Exploring the Use of MODIS NDVI-Based Phenology Indicators for Classifying Forest General Habitat Categories
Abstract
:1. Introduction
2. Materials and Methods
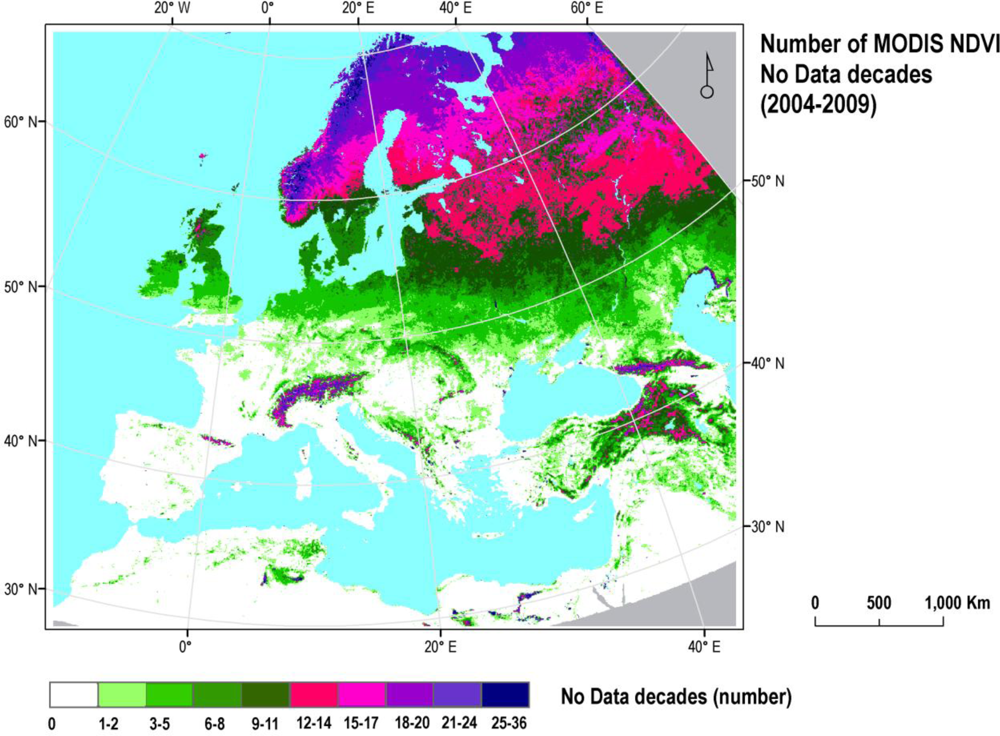
2.1. MODIS NDVI Data and Pre-Processing
2.2. Extraction of Phenological Information
2.3. Classifications Using Random Forests
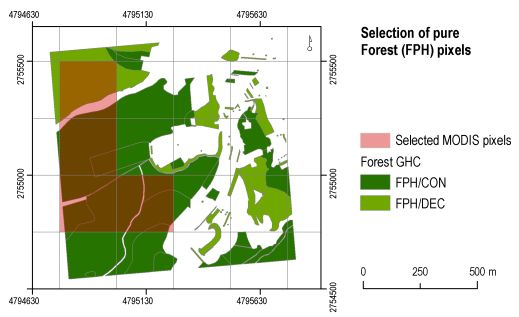
2.3.1. Field Data and Reference Pixels Selection
- JFM2006 data were re-gridded to match the spatial resolution of the MODIS NDVI data by summarizing the proportion of 25 m forest pixels present within each 250 m pixel;
- The 250 m pixels characterized by a proportion of either coniferous or broad-leaved forest ≥ 70% were selected.
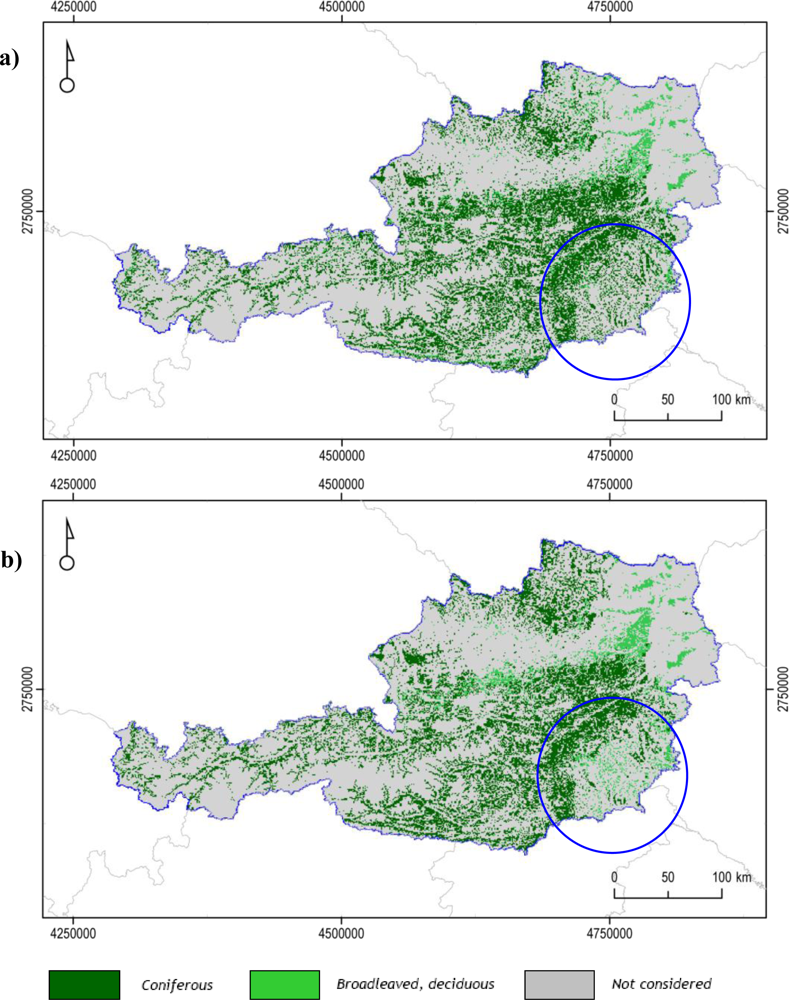
2.3.2. Classifications: Austria
2.3.3. Classifications: Mediterranean Environmental Zone
2.3.4. Classifications: The Impact of Data Gaps in VI Time Series
- Introduction of 10 consecutive data gaps (i.e., 10 contiguous no-data decades) per year across the full 6 years of MODIS NDVI time series;
- Extraction of the FPH/CON and FPH/DEC reference (pure) pixels from the NDVI time series with added data gaps;
- RF classifications, using all the phenology metrics, of the NDVI time series with added data gaps;
- Accuracy assessment and comparison with classification accuracy using the original gap-free NDVI data.
3. Results and Discussion
4. Conclusions
Acknowledgments
References
- Convention on Biological Diversity (CBD). COP 10 Decisions. Proceedings of Tenth Meeting of the Conference of the Parties to the Convention on Biological Diversity, Nagoya, Japan, 18–29 October 2010.
- Environment Council. Conclusions of 19 December on the EU 2020 Biodiversity Strategy; 18862/11. Council of the European Union: Brussels, Belgium, 2011. Available online: http://consilium.europa.eu/media/1379139/st18862.en11.pdf (accessed on 20 March 2012).
- Muchoney, M.M. Earth observations for terrestrial biodiversity and ecosystems. Remote Sens. Environ 2008, 112, 1909–1911. [Google Scholar]
- Pereira, H.M.; Cooper, H.D. Towards the global monitoring of biodiversity change. Trends Ecol. Evol 2006, 21, 123–129. [Google Scholar]
- Clerici, N.; Bodini, A.; Eva, H.; Grégoire, J-M.; Dulieu, D.; Paolini, C. Increased isolation of two Biosphere Reserves and surrounding protected areas (WAP ecological complex, West Africa). J. Nat. Conserv 2007, 15, 26–40. [Google Scholar]
- Bunce, R.H.G.; Metzger, M.J.; Jongman, R.H.G.; Brandt, J.; de Blust, G.; Elena Rossello, R.; Groom, G.B.; Halada, L.; Hofer, G.; Howard, D.C.; et al. A Standardized Procedure for Surveillance and Monitoring European Habitats and provision of spatial data. Lands. Ecol 2008, 23, 11–25. [Google Scholar]
- Fuller, R.M.; Wyatt, B.K.; Barr, C.J. Countryside survey from ground and space: Different perspectives, complementary results. J. Environ. Manage 1998, 54, 101–126. [Google Scholar]
- Halada, L.; Jongman, R.H.G.; Gerard, F.; Whittaker, L.; Bunce, R.H.G.; Bauch, B.; Schmeller, D.S. The European Biodiversity Observation Network—EBONE. In Proceedings of the European conference of the Czech Presidency of the Council of the EU TOWARDS eENVIRONMENT Opportunities of SEIS and SISE: Integrating Environmental Knowledge in Europe; Masaryk University: Brno, Czech, 2009. [Google Scholar]
- Lengyel, S.; Kobler, A.; Kutnar, L.; Framstad, E.; Henry, P.-Y.; Babij, V.; Gruber, B.; Schmeller, D.; Henle, K. A review and a framework for the integration of biodiversity monitoring at the habitat level. Biodivers. Conserv 2008, 17, 3341–3356. [Google Scholar]
- Lucas, R.; Medcalf, K.; Brown, A.; Bunting, P.; Breyer, J.; Clewley, D.; Keyworth, S.; Blackmore, P. Updating the Phase 1 habitat map of Wales, UK, using satellite sensor data. ISPRS J. Photogramm 2011, 66, 81–102. [Google Scholar]
- Thomson, A.G.; Fuller, R.M.; Yates, M.G.; Brown, S.L.; Cox, R.; Wadsworth, R.A. The use of airborne remote sensing for extensive mapping of intertidal sediments and saltmarshes in eastern England. Int. J. Remote Sens 2003, 24, 2717–2737. [Google Scholar]
- Wessels, K.; Steenkamp, K.; von Maltitz, G.; Archibald, S. Remotely sensed vegetation phenology for describing and predicting the biomes of South Africa. Appl. Veg. Sci 2010, 14, 1–19. [Google Scholar]
- Fuller, R.M.; Cox, R.; Clarke, R.T.; Rothery, P.; Hill, R.A.; Smith, G.M.; Thomson, A.G.; Brown, N.J.; Howard, D.C.; Stott, A.P. The UK land cover map 2000: Planning, construction and calibration of a remotely sensed, user-oriented map of broad habitats. Int. J. Appl. Earth Obs 2005, 7, 202–216. [Google Scholar]
- Reese, H.M.; Lillesand, T.M.; Nagel, D.E.; Stewart, J.S.; Goldmann, R.A.; Simmons, T.E.; Chipman, J.W.; Tessar, P.A. Statewide land cover derived from multiseasonal Landsat TM data—A retrospective of the WISCLAND project. Remote Sens. Environ 2002, 82, 224–237. [Google Scholar]
- Dechka, J.A.; Franklin, S.E.; Watmough, M.D.; Bennett, R.P.; Ingstrup, D.W. Classification of wetland habitat and vegetation communities using multi-temporal Ikonos imagery in southern Saskatchewan. Can. J. Remote Sens 2002, 28, 679–685. [Google Scholar]
- Hüttich, C.; Gessner, U.; Herold, M.; Strohbach, B.J.; Schmidt, M.; Keil, M.; Dech, S. On the suitability of MODIS time series metrics to map vegetation types in dry savanna ecosystems: A case study in the Kalahari of NE Namibia. Remote Sens 2009, 1, 620–643. [Google Scholar]
- Van Leeuwen, W.J.; Davison, J.E.; Casady, G.M.; Marsh, S.E. Phenological characterization of desert sky island vegetation communities with remotely sensed and climate time series data. Remote Sens 2010, 2, 388–415. [Google Scholar]
- Hill, R.; Wilson, A.; George, M.; Hinsley, S. Mapping tree species in temperate deciduous woodland using time-series multi-spectral data. Appl. Veg. Sci 2010, 13, 86–99. [Google Scholar]
- Stockli, R.; Vidale, P.L. European plant phenology and climate as seen in a 20-year AVHRR land-surface parameter dataset. Int. J. Remote Sens 2004, 25, 3303–3330. [Google Scholar]
- McCloy, K.R. Development and evaluation of phenological change indices derived from time series of image data. Remote Sens 2010, 2, 2442–2473. [Google Scholar]
- Townshend, J.R.G.; Goff, T.E.; Tucker, C.J. Multitemporal dimensionality of images of normalized difference vegetation index at continental scales. IEEE Trans. Geosci. Remote Sens 1985, 23, 888–895. [Google Scholar]
- Geerken, R.; Zaitchik, B.; Evans, J.P. Classifying rangeland vegetation type and coverage from NDVI time-series using Fourier Filtered Cycle Similarity. Int. J. Remote Sens 2005, 26, 5535–5554. [Google Scholar]
- Verstraete, M.M.; Gobron, N.; Aussedat, O.; Robustelli, M.; Pinty, B.; Widlowski, J.-L.; Taberner, M. An automatic procedure to identify key vegetation phenology events using the JRC-FAPAR Products. Adv. Space Res 2008, 41, 1773–1783. [Google Scholar]
- Myneni, R.B.; Keeling, C.D.; Tucker, C.J.; Asrar, G.; Nemani, R.R. Increased plant growth in the northern high latitudes from 1981 to 1991. Nature 1997, 386, 698–702. [Google Scholar]
- Bradley, B.A.; Mustard, J.F. Comparison of phenology trends by land cover class: a case study in the Great Basin, USA. Glob. Change Biol 2008, 14, 334–346. [Google Scholar]
- Lu, H.; Raupach, M.R.; McVicar, T.R.; Barrett, D.J. Decomposition of vegetation cover into woody and herbaceous components using AVHRR NDVI time series. Remote Sens. Environ 2003, 86, 1–18. [Google Scholar]
- Jakubauskas, M.; Legates, D.; Kastens, J. Crop identification using harmonic analysis of time series AVHRR NDVI data. Comput. Electron. Agr 2002, 37, 127–139. [Google Scholar]
- Jung, M.; Verstraete, M.; Gobron, N.; Reichstein, M.; Papale, D.; Bondeau, A.; Robustelli, M.; Pinty, B. Diagnostic assessment of European gross primary production. Glob. Change Biol 2008, 14, 2349–2364. [Google Scholar]
- Holben, B.N. Characteristics of maximum-value composite images from temporal AVHRR data. Int. J. Remote Sens 1986, 7, 1417–1437. [Google Scholar]
- Rahman, H.; Dedieu, G. SMAC: A simplified method for the atmospheric correction of satellite measurements in the solar spectrum. Int. J. Remote Sens 1994, 15, 123–143. [Google Scholar]
- Paola, J.D.; Schowengerdt, R.A. A review and analysis of Backpropagation neural networks for classification of remotely sensed multispectral imagery. Int. J. Remote Sens 1995, 16, 3033–3058. [Google Scholar]
- Klisch, A.; Royer, A.; Lazar, C.; Baruth, B.; Genovese, G. Extraction of phenological parameters from temporally smoothed vegetation indices. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci 2005, 36, 91–96. [Google Scholar]
- Lohninger, H. Teach/Me Data Analysis; Springer-Verlag: Berlin, Germany, 1999. [Google Scholar]
- Chen, J.; Jönsson, P.; Tamura, M.; Gu, Z.; Matsushita, B.; Eklundh, L. A simple method for reconstructing a high-quality NDVI time-series data set based on the Savitzky–Golay filter. Remote Sens. Environ 2004, 91, 332–344. [Google Scholar]
- Jönsson, P.; Eklundh, L. TIMESAT—A program for analysing time-series of satellite sensor data. Comput. Geosci 2004, 30, 833–845. [Google Scholar]
- Weissteiner, C.; Sommer, S.; Strobl, P. Time Series Analysis of NOAA AVHRR Green Vegetation Fraction as a Means to Derive Permanent and Seasonal Vegetation Component. Proceedings from the EARSeL Workshops in the Framework of the 27th Symposium, Bozen, Italy, 7–9 June 2007.
- Ivits, E.; Cherlet, M.; Mehl, W.; Sommer, S. Estimating the ecological status and change of riparian zones in Andalusia assessed by multi-temporal AVHHR datasets. Ecol. Indic 2009, 9, 422–431. [Google Scholar]
- Ivits, E.; Cherlet, M.; Mehl, W.; Sommer, S. Spatial Assesment of the Status of Riparian Zones and Related Effectiveness of Agri-Environmental Schemes in Andalusia, Spain; EUR Report; EUR 23299 N1018-5593; JRC: Luxembourg, 2008. [Google Scholar]
- White, M.A.; De Beurs, K.M.; Didan, K.; Inouye, D.W.; Richardson, A.D.; Jensenk, O.P.; O’Keefe, J.; Zhang, G.; Nemani, R.R.; Van Leeuwen, W.J.D.; Brown, J.F.; et al. Intercomparison, interpretation, and assessment of spring phenology in North America estimated from remote sensing for 1982–2006. Glob. Change Biol 2009, 15, 2335–2359. [Google Scholar]
- Reed, B.C.; Brown, J.F.; Vander Zee, D.; Loveland, T.R.; Merchant, J.W.; Ohlen, D.O. Measuring phenological variability from satellite imagery. J. Veg. Sci 1994, 5, 703–714. [Google Scholar]
- Hill, M.J.; Donald, G.E. Estimating spatio-temporal patterns of agricultural productivity in fragmented landscapes using AVHRR NDVI time series. Remote Sens. Environ 2003, 84, 367–384. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn 2001, 45, 5–32. [Google Scholar]
- Bunce, R.G.H.; Roche, P.; Bogers, M.M.B.; Walczak, M.; de Blust, G.; Geijzendorffer, I.R.; Van den Borre, J.; Jongman, R.H.G. Handbook for Surveillance and Monitoring of Habitats, Vegetation and Selected Species; EBONE Deliverable Report. Alterra: Wageningen, The Netherlands, 2010. Available online: http://www.ebone.wur.nl/UK/Publications/Deliverables/ (accessed on 1 February 2012).
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random Forests for land cover classification. Pattern Recogn. Lett 2004, 27, 294–300. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and regression by RandomForests. R News 2002, 2, 18–22. [Google Scholar]
- Metzger, M.J.; Bunce, R.G.H.; Jongman, R.H.G.; Mücher, C.A.; Watkins, J.W. A climatic stratification of the environment of Europe. Glob. Ecol. Biogeogr 2005, 14, 549–563. [Google Scholar]
- Kempeneers, P.; Sedano, F.; Seebach, L.; Strobl, P.; San-Miguel-Ayanz, J. Data fusion of different spatial resolution remote sensing images applied to forest-type mapping. IEEE Trans. Geosci. Remote Sens 2011, 99, 1–10. [Google Scholar]
- Kempeneers, P.; Sedano, F.; Seebach, L.; San-Miguel-Ayanz, J. A High Resolution PAN-European Forest Type Map Based on Multispectral and Multi-Temporal Remote Sensing Data. Proceedings of ForestSat2010 Conference: Operational Tools in Forestry Using Remote Sensing Techniques, Lugo and Santiago de Compostela. Spain, 7–10 Semptember 2010.
- Soudani, K.; Hmimina, G; Delpierre, N.; Pontailler, J.Y.; Aubinet, M.; Bonal, D.; Caquet, B.; de Grandcourt, A.; Burban, B.; Flechard, C.; et al. Ground-based Network of NDVI measurements for tracking temporal dynamics of canopy structure and vegetation phenology in different biomes. Remote Sens. Environ 2012, 123, 234–245. [Google Scholar]
- Doktor, D.; Bondeau, A.; Koslowski, D.; Badeck, F.W. Influence of heterogeneous landscapes on computed green-up dates based on daily AVHRR NDVI observations. Remote Sens. Environ 2009, 113, 2618–2632. [Google Scholar]
- Lobell, D.B.; Asner, G.P. Cropland distributions from temporal unmixing of MODIS data. Remote Sens. Environ 2004, 93, 412–422. [Google Scholar]
- Benhadj, I.; Duchemin, B.; Maisongrande, P.; Simonneaux, V.; Khabba, S.; Chehbouni, A. Automatic unmixing of MODIS multitemporal data for inter-annual monitoring of land use at a regional scale (Tensift, Morocco). Int. J. Remote Sens 2012, 33, 1325–1348. [Google Scholar]


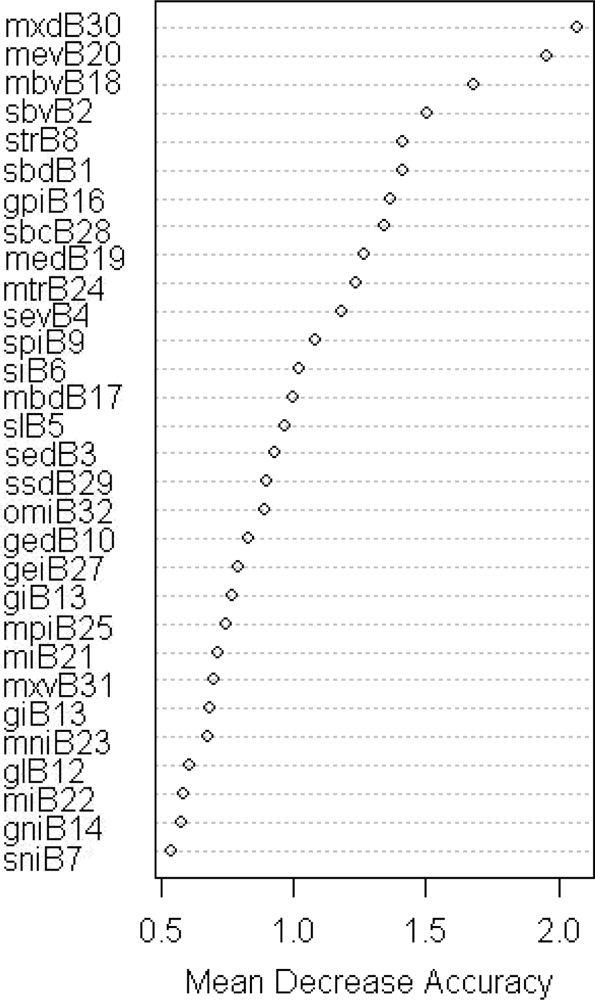

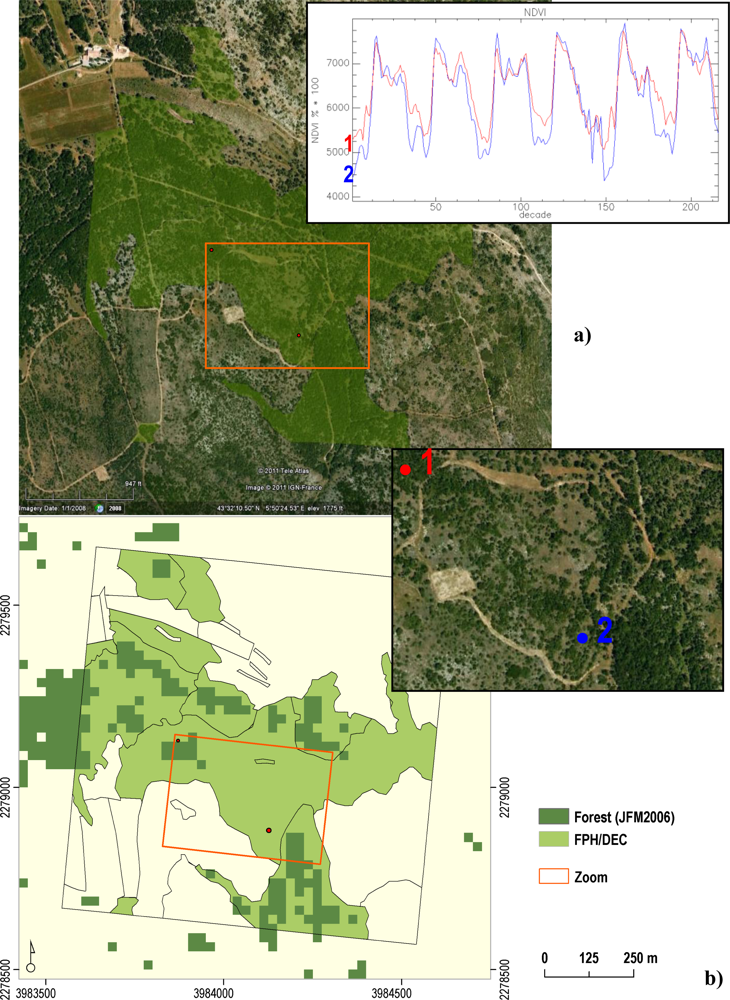
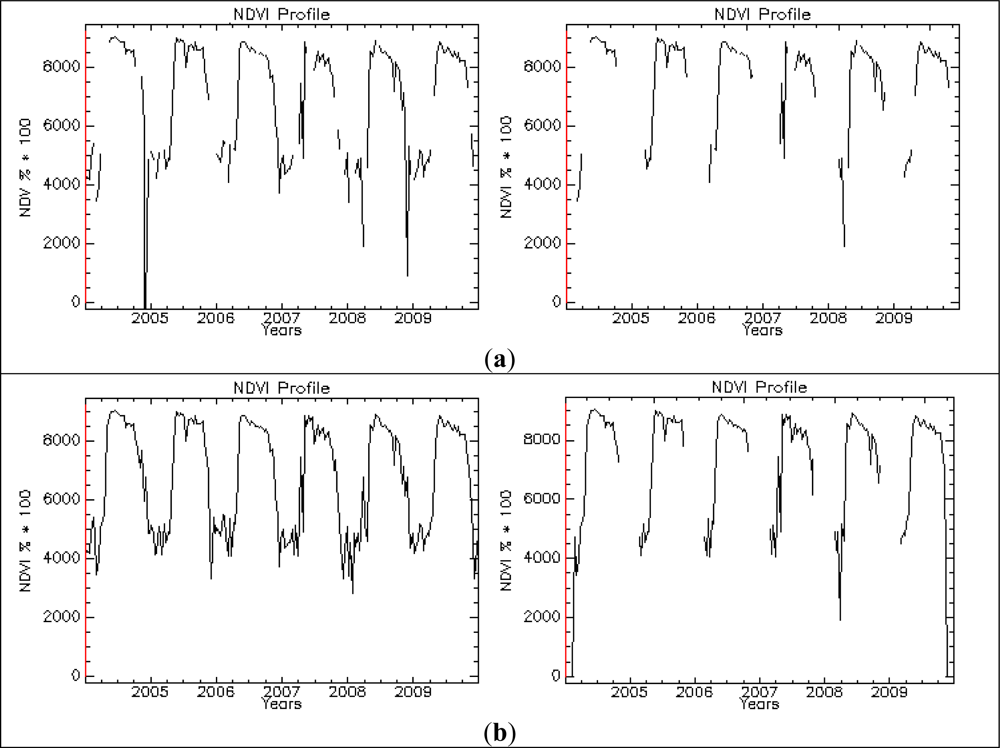
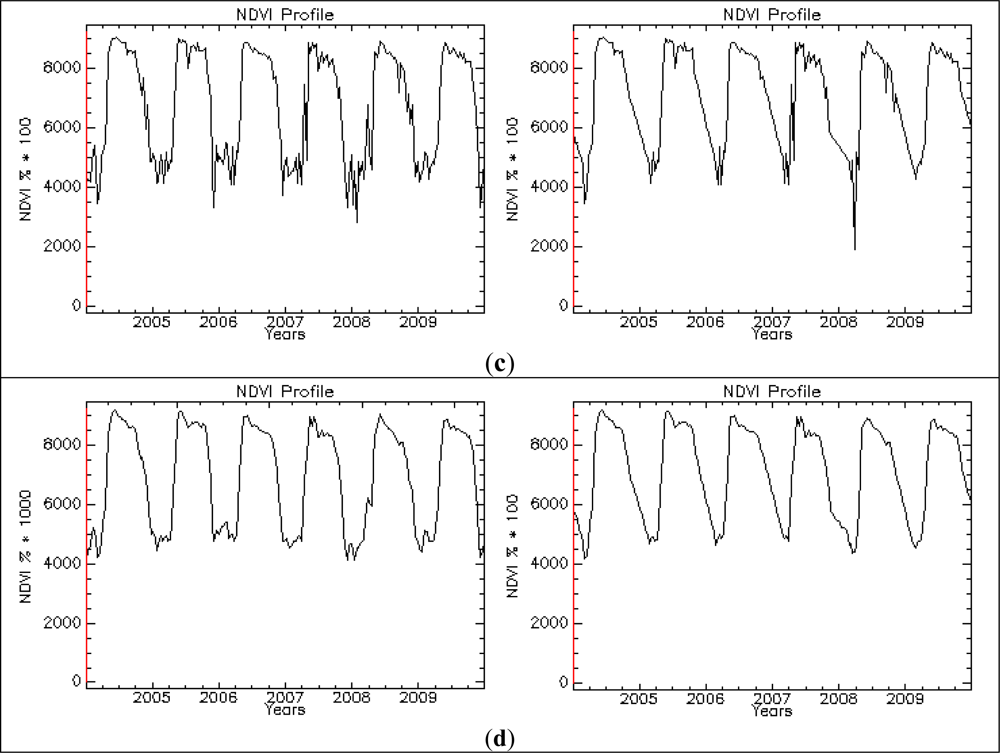
| Phenology Indicator | Acronym in Phenolo |
|---|---|
| Start of Season, SOS (Day) | SBD |
| Start of Season, SOS (Value) | SBV |
| End of Season, EOS (Day) | SED |
| End of Season, EOS (Value) | SEV |
| Season Length (EOS-SOS) | SL |
| Season Integral: the integral under the vegetation signal curve delimited by EOS and SOS | SI |
| Normalized Season Integral | SNI |
| Seasonal Permanent Fraction: the area below the line connecting SOS with EOS, and the x axis. | SPI |
| Season Total Ratio [SI/(SI+SPF)] | STR |
| Growing Season End, GE (day) | GED |
| Growing Season End, GE (value) | GEV |
| Growing Season Length | GL |
| Growing Season Integral | GI |
| Normalized Growing Season Integral | GNI |
| Growing Season Total Ratio*: [GI/(GI+SPF)] | GTR |
| Growing Season Permanent Fraction: the permanent area fraction below the curve connecting SOS with Growing Season End | GPI |
| Minimum before SOS (Day) | MBD |
| Minimum before SOS (Value) | MBV |
| Minimum after EOS (Day) | MED |
| Minimum after EOS (Value) | MEV |
| Total Length: Length in time between minima (Days) | ML |
| Total Integral, TI: the area under the vegetation signal curve delimited by the two minima. | MI |
| Normalized Total Integral | MNI |
| Above Minima Total Ratio: above minima integral over TI | MTR |
| Total Permanent Fraction, TPF: the area below the line connecting the vegetation signal minima and the x axis. | MPI |
| Season Exceeding Integral: (TI-SI) | SEI |
| Growing Season Exceeding Integral: (TI-GI) | GEI |
| Season Barycentre | SBC |
| Standard Deviation of the Season vegetation curve | SSD |
| Peak of Season, POS (Day) | MXD |
| Peak of Season, POS (Value) | MXV |
| Output minus Input Length (365 – GL) | OMI |
| Phenology Metrics Configuration | |||
| Forest Class | A | B | C |
| Coniferous – FPH/CON | 82.18 | 82.39 | 76.70 |
| Deciduous – FPH/DEC | 37.31 | 36.55 | 29.65 |
| Forest Class | - | B | - |
| Coniferous – FPH/CON | - | 79.03 | - |
| Deciduous – FPH/DEC | - | 44.54 | - |
| Mixed – MIX | - | 21.31 | - |
| Phenology Metrics Configuration | ||
|---|---|---|
| Forest Class | A | All |
| Coniferous – FPH/CON | 34.84 | 47.65 |
| Deciduous – FPH/DEC | 52.96 | 58.01 |
Share and Cite
Clerici, N.; Weissteiner, C.J.; Gerard, F. Exploring the Use of MODIS NDVI-Based Phenology Indicators for Classifying Forest General Habitat Categories. Remote Sens. 2012, 4, 1781-1803. https://doi.org/10.3390/rs4061781
Clerici N, Weissteiner CJ, Gerard F. Exploring the Use of MODIS NDVI-Based Phenology Indicators for Classifying Forest General Habitat Categories. Remote Sensing. 2012; 4(6):1781-1803. https://doi.org/10.3390/rs4061781
Chicago/Turabian StyleClerici, Nicola, Christof J. Weissteiner, and France Gerard. 2012. "Exploring the Use of MODIS NDVI-Based Phenology Indicators for Classifying Forest General Habitat Categories" Remote Sensing 4, no. 6: 1781-1803. https://doi.org/10.3390/rs4061781
APA StyleClerici, N., Weissteiner, C. J., & Gerard, F. (2012). Exploring the Use of MODIS NDVI-Based Phenology Indicators for Classifying Forest General Habitat Categories. Remote Sensing, 4(6), 1781-1803. https://doi.org/10.3390/rs4061781