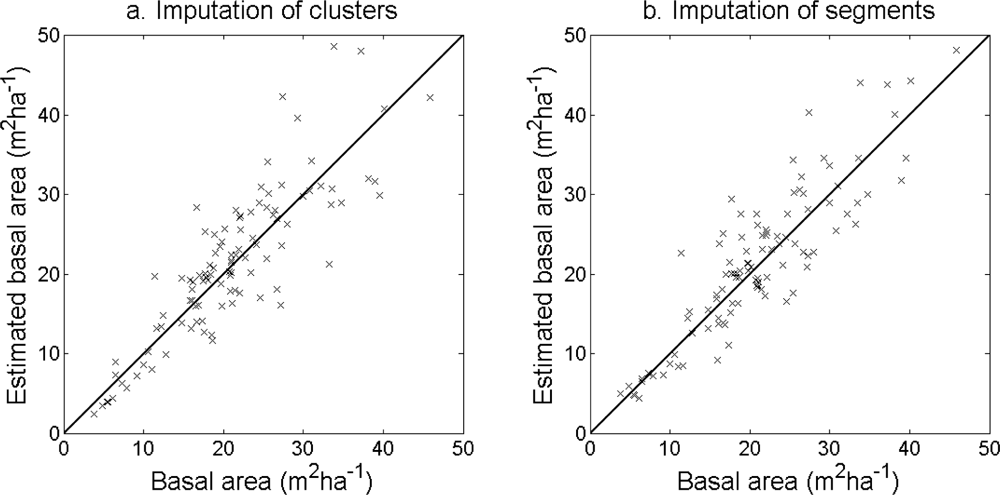
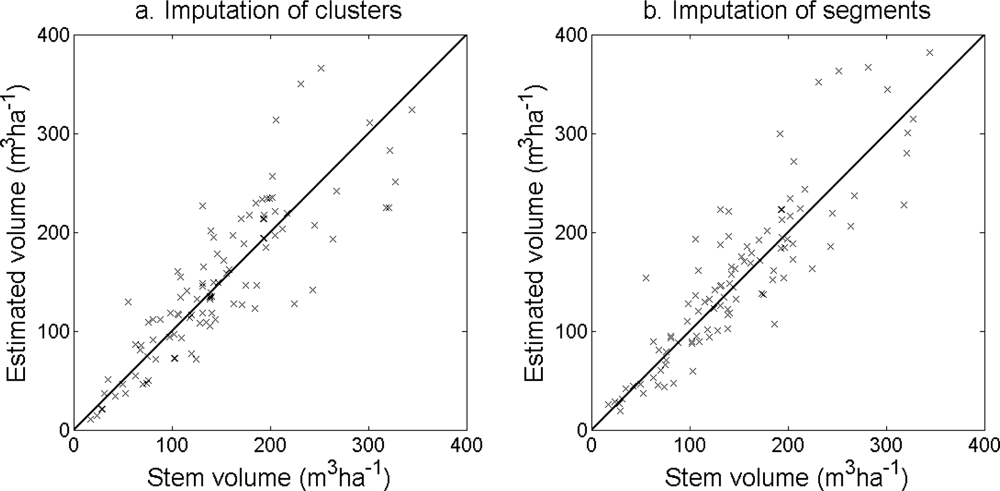
The accuracy of the estimated stem density (
Figure 10) and tree height (
Figure 11) was slightly higher for the imputation of clusters than for the imputation of segments (
Table 6). The accuracy of the estimated basal area (
Figure 12) and stem volume (
Figure 13) was slightly higher for the imputation of segments than for the imputation of clusters. The EI were slightly better for the imputation of segments than for the imputation of clusters. Examples of three field plots with field-measured trees and trees imputed from clusters and segments are shown in
Figure 14.
5. Discussion
The tree crowns were delineated by segmentation of a correlation surface model followed by 3D analysis with a new tree model clustering approach. The linking of segments delineated from the CS had a high success rate for trees with a DBH ≥ 20 cm. Since the tree model clustering was based on the segmentation, the success rate was equally high in that case. However, more trees with a DBH < 20 cm could be linked to the result from the tree model clustering than to the segments delineated from the CS, especially in field plots with a higher basal area-weighted mean height where those trees were part of the understory below the tallest canopy layer. The tree model clustering appears to be successful at identifying tree crowns also for trees in the understory below the tallest canopy layer.
The accuracy at plot level of the estimated forest variables was similar for the tree model clustering and for the segmentation after
k-MSN imputation. The segmentation typically identified the largest trees that contributed most to the stem volume and basal area, which resulted in accurate estimates for those forest variables. The tree model clustering divided some large tree crowns into several clusters and information was lost about those large trees. The segmentation is most successful for larger trees while methods that identify smaller trees may be less successful in delineating the tree crowns of the larger trees. The accuracy was comparable to previous studies with similar ALS data densities and forest conditions [
19,
20,
44,
45].
Tests with standard
k-means clustering of the ALS returns resulted in much lower accuracy than obtained with segmentation of the CS. This was not improved by trying different parameters of the
k-means clustering (e.g., different weights vertically and horizontally). For managed boreal forest, most trees can be identified successfully with segmentation of a surface model [
10]. To utilize this, a tree model clustering approach was developed to combine the information derived from the CS with the 3D distribution of the ALS data below the surface model, in order to delimit the tallest tree crowns from lower vegetation and derive information about trees in the understory.
The k-means clustering divides the data into clusters based on the Euclidean distance to the cluster centres. The distances in horizontal and vertical directions had an equal weight, which means that the clusters resembled spheres. To model elongated (ellipsoid) tree crowns, different weights in horizontal and vertical directions could be used. However, tests with relative weights of 1.5–2 in the vertical direction made the result worse.
The delineation methods used in this study depend on several parameter values. Most existing methods for delineation of tree crowns from ALS data depend on parameter settings (e.g., height thresholds, raster cell sizes and filter sizes) selected manually by the operator in order to optimize the delineation [
8–
11,
14,
24,
26]. Automated optimization of the parameters based on field data with known tree positions [
25] could possibly be done for different forest types (e.g., coniferous forest or beech forest); however, this would require further research.
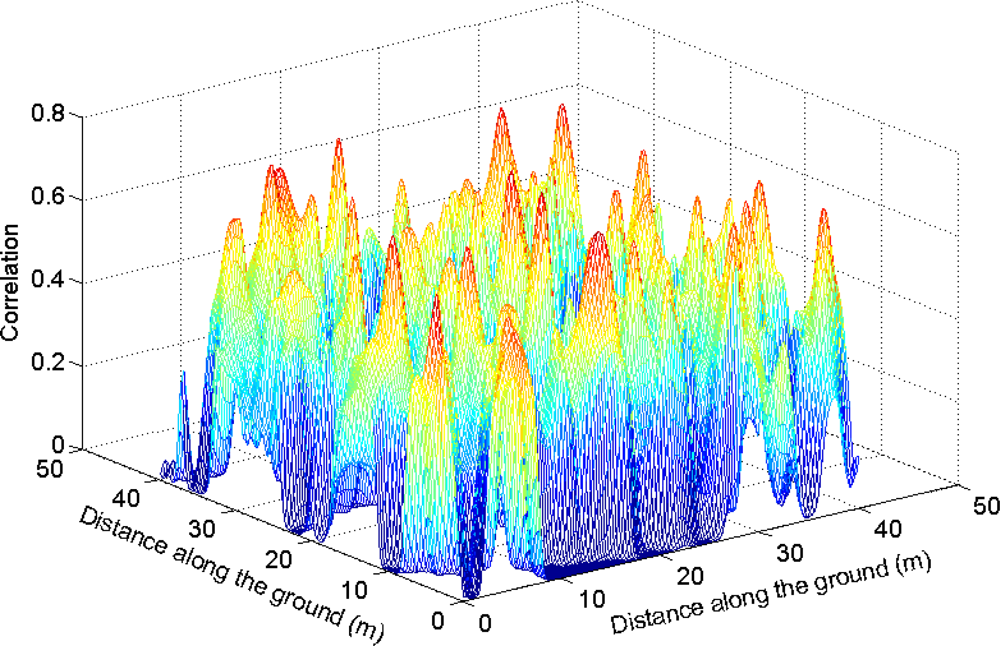
The CS was based on a priori knowledge of the shapes and proportions of tree crowns. Three-dimensional delineation of tree crowns may also benefit from using assumptions about the shapes and proportions of the tree crowns. In this study, this was achieved by fitting a parabolic surface to the top of each cluster and by joining clusters along a vertical axis if they were close enough.
The tree model clustering resulted in a large number of small clusters that could not be linked to any field-measured trees. Those un-linked clusters may correspond to parts of larger trees or to trees with a DBH smaller than the criterion to measure a tree in the field [
20]. However, the result of the imputation was not impaired by the un-linked clusters, probably because the properties of the un-linked clusters differed from the linked clusters.
The segmentation method used in this study was the same as in Holmgren
et al.[
18] with the exception that the expected ratio of radius to model height was fixed (
i.e., no training phase was used). The result of the segmentation may be improved by using a training phase to predict optimal parameter settings as a function of variables that can be derived from the ALS data. However, no training phase was used to set the parameters for the tree model clustering in this study, which means that the tree model clustering and the segmentation were based on the same conditions. The segmentation method has proved to perform well in a recent comparison with other segmentation methods for forests in Norway, Sweden, Germany, and Brazil [
46].
Only parts of the laser light can pass through the higher layers of the canopy and the measurements will not cover the area completely [
47]. Due to this occlusion effect, some suppressed trees will give rise to very few or no ALS returns [
48]. Hence, they cannot be delineated from the ALS data and it is difficult to estimate the complete tree height distribution in such cases.
The ALS data consisted of first and last discrete returns. The last return is typically from the ground, and ALS data with intermediate returns might provide more information about the understory. Another option is to use waveform ALS data. Waveform ALS data describe the whole backscattered signal and allow for detailed processing, such as derivation of returns from the waveforms using more advanced algorithms [
49] and measurements of the scattering properties of vegetation and terrain surfaces [
50].
6. Conclusions
Delineation of tree crowns from a surface model based on a priori knowledge about the shape and proportion of the tree crowns identifies most of the trees in the tallest canopy layer of coniferous-dominated boreal forest and has been shown to perform at least as well as some 3D methods. Three-dimensional methods may also benefit from using a priori knowledge about the tree crowns. In this study, this was achieved with a tree model clustering approach by fitting a parabolic surface to the top of each cluster and by joining clusters along a vertical axis if they were close enough.
Segmentation of a CS (
i.e., a surface model) identified 1,960 trees out of a total of 3,757, while the tree model clustering identified 2,169 trees. The results from the segmentation together with a model to estimate several trees for each delineated tree crown resulted in unbiased estimates of forest variables with a low RMSE (stem density RMSE 33.6% and bias −1.8%; stem volume RMSE 26.1% and bias 3.5%). The segmentation identified most trees with a DBH ≥ 20 cm. The tree model clustering approach was more successful than the segmentation in delineating trees with a DBH < 20 cm but did not improve the accuracy of the estimated forest variables at plot level (stem density RMSE 32.7% and bias 0.5%; stem volume RMSE 28.3% and bias 2.1%). Results from previous comparisons of segmentation of a surface model with tree crown models and clustering have shown higher accuracy for the segmentation [
46,
51,
52]. The tree model clustering used the results from the segmentation for trees close to the top of the canopy and knowledge about the shapes and proportions of the tree crowns for all trees, which has not been done before. Three-dimensional analysis of ALS data may produce better results in forests with a large number of understory trees below the tallest canopy layer.