Abstract
High-resolution NDVI maps derived from UAV imagery are valuable in precision agriculture, supporting vineyard management decisions such as disease risk and vigor assessments. However, the expense and complexity of multispectral sensors limit their widespread use. In this study, we evaluate Generative Adversarial Network (GAN) approaches—trained on either multispectral-derived or true RGB data—to convert low-cost RGB imagery into NDVI maps. We benchmark these models against simpler, explainable RGB-based indices (RGBVI, vNDVI) using Botrytis bunch rot (BBR) risk and vigor mapping as application-centric tests. Our findings reveal that both multispectral- and RGB-trained GANs can generate NDVI maps suitable for BBR risk modelling, achieving R-squared values between 0.8 and 0.99 on unseen datasets. However, the RGBVI and vNDVI indices often match or exceed the GAN outputs, for vigor mapping. Moreover, model performance varies with sensor differences, vineyard structures, and environmental conditions, underscoring the importance of training data diversity and domain alignment. In highlighting these sensitivities, this application-centric evaluation demonstrates that while GANs can offer a viable NDVI alternative in some scenarios, their real-world utility is not guaranteed. In many cases, simpler RGB-based indices may provide equal or better results, suggesting that the choice of NDVI conversion method should be guided by both application requirements and the underlying characteristics of the subject matter.
Keywords:
generative AI; generalization; UAV imagery; GANs; NDVI; precision agriculture; explainable AI; domain shift 1. Introduction
Paradigms like Agriculture 5.0 aim to upend a cycle of increasing agricultural yield at the cost of global environmental, social, and financial stability [1]. One aspect to achieve this vision is a demand for further technological innovation, to better understand and operate agricultural areas. An increasingly widely adopted technology within Precision Agriculture (PA) is the use of remote sensing technologies from unmanned aerial vehicles (UAVs) in precision agriculture [2]. The sensing technology, in combination with the flexibility and high resolution of the UAV, offers non-destructive measurement of a wide range of variables of a crop [3]. Applications for UAVs range from irrigation monitoring [4], phenotyping [5,6] to grape-bunch rot [7], vine disease detection [8] and vigor mapping [9].
These applications often make use of multispectral (MS) sensors [10]. A MS sensor captures the light intensity in reflectance at specific wavelengths of light, instead of the color representation of a ‘standard’ camera. These different reflectance values can be correlated to features such as plant health, disease, water stress, etc. Furthermore, standard RGB cameras lack the near-infrared band that is often used by these applications.
Most common in MS UAV applications, is the use of the near-infrared (NIR) band to calculate NDVI (Normalized Difference Vegetation Index). NDVI was developed in the 1970s for analyzing satellite imagery [11]. NDVI correlates to many plant variables, such as raw biomass, leaf area, potential yield, and water stress [12,13]. Combined with other structural parameters, such as canopy height, the vegetation index is an invaluable tool in the PA processing pipeline [14].
However, multispectral images come at an increased cost and complexity [3] compared to a standard RGB camera. The cost difference between an RGB sensor and an MS sensor is around a factor of 10. Additionally, the MS sensor requires radiometric calibration, which converts the digital numbers to reflectance values, based on the sun incidence angle, time of day, and cloud cover [15]. Finally, this radiometric calibration also requires additional focus on operational conditions [15]. Considering the Agriculture 5.0 paradigm, it is therefore not only important to develop multispectral UAV analysis techniques but also to increase access to such techniques [1]. This framing emphasizes affordable and easy-to-use tools for a wide range of audiences, making cost-effective RGB-based sensors the logical endpoint [3].
Estimating multispectral information through more accessible means is a growing area of interest. RGBVI is the Red-Green-Blue Vegetation Index which captures vegetation, without requiring a Near-Infrared band like NDVI. Ashapure et al. [16] compared visible-range vegetation indices to NDVI to identify canopy cover in cotton fields. The results indicated that the multispectral sensor-based canopy cover model provided more stable and accurate estimations. In contrast, the RGB-based model exhibited higher variability, particularly after canopy maturation when leaf color changed. Agapiou [17] compared various visible-range vegetation indices in diverse UAV orthophotos, such as the Green Leaf Index (GLI) and RGBVI. Costa et al. [18], identified a new visible-range estimation of NDVI using a genetic algorithm trained and tested on citrus, sugarcane and grapes. More recent work focuses on Deep Learning (DL) approaches to estimate NDVI. In Suárez et al. [19] a Cyclic Generative Adversarial Network (CycleGAN) is used to convert RGB imagery to NDVI images, this network outperformed other selected Deep Learning approaches for RGB to NDVI conversion. Similar results were found in Farooque et al. [20], Boroujeni and Razi [21], Davidson et al. [22].
Beyond RGB to NDVI conversion, Deep Learning has found many applications within remote sensing, vineyard analysis and UAVs. Image-based Deep Learning models used in these applications are highly task-specific, built for detection, segmentation, domain adaptation and super-resolution [23,24]. Mahara and Rishe [25] present a model to generate satellite imagery from one band to another band. They propose a cross-attention module inside the GAN model. This cross-attention module is adapted from the Transformer architecture [26]. Their method outperforms existing GAN models in generating the satellite band. Furthermore, in Ahmedt-Aristizabal et al. [27], a multi-model approach is introduced for vineyard yield prediction, utilizing a pipeline of three models. First, the input image is processed by a detector model that identifies likely regions of interest for further analysis. These regions are then passed to a segmentation model, which refines the identification of relevant areas. Finally, the segmented output is fed into a tracking model to count individual berries. While the authors acknowledge that the approach is not perfectly accurate due to occlusions and dataset limitations, it significantly enhances the efficiency of yield prediction. Compared to labor-intensive human counting, the proposed system offers greater consistency and reliability. Neither study provided the code necessary for independent verification of their results.
In the RGB to NDVI conversion study from Farooque et al. [20] the Pix2Pix GAN and the updated Pix2PixHD model from Wang et al. [28] were used. The models were trained on 500 images of potatoes at different moments during the 2021 growing stage. They evaluated the model by holding out a random 10% of the images as a testing set. The accuracy measurements used are Structural Similarity (SSIM), Root Mean Squared Error (RMSE), discriminator loss, and visual inspection. Across these metrics, the Pix2PixHD model outperformed the Pix2Pix model.
In Davidson et al. [22] the Pix2Pix GAN [29] was deployed to train RGB to NDVI conversion. They tested the effect of dataset size on accuracy, by training the GAN model on 500 images, instead of 3000, the results indicate only a slight increase in accuracy, indicating that dataset variety is more important than dataset size. In their experiments, they covered three agricultural crops: corn, soybeans, and cotton. These three crops were the basis for seven different permutations of training and testing: e.g., training and testing on all crops, training on crop A and testing on crop B, etc. The numeric measurements cover Structural Similarity (SSIM), Mean Square Error (MSE) and Peak Signal to Noise Ratio (PSNR). When Pix2Pix was trained on a different crop to the testing data, the model performed much poorer across all accuracy metrics (). It is not shared which crops were selected for training or testing. Finally, three other DL approaches for index conversion were chosen for comparison against Pix2Pix: CycleGAN, CUT and a custom ANN. The results indicate that Pix2Pix outperforms all other GAN models in the accuracy metrics by a ‘convincing margin’ [22].
The above studies deployed GANs to solve RGB to NDVI conversion to varying degrees of success. However, three shortcomings in previous work limit the adoption of the methods. (1) NDVI is never a final result. NDVI is often one of the inputs for downstream precision agricultural applications [14]. Therefore testing the usability of generated NDVI maps, requires the development of more elaborate evaluation scenarios [30]. (2) the generalization of GAN models across subject matter remains poor [31]. Deep Learning models tend to overfit the data and are sensitive to out-of-training examples [23,32]. Additionally, it is not well understood how these models cope with variance in sensor or spatiotemporal variation [30]. (3) the studies did not compare GANs to other non-DL approaches for RGB to NDVI conversion. GANs are an interesting and novel approach for an image-based Precision Agricultural application. It is important to reflect on whether the complex DL black-box models even outperform simpler and/or explainable approaches such as RGBVI.
This paper builds upon previous work on RGBtoNDVI conversion of UAV imagery by evaluating GAN conversion methods and explainable conversion methods in practical evaluation scenarios. This addresses the usability of the artificially generation NDVI in real-world applications. It also addresses whether the conversion can generalize beyond the training datasets. This generalization is divided into two dimensions: 1. different location to training data, and 2. different sensor type to training data. The contribution of this paper is therefore as follows:
2. Methodology
The methodology comprised four steps: dataset preprocessing, GAN training, Artificial NDVI reconstruction, and evaluation, as shown in Figure 1 (The complete methodology, including data preprocessing, model training, reconstruction and evaluation scripts, as well as the trained model-weights can be found on https://www.github.com/jurriandoornbos/RGBtoNDVIconversion, accessed on 15 December 2024). The evaluation used the generated artificial NDVI maps as input for direct accuracy assessment (pixel-level), and two applications using NDVI.
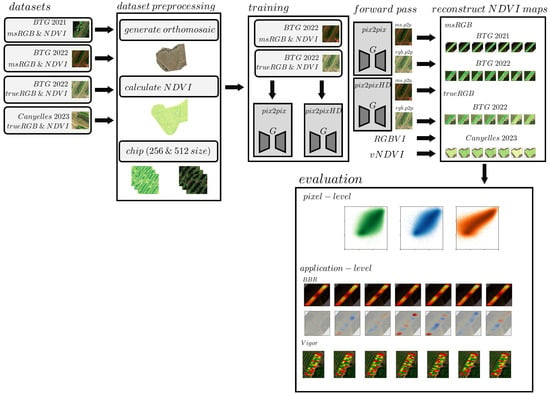
Figure 1.
Overview of the Methodology. The process began with dataset preprocessing (Section 2.2), followed by GAN model training (Section 2.3), artificial NDVI map reconstruction (Section 2.4), and finally, model evaluation. The btg2021 and btg2022 datasets were acquired at the Bodegas Terras Gaudas vineyard in Spain, with a MicaSense RedEdge, MicaSense Altum-PT and DJI Phantom 4-RTK (RGB) cameras. can2023, collected at the Canyelles vineyard in Spain using a DJI Mavic 3M multispectral system. Model training resulted in four distinct models, Pix2Pix and Pix2PixHD, trained on either RGB and NDVI pairs, or multispectral composite RGB and NDVI pairs. The evaluation stage included direct pixel-level accuracy assessments (Section 3.1) across these datasets as well as Botrytis (Section 3.2) from Ariza-Sentís et al. [33] and Vigor mapping (Section 3.3) from Matese et al. [4].
2.1. Artificial NDVI Models
Four leading approaches were chosen to convert RGB to NDVI values: two GAN models, Pix2Pix and Pix2PixHD and two explainable approaches, RGBVI and vNDVI.
The Pix2Pix model, first developed by Isola et al. [29], provides a general solution for conditional image-to-image translation by using an adversarial loss function combined with L1 regularization. This approach ensures realistic image generation by balancing noise and preventing overfitting. The Pix2Pix architecture is based on a U-Net Convolutional Neural Network with skip-connections, preserving spatial information throughout the image translation process. This model was also employed in various earlier work on RGB to NDVI conversion [20,21,22]. A visual representation of the model is presented in Figure A1 in Appendix A.
Pix2PixHD, introduced by [28], enhances Pix2Pix by adding a second generator network (G2) and two additional discriminator networks (D2 and D3). G2 downsamples a high-resolution image for G1 to translate, then upsamples the output back to a larger resolution. D2 and D3 evaluate the generated image at multiple resolutions. Additionally, a feature-matching loss is incorporated to ensure that generated images not only deceive the discriminator but also match the real data intermediate feature statistics. This model was also employed in earlier work, outperforming Pix2Pix [20]. A visual representation of the model is presented in Figure A2 in Appendix A.
Previous research did not publish the model weights, therefore Pix2Pix and Pix2PixHD required training on aligned RGB to NDVI images.
RGBVI, a vegetation index derived from RGB image bands, was initially developed for assessing barley using UAVs [34]. However, it is also observed as a useful replacement for NDVI in vineyards [35]. It is calculated as follows: , with the green values mimicking the effect of chlorophyll and infrared reflectance. A limitation of RGBVI is that it accentuates all green objects, not just those with chlorophyll, though this is less problematic in the controlled conditions of UAV-based remote sensing [15].
vNDVI is a vegetation index derived from RGB and NDVI training data, and using genetic algorithms to assess the best equation to derive NDVI from RGB imagery [18]. The genetic algorithm was trained on 1,000,000 pixels of RGB and NDVI containing vegetation, buildings, roads and lakes in UAV imagery. The genetic algorithm found to be the best performing, with an overall mean percentage error of 6.9% and a mean average error of 0.05.
2.2. Datasets Employed
Three different datasets were used for the analysis. The first dataset was a multispectral dataset acquired in the Bodegas Terras Gauda vineyard, acquired in 2021 [7]. A DJI Matrice M210-RTK UAV was flown at a height of 15 m with a MicaSense RedEdge multispectral sensor. The flight was performed with 80% overlap between images. The red, green and blue bands of the image were used for the RGB data. This study refers to this dataset as btg2021. This dataset was exclusively used for generating artificial NDVI from RGB images and evaluating the results.
The second dataset consisted of an RGB and multispectral dataset acquired on 12 July 2022, in the same Bodegas Terras Gauda vineyard, referred to in this study as btg2022. In this dataset, a DJI Matrice M300-RTK was flown with a MicaSense Altum-PT multispectral sensor at a height of 15 m. In addition, a DJI Phantom 4-RTK was also flown at 15 m to acquire true RGB images. The flights were performed with 80% overlap between images. btg2022-train and btg2022-val were used for training and validating the GAN networks before deploying them on the btg2022-test set for evaluations. The train/test/validation split of this dataset is shown in Figure 2.
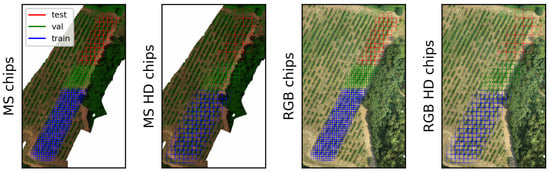
Figure 2.
The btg2022 training, testing and validation chips subsets. Left shows the Pix2Pix chips, at a size of 256 by 256 pixels. The right image shows the Pix2PixHD chips at 512 by 512 pixels. The blue and green squares are the training and validation chips, whereas the red squares are the testing chips. Smaller-looking squares in train and val are due to overlapping chips.
The third dataset consisted of an RGB and multispectral dataset acquired in the Canyelles vineyard in 2023 [36], referred to as can2023. A DJI Mavic 3M was flown at 10 m height with 80% image overlap. This dataset was exclusively used for generating artificial NDVI from RGB images and evaluating the results.
The image datasets from the UAV flights were processed into orthomosaics using Agisoft Metashape [37], all settings on high, with ground-control points for alignment. The output orthomosaics are visualized in Figure 3. Additionally, for the RGB image sets, a digital surface model (DSM) was reconstructed from the dense point cloud. The multispectral data was used to calculate the NDVI values, using the Red and NIR bands. To ensure alignment between RGB and MS sensor observations in the btg2022 dataset, the QGIS georeferencer tool was employed [38]. Blue tags, visible in both RGB and MS orthomosaics, were used as reference points for alignment (see Figure 4). The differing spatial resolutions were harmonized using bilinear interpolation to match the multispectral resolution. For a summary on all the datasets, refer to Table 1 and Figure 3.
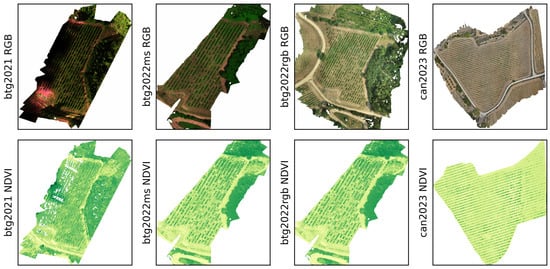
Figure 3.
Visual overview of the btg2021, btg2022ms, btg2022rgb and can2023 orthomosaics before alignment and chipping. The top row visualizes the composite or true RGB for btg2021, btg2022 and can2023. The real NDVI maps are shown on the bottom row.

Figure 4.
Example blue tags visible in both btg2022 RGB and MS orthomosaics were used as alignment points. The spatial resolution of the RGB sensor was bi-linearly interpolated to match the MS resolution.

Table 1.
Overview of the employed datasets: btg2021, btg2022 and can2023. WGS84 refers to the coordinates in WGS84 format. doi is the open-access URL to the dataset. AOP is the Area of Appelation of the vineyard.
A key consideration in our data preparation is the spatial separation of training and testing chips. First, spatial separation enables the inclusion of overlapping chips within the training dataset while ensuring that none of these areas appear in the test set, preventing unintended information leakage. Second, as underscored by Diez et al. [32], spatially mixing chips can result in artificially inflated performance metrics due to spatial autocorrelation. By rigorously separating chips based on their locations, we avoid region-specific biases and ensure that the evaluation of our models reflects genuine generalization capabilities rather than the exploitation of localized features.
2.3. Training Approach
The GAN models are limited in input resolution to for Pix2Pix and for Pix2PixHD. The orthomosaics are much higher resolution, requiring the orthomosaics to be ‘chipped’ into many smaller, square images. These chips fit the input resolution of the GAN model. The chips in this study were set to for Pix2Pix and for Pix2PixHD. In Figure 2 the training and validation chips for training are visualized. The test chips were exclusively used for the evaluation. Additionally, the training and validation chips were also rotated and mirrored, to augment the training dataset size. This resulted in the total amount of chips reflected in Table 2. We evaluated the trade-offs between computational efficiency, spatial information preservation, and model performance for and chip sizes, summarized in Table 2. The chips include 87.3 M training pixels (26% more than ), offering greater spatial context but requiring 240 s per epoch—a 20% increase over the chips, which cover 69.0 M pixels and provide higher spatial diversity with 1053 training chips versus 333.

Table 2.
Training overview for the selected chip sizes. The table summarizes the number of training chips, total training pixels, and epoch time for each model configuration. These numbers are identical for both the MS and RGB subsets.
The decision was made not to retrain or fine-tune the GAN models on the btg2021 or can2023 datasets to preserve the generalizability of the method. Requiring true NDVI observations for each new location would negate the need for an RGB-to-NDVI conversion method, as the utility of such a method lies in its ability to generalize across different locations without the need for specific training data for each site.
The Pix2Pix and Pix2PixHD models were both trained for 100 epochs on the btg2022 dataset. The learning rate was set to 0.0002 which decayed to 0 in the last 50 epochs, these settings were the same as in the original Pix2Pix paper by Isola et al. [29]. During training, GAN losses were monitored to assess the stability of the discriminator and generator. While discriminator loss is expected to fluctuate, a stable generator loss over time is desirable; however, a completely static generator loss may indicate mode collapse. Mode collapse occurs when the generator only generates a single output over a diverse range of inputs, it finds a local minimum that fools the discriminator. These losses, though informative, are not suitable for model selection. MSE was employed as a regression-based evaluation metric, enabling the identification of the best-performing and stable model. This process involved 5-fold cross-validation on the validation dataset, focusing on minimizing variance and utilizing a running average (sliding window of 5 epochs). The selection is visualized in Figure 5.
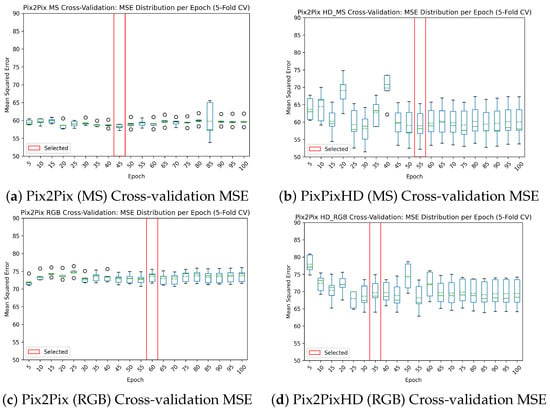
Figure 5.
Cross-validation results and model selection based on MSE results on the validation dataset. The green lines indicate the mean. The red box shows the selected model from the training procedure. Sub-figures (a–d) show the performance of four candidate models during cross-validation, highlighting the differences in MSE values. These results were used to guide model selection for optimal performance.
2.4. NDVI Reconstruction
The trained Pix2Pix and Pix2PixHD models were applied to the RGB chips from the btg2021-test, btg2022, and can2023 datasets. The testing chips for btg2022 and can2023 are visualized in red in Figure 6 and Figure 7, respectively. This process produced artificial NDVI chips for each dataset, which were then reconstructed into their original locations in the orthomosaic using the location information created during the chipping step. This resulted in artificial NDVI maps generated by Pix2Pix and Pix2PixHD, respectively. Additionally, these outputs were masked by the respective vineyard shapes. RGBVI and vNDVI were calculated using the Red, Green, and Blue layers from the RGB orthomosaics with the equations presented above.
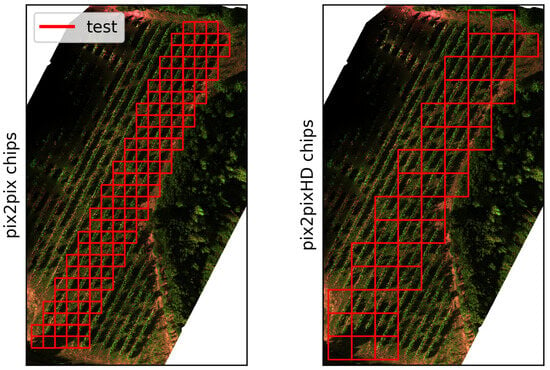
Figure 6.
The location of the testing chips in the btg2021 dataset. Left shows the smaller Pix2Pix chips, with chips at a resolution of 256 by 256 pixels. Right shows the Pix2PixHD chips, with chips at a resolution of 512 by 512 pixels.
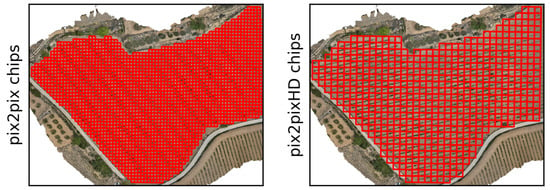
Figure 7.
The location of the testing chips in the can2023 dataset. Left shows the smaller Pix2Pix chips, with chips at a resolution of 256 by 256 pixels. Right shows the Pix2PixHD chips, with chips at a resolution of 512 by 512 pixels.
2.5. Evaluation Approach
Inspired by the multiple evaluations from [20], three evaluations were performed on the created orthomosaics: one evaluation at the pixel-level, and two evaluations using a downstream NDVI application: Botrytis Bunch Rot mapping (BBR) and Vigor mapping.
In the pixel-level evaluation, the generated NDVI maps and RGBVI were directly compared to the true NDVI. The pixel-level comparison comprised of four metrics. These metrics were also used in previous work. The metrics were r-squared (), mean-squared error (MSE), structural similarity (SSIM) and peak-signal-to-noise-ratio (PSNR). R-squared is a statistical measure of how well the regression line approximates the real data points, calculated as:
where is the i-th element of the observed values array Y or true NDVI, is the i-th element of the predicted values array or artificial NDVI. is the mean of the observed values Y or true NDVI. A value of 1 indicates that the regression line perfectly fits the data, meaning that all variability in the dependent variable is explained by the independent variable. The flowchart for this pixel-level evaluation step is also presented in Figure 8.
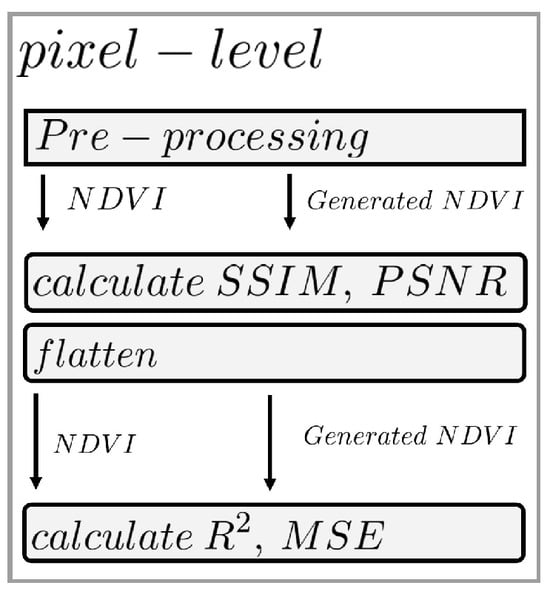
Figure 8.
Processing flowchart for pixel-level accuracies. In the pre-processing step, the earlier steps are performed, and the real and artificial NDVI orthomosaics are generated. Which is directly used to calculate the SSIM and PSNR values. These arrays are then flattened into a list for MSE and R-squared.
For MSE, it assumes two arrays Y and of length n, where Y or true NDVI, represents the observed values and or artificial NDVI represents the predicted values. MSE is a measure of the average error, meaning it gives equal weight to all errors regardless of their magnitude. A smaller MSE value indicates less discrepancy between the two arrays, implying better agreement or similarity between the observed and predicted values. Conversely, a larger MSE value indicates greater discrepancy or error.
Structural similarity is used to describe the similarity between two images. Developed to evaluate the structure, brightness and contrast between two images. This is operationalized in the following equation:
In this equation x is assumed to be true NDVI, whereas y is artificial NDVI. and are the means of the two images (brightness), and are the variances of the two images (contrast), is the covariance of the two images (structure), and are constants to stabilize the division with weak denominator.
PSNR is used to describe the peak signal-to-noise ratio between two images, which is calculated as:
where is the maximum possible pixel value, is the mean squared error between the two images. A higher PSNR value indicates less distortion in the reconstructed/compressed image, as it means that the signal (original image) is much stronger compared to the noise (distortion). PSNR is expressed in decibels (dB), and higher values indicate better image quality.
In the application-level evaluation, the true and generated NDVI maps were used in a Botrytis bunch rot risk assessment pipeline presented in Ariza-Sentís et al. [33]. Botrytis develops in the vine bunches and is more difficult to detect at an early stage using object detection algorithms, as the grapes themselves are impossible to detect from top-view imagery in a vineyard. However, certain vineyard features are a good determinant of Botrytis cinerea proliferation. This pipeline requires a DTM, CHM, NDVI and plant shadows as input, and calculates a risk heatmap as output. For this evaluation, the different input NDVI maps were varied: True, Pix2Pix, and Pix2PixHD. Resulting in three different heatmaps. These heatmaps were evaluated using and MSE. The accuracy assessment is therefore whether the generated NDVI has a significant impact on the heatmap. This evaluation step is also shown in Figure 9.
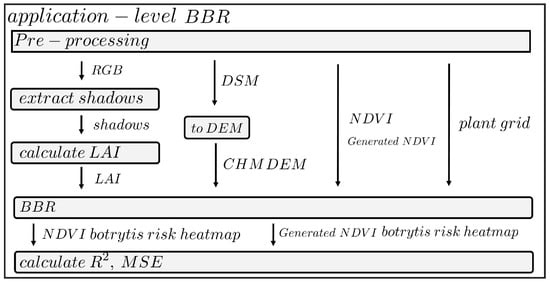
Figure 9.
Processing flowchart for Botrytis Bunch Rot Risk (BBR) mapping. In the pre-processing step, the orthomosaic photogrammetric reconstruction, digital surface model (DSM) creation and the real and artificial NDVI orthomosaics were generated. The red band was used to identify the shadow-pixels and is a direct measure of leaf area index, the DSM was further processed to acquire a canopy height model. These products were then placed into the BBR algorithm from Ariza-Sentís et al. [33].
Various vineyard health assessment studies with UAVs make use of the Vigor indicator [4,9,35]. This indicator aims to give an overview of the health across the vineyard. The Vigor indicator is correlated to harvest stock biomass, grape yield, and total soluble solids [9]. The processing steps to acquire Vigor are presented in Figure 10, and a direct adaption from the process described in Matese et al. [4]. This method classifies the health of the vineyard in a predefined 2x2m grid, the NDVI values are extracted to this grid, and classified in tertiles, resulting in a low, medium and high vigor classification. This classification is evaluated in a confusion matrix and corresponding weighted F1-score, using the Vigor map created from NDVI as the comparative example.
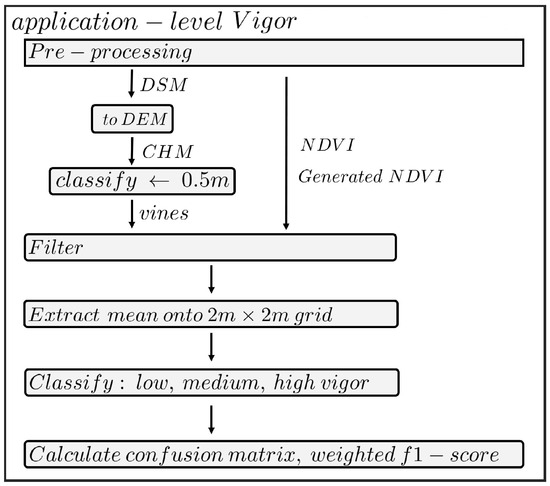
Figure 10.
Processing flowchart for Vigor mapping. In the pre-processing step, the orthomosaic photogrammetric reconstruction, DSM creation and the real and artificial NDVI orthomosaics were generated. The DSM was further processed to acquire a canopy height model (CHM). Then the steps were followed as described in Matese et al. [4]. Consisting of thresholding to only include canopy rows, mean-extraction onto a 2-by-2-meter grid and classifying the values into tertiles.
3. Results
The real NDVI maps were compared to the Pix2Pix, Pix2PixHD, RGBVI and vNDVI maps using evaluation approaches presented in the methodology: pixel-level, BBR mapping and Vigor mapping. These evaluations were performed on four vineyard-datasets: btg2021, btg2022ms, btg2022rgb and can2023. The zoomed images are shown in Figure 11, Figure 12, Figure 13 and Figure 14 respectively. This resulted in twelve distinct evaluations: three evaluation approaches on four datasets. The btg2021 dataset consists of generated NDVI based on the Bodegas Terras Gauda vineyard in 2021. This dataset is expected to yield good metrics for the Multispectral models as it closely resembles the training dataset, differing only by location within the same vineyard and sensor type. The btg2022 data would result in the best results for both the RGB and MS subsets. Because the models trained on the southern section of the vineyard have seen highly similar examples during training. The can2023 dataset, obtained from a different vineyard with a different sensor (the true RGB sensor from the DJI Mavic 3M) and in a different year, poses the most significant challenge for all the models, although the RGB-trained models are expected to perform well, as the sensor characteristics are very similar.

Figure 11.
btg2021 zoomed in. VI color range from (−0.2, 1) to enhance the differences between the images. Green is vegetation, whereas orange is the bare soil, yellow colors indicate grass. The NDVI and generated NDVI maps show a high similarity across both canopy and in-between rows. the RGBVI map has lower values for the bare soil. vNDVI has a lower range of values altogether.

Figure 12.
btg2022ms zoomed in, VI values colored from (−0.2, 1) to enhance the differences between the images. Green is vegetation, whereas orange is the bare soil, yellow colors indicate grass. The NDVI map indicates minimal bare soil, the Pix2Pix maps show less intensity in the canopy vegetation and less grass between rows, RGBVI indicates lower values overall. Pix2Pix has slightly higher values for canopy compared to Pix2PixHD.

Figure 13.
btg2022rgb zoomed in, VI values colored from (−0.2, 1) to enhance the differences between the images. Green is vegetation, whereas orange is the bare soil, yellow colors indicate grass. The NDVI map indicates minimal bare soil, the Pix2Pix maps show less intensity in the canopy vegetation and less grass between rows, and RGBVI indicates lower values overall. Pix2Pix has slightly higher values for canopy compared to Pix2PixHD.

Figure 14.
can2023 zoomed in on tractor tracks, VI color range from (−0.2, 1) to enhance the differences between the images. Green is vegetation, whereas orange is the bare soil, yellow colors indicate grass. The NDVI and RGBVI maps indicate a pattern of tractor tracks and canopy vegetation, Pix2Pix shows more vegetation over the image and hard boundaries between reconstructed chips. Pix2PixHD shows less vegetation than Pix2Pix, although more than the true NDVI image.
3.1. Pixel-Level Results
The evaluation of pixel-level performance across models and datasets, based on R-squared and MSE show that Pix2PixHD (MS) model is the best performing (see Table 3 and Table 4. For the btg2021 dataset, the highest R-squared value (0.281) is achieved by Pix2PixHD (MS), while other models underperform, including negative scores for Pix2Pix (RGB) (−0.460). Similarly, the lowest MSE (0.029) is recorded for Pix2PixHD (MS). In the btg2022ms dataset, Pix2Pix (MS) achieves the best R-squared (0.837) and MSE (0.009), Vegetation indices like RGBVI and vNDVI perform poorly, with higher MSE values (e.g., 0.043 for RGBVI). As expected, the btg2022rgb dataset favours RGB-trained models, with Pix2Pix (RGB) achieving the highest R-squared (0.637) and the lowest MSE (0.020), closely followed by Pix2PixHD (RGB) (R-squared of 0.617, MSE of 0.021). Multispectral models and vegetation indices show substantially poorer performance, with negative R-squared values (e.g., −1.506 for RGBVI) and higher MSE values. In the can2023 dataset, all models struggle, with Pix2PixHD (MS) achieving the least negative R-squared (−0.021) and the lowest MSE (0.028).

Table 3.
Pixel-level R-squared values for all models and datasets. Bold values indicate the best scoring model.

Table 4.
Pixel-level MSE values for all models and datasets. Bold values indicate the best scoring model.
For SSIM, in Table 5, vegetation indices consistently outperform deep learning models. In btg2021, RGBVI achieves the highest score (0.677), followed by vNDVI (0.596). Similarly, in btg2022ms, vNDVI performs best (0.746), with Pix2Pix (MS) close behind (0.707). In RGB-focused datasets like btg2022rgb and can2023, vNDVI also performs best with SSIM of 0.676 and 0.514. For PSNR, in Table 6, the performance varies with dataset characteristics. In btg2021, RGBVI achieves the highest score (21.167), while deep learning models like Pix2Pix (RGB) lag behind (18.507). In btg2022ms, Pix2Pix (MS) shows superior fidelity with a PSNR of 26.625, surpassing vegetation indices. For btg2022rgb, RGB-trained Pix2Pix (RGB) achieves the best PSNR (23.176), with vegetation indices performing poorly. In can2023, RGBVI achieves the highest PSNR (21.317), followed by vNDVI (20.234).

Table 5.
Structural Similarity values for all models and datasets. Bold values indicate the best scoring model.

Table 6.
Peak signal-to-noise ratiovalues for all models and datasets. Bold values indicate the best scoring model.
In the pixel-level comparison, R-squared and MSE of NDVI estimation, generative models perform best, estimating more accurate values for NDVI. However, the vegetation indices (RGBVI, vNDVI) excel in structural similarity and peak-signal-to-noise, indicating that these approaches maintain the structure of leaves, and soil better than the Pix2Pix approach.
3.2. Botrytis Bunch Rot Results
Figure 15, Figure 16, Figure 17 and Figure 18 show the BBR risk heatmaps generated using multiple input vegetation indices across btg2021, btg2022ms, btg2022rgb, and can2023. Each figure provides a comparison of the predicted BBR risk when using the true NDVI map against Pix2Pix (MS), Pix2PixHD (MS), Pix2Pix (RGB), Pix2PixHD (RGB), RGBVI, and vNDVI. Differences between the predicted BBR risk maps and the baseline NDVI-driven predictions are further explored in the corresponding difference maps (Figure A7, Figure A8, Figure A9 and Figure A10). For instance, in the can2023 dataset, the generative approaches show broadly similar patterns, whilst RGBVI and vNDVI underestimate the risk compared to the true NDVI reference.

Figure 15.
btg2021 Botrytis bunch rot risk (BBR) heatmap outputs using the different vegetation maps as input.

Figure 16.
btg2022ms Botrytis bunch rot risk (BBR) heatmap outputs using the different vegetation maps as input.

Figure 17.
btg2022rgb Botrytis bunch rot risk (BBR) heatmap outputs using the different vegetation maps as input.
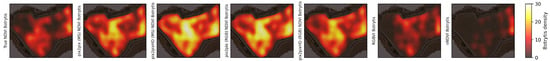
Figure 18.
can2023 Botrytis bunch rot risk (BBR) heatmap outputs using the different vegetation maps as input. There is a high similarity between all the BBR outputs. With the largest difference in risk being shown in the top-left section of the vineyard. In this section, by using the Pix2Pix generated NDVI, the BBR model shows distinct hotspots, Pix2PixHD closely represents the real NDVI map, and RGBVI underestimates the risk, in comparison to using the true NDVI map.
To quantitatively evaluate these modelling approaches, Table 7 and Table 8 present the R-squared and mean squared error (MSE) values, respectively, for each dataset and model combination. Across all datasets, the top-performing models in terms of R2 also tend to have the lowest MSE.

Table 7.
Botrytis bunch rot R-squared values for all models and datasets. Bold values indicate the best scoring model on the dataset.

Table 8.
Botrytis bunch rot MSE values for all models and datasets. Bold values indicate the best scoring model on the dataset.
In the multispectral datasets btg2021 and btg2022ms all approaches perform well. In the btg2021 set, the Pix2PixHD (MS) model achieves the highest R2 (0.974). This indicates an exceptionally strong alignment between predicted and observed BBR risk. All models, however, maintain relatively high R2 scores (above 0.85), suggesting that for this vineyard, the BBR risk patterns are relatively straightforward to approximate from various vegetation inputs. In btg2022ms the standout performance is from Pix2PixHD (RGB) and Pix2Pix (RGB), each producing R2 values above 0.99 (0.993 and 0.992, respectively). Again, generative models prevail: both Pix2PixHD (MS) and Pix2Pix (RGB) achieve near-perfect R2 scores (0.992). In stark contrast, the RGB datasets are not as straightforward to model. In btg2022rgb the Pix2PixHD models perform well with R-squared values at 0.992. It is vNDVI and RGBVI that yield highly negative R2 values (−16.906 and −5.788, respectively), signifying a catastrophic mismatch between these indices and the observed disease patterns. The extreme negativity indicates that in this vineyard, a naïve guess (e.g., predicting the mean risk everywhere) would outperform using the modelled approaches. The can2023 site also poses a more challenging scenario. All generative models yield negative R2 values, meaning they struggle to explain the variance in BBR risk at this site. The best performer here is the simpler RGBVI approach, with a modest R2 of 0.213. Although 0.213 is not a particularly high correlation, it is substantially better than negative values and indicates that RGBVI captures at least some vegetation information. The poor generative model performances suggest that factors unique to this dataset cause a domain shift not captured in the training data.
3.3. Vigor Results
The vigor maps presented in Figure A11, Figure A12, Figure A13 and Figure A14 illustrate how different vegetation map inputs—ranging from true NDVI to various generative and index-based approaches—affect the classification of vine vigor. In each map, green indicates high-vigor areas, yellow represents medium vigor, and red denotes low vigor. Table 9 presents weighted F1-scores for vigor classification, providing a statistical benchmark against which the visual assessments can be verified. Higher F1-scores indicate better overall classification performance across the three vigor classes. Figure A12 and Figure A13 reveal a distinct spatial pattern where the northern section of the vineyard consistently exhibits lower vigor, a trend also observed in the lower portion of the maps. In Figure A14, the reduced vigor in the eastern section is well-modelled by all approaches. However, the top-left portion of the vineyard shows lower vigor that is primarily captured by RGB-based indices. This indicates that RGB-derived vegetation indices may offer finer-scale sensitivity to variations in canopy structure.

Table 9.
Vigor mapping weighted F1-scores for all models and datasets.
In btg2021, visualized in Figure A11, the best performance is achieved by RGBVI (0.847), followed closely by Pix2Pix (MS) (0.805) and vNDVI (0.786). This aligns well with the visual observation that RGBVI produced sharply delineated vigor patterns. By contrast, Pix2Pix (RGB) and Pix2PixHD (RGB) lag behind, with F1-scores around 0.55, reflecting their difficulty in capturing subtle vigor gradients. In the other multispectral dataset btg2022ms, visualized in Figure A12, RGBVI again tops the chart with 0.904, a notably high F1-score, and vNDVI follows with 0.860. Among the generative models, Pix2PixHD (MS) is the best performer (0.807), demonstrating that multispectral-driven generative models can also yield robust vigor maps. For btg2022rgb, visualized in Figure A13, sees generative models narrowly outperforming simpler indices. The top scorers are Pix2PixHD (MS) at 0.789 and Pix2Pix (RGB) at 0.782, narrowly outperforming even vNDVI and RGBVI. At can2023, visualized in Figure A14, the F1-scores show a relatively tight cluster. RGBVI and vNDVI both achieve 0.661, tied for the best performance and closely aligned with Pix2Pix (MS) (0.660). The Pix2PixHD (MS) model, however, yields a lower score (0.483), indicating it is less effective at capturing critical vigor variations, such as the low-vigor zones in the top-left. This performance gap highlights that model architecture and training nuances significantly influence outcomes, and a model trained in one vineyard might not do so in another.
4. Discussion
This study presented an evaluation approach for RGB to NDVI conversion, placing real-world application of the trained GAN models at the centre, as operationalized through Botrytis bunch rot risk mapping and Vigor mapping. Whilst maintaining previously used accuracy metrics of SSIM, PSNR, MSE and R-squared [20,22,30]. The performed assessment of application-level evaluation indicates nuance for evaluating NDVI beyond standard accuracy metrics. High pixel-level metrics such as MSE and SSIM do not automatically translate to high application-level scores as well. This new approach gives a new dimension to assessing the applicability of the results in the real world. The used evaluation approach, introduced a wide variety of sensors and locations, within the same subject matter of vineyards. This approach aimed to further explore domain shifts between sensors and locations within UAV research.
4.1. Explainable Outperforms GAN
The previously used accuracy metrics were operationalized in this study as pixel-level evaluations. This evaluation suggests that vNDVI or RGBVI has the best structural similarity, compared to the Pix2Pix NDVI maps. This is observed in all four evaluation datasets (see Table 5). The accuracy of these maps in terms of R-squared and MSE were always best for Pix2Pix or Pix2PixHD. However, the Pix2Pix-generated orthomosaics exhibited noticeable square artefacts due to the model’s use of smaller input patches, as seen in the NDVI maps (Figure 11). While Pix2PixHD mitigated this issue somewhat by utilizing larger patches, seamlines remained visible. These artefacts help explain the lower SSIM and PSNR scores for both Pix2Pix-based methods compared to the vegetation index approaches. The chip size optimizes computational efficiency while maintaining spatial diversity due to reconstruction into orthomosaic, making it suitable for resource-constrained environments. Conversely, the chips have slightly lower R-squared and MSE, albeit at a computational cost. However, when deploying these maps in an application such as Botrytis bunch rot mapping, RGBVI was very usable with R-squared values of 0.942 and 0.857 for btg2021 and btg2022ms respectively. The BBR model from [33] assumes the use of NDVI. Therefore, the lower values for vegetation in both vNDVI and RGBVI will usually result in a lower estimation of Botrytis risk. A new version of BBR could be adjusted to fit RGBVI. Vigor is a relative measure derived from three vigor classes, which are separated by tertiles relative to the observed values of NDVI. Whilst RGBVI and vNDVI might have lower estimations for the vegetation, they do align with their structure to NDVI. This is evidenced by the higher SSIM values across the pixel-level evaluation. This structural resemblance makes RGBVI and vNDVI more suitable for capturing the relative differences required for vigor mapping. Additionally, the explainable nature of these indices allows for better alignment with the relative thresholds used to define vigor classes, making them inherently more appropriate for this application compared to the Pix2Pix method. These results are comparable to previous work on RGBVI that indicates that RGBVI represents many of the same aspects as NDVI [4,34].
4.2. GAN Generalization
Pix2Pix is claimed to be a robust and inexpensive alternative to the multispectral sensors [20,22]. A multispectral sensor, in contrast, can evaluate different crops, different lighting conditions and different locations. However, previous work falls short of supporting these claims: their testing datasets are highly similar to the training datasets. Previous work used the same sensor type, same location, and same flight being used for training and testing. This experimental setup ensures a high reporting accuracy, as the model has been trained on very similar samples. To claim Pix2Pix to be a robust and inexpensive alternative to multispectral sensors, a single trained model should be able to perform a wide range of RGB to NDVI conversions [30].
4.2.1. RGB Type Matters
Previous research has demonstrated two options for training a conversion model. The first is to use a single multispectral sensor. which trains on multispectral data by extracting RGB and NDVI values, such as in Farooque et al. [20]. This first approach may not perfectly recreate the spectral properties of true RGB imagery, which would be the envisioned application. The second option therefore is to align RGB observations from a RGB sensor and separate NDVI values from a multispectral sensor, such as in [22]. Which would presumably result in better NDVI prediction when using a more affordable RGB sensor. This study found a strong correlation between the similarity of the training and evaluation datasets and the resulting model accuracy. Specifically, models trained on the btg2022ms dataset exhibit significantly higher scores compared to the alternatives. Pix2Pix (MS) has an R-squared of 0.837 (0.449 higher than Pix2Pix (RGB)), and a PSNR of 26.625 (6 higher than Pix2Pix (RGB)). A similar trend is observed for the RGB-based model tested on the btg2022rgb test set.
Furthermore, the two Pix2Pix RGB models evaluated on the btg2022ms test set were better than btg2021 and can2023 in R-squared, MSE, SSIM, Botrytis and Vigor. This indicates that the sensor type plays a limited role when trained on true RGB imagery. However, when trained on composite RGB imagery from the Multispectral sensor, the Pix2PixHD (MS) model scores better in btg2021 than on btg2022rgb in R-squared, MSE, SSIM and Botrytis. This indicates that the domain shift between years is less significant between years than from sensor to sensor, because btg2021 is also a multispectral composite RGB dataset. Furthermore, the direction of domain shift is important: multispectral models performed better on the RGB cases than the RGB models on the composite RGB cases.
The most difficult dataset is can2023. R-squared was negative for both the pixel-level and Botrytis evaluations. Here the domain shift from the training data is not easily explained by just the sensor. Both the RGB and MS-trained models trade blows and are systematically outperformed by simple RGB vegetation indices. Several factors may account for these differences. First, the vineyard row structure in the Canyelles dataset (as shown in Figure 14) diverges substantially from that of the Bodegas Terras Gaudas data. In addition, the soil differs in coloration and supports less vegetation between rows. Third, the multispectral data were acquired using a DJI Mavic 3 Multispectral system rather than a MicaSense sensor. These observations align with the findings of Kasimati et al. [14], suggesting that variability in NDVI output maps may stem as much from the specific sensor used as from the vineyard’s location or the year in which data are collected.
4.2.2. Evaluation Approach Matters
Interestingly, performance variations observed across datasets are often as striking as those across modelling approaches or vegetation indices. In other words, the dataset itself can be as influential a factor as the specific method employed. For example, with the Botrytis bunch rot (BBR) predictions, the Pix2Pix (MS) model achieves an MSE of just 1.464 on the btg2021 dataset, whereas on can2023, the same model’s MSE skyrockets to 41.476—an increase of nearly 30 times, indicating substantial challenges in capturing the vineyard’s unique conditions. Additionally, for vigor classification, the RGBVI-based approach achieves a strong weighted F1-score of 0.904 on btg2022ms, clearly outperforming all other approaches on that dataset. However, when applied to btg2022rgb, the F1-score drops to 0.693, indicating a marked decline in its ability to accurately classify vigor. Whilst Pix2PixHD (MS) scores well for Vigor classification in btg2022rgb, but worse compared to RGBVI in btg2022ms. This contrast suggests that what works exceptionally well in one context may fail to deliver comparable accuracy in another, even if the model and input type remain unchanged. It underscores that the intrinsic characteristics of the vineyard environment—such as canopy structure, soil coloration, vine spacing, and sensor variations—can heavily influence predictive accuracy. As such, improving model performance is not solely about refining algorithms or indices; it also requires careful attention to the distinct conditions and data distributions inherent to each vineyard location and dataset.
4.3. Next Steps: Scenario Building
Further generalization testing is required to achieve great accuracy for RGB to NDVI conversion and therefore the envisioned low-cost alternative to the multispectral sensor. The directionality of generalization is important to take into account when looking at domain shifts, as multispectral RGB more easily translates to RGB than vice versa. UAV-related variation needs to be explored, such as the angle of the sensor and flight height [3]. Furthermore, models should also be evaluated according to different lighting conditions on the day itself. Finally, various crops and applications ought to be included in the test datasets. Luckily, relatively few images are required for a well-performing Pix2Pix(HD) model [22]. Therefore, it can be explored whether augmenting the training dataset with different light conditions and angles is a fruitful endeavour to make Deep Learning models more robust. This could also be a further application for style-transfer generative models. See Table 10 for a starting point within the area. Open data would be a necessity to enable further domain-shift research.

Table 10.
Domain Shift Scenarios within UAV Remote Sensing.
4.3.1. Alternative Models
However, different Deep Learning models could be included for further steps, such as denoising diffusion probabilistic models (DDPMs) [40]. DDPMs have shown better generalization and higher-fidelity results, as well as more predictable training over GANs [40]. Although many of these approaches are unconditional or are not precise in their spatial continuity, therefore models such as DDIB [41], I2SB [42] and MedFusion [43] seem like the best start. Furthermore, Vision Transformers (ViTs) have also been shown to outperform convolutional neural networks in a variety of tasks [44]. There are very few implementations of conditional image generation using ViTs however and almost no published articles using such models in real-world applications. Models such as DiFFiT [44] could be a start. Problematically however, DDPMs and ViTs tend to be much larger networks than Pix2Pix (HD), larger networks take more resources to train and deploy and due to their recent development, there is little documentation compared to Pix2Pix. Even though DDPM and ViT architectures show impressive generalization [44], an enormous variety of input training data is required [45] in the order of multiple terabytes [46]. These limitations in documentation, compute resource requirements and dataset sizes also hinder the imagined low-cost alternative to multispectral data [23].
4.3.2. Alternative Indices
However, a potential further application of the conversion models could be more complex indices or direct estimation of short-wave and/or long-wave infrared bands. Examples such as the water-extraction index AWEinsh [47], or soil moisture [48]. However, the development and understanding of such indices at an ultra-high spatial resolution such as UAVs is still an open topic in Remote Sensing [48]. Furthermore, the applications of such measurements for viticulture and beyond remain unexplored, although could see more traction in the coming years [48]. Only after these requirements are satisfied would it be an interesting topic to generate these indices directly from RGB images in the vineyard or beyond.
5. Conclusions
This study presents a new evaluation approach for artificial NDVI models. It goes beyond previous work by proposing a set of application-specific evaluations using Botrytis bunch rot mapping and vigor mapping. These go beyond directly comparing RMSE, SSIM and R-squared to measured NDVI. Instead, it uses the NDVI maps as input for Botrytis mapping, and for Vigor mapping, comparing them to using the true NDVI map. Especially in the Botrytis mapping evaluation, it becomes clear that the Pix2PixHD model generates a usable output for the model, across all evaluation datasets. Whilst RGBVI indicates a good drop-in replacement as well. In Vigor mapping the vNDVI explainable approach scores the highest. The proposed evaluation methodology is able to better define the usefulness of the output maps, as it goes beyond the direct comparisons. This extension of evaluation enables explainable approaches such as RGBVI and vNDVI to show that they perform as well as GAN models.
Perhaps difficult to acquire (hyper) spectral indices such as soil moisture or water content is a next step for RGB conversion GANs. Deep Learning is clearly capable of capturing complex features in data. Generalization continues to be a challenge for many Deep Learning approaches with UAV data. Testing on a different area of the same flight is not sufficient to evaluate a trained Deep Learning model. By formalizing generalization into distinct categories of location and sensor, the trained Pix2Pix and Pix2PixHD models were evaluated using highly varying evaluation datasets. While Pix2Pix-based GAN methods can achieve superior accuracy metrics (R-squared, MSE) compared to simpler RGB-based vegetation indices, their outputs often contain artefacts, resulting in lower SSIM and PSNR values. In contrast, indices like RGBVI and vNDVI maintain better structural consistency, making them more suitable for tasks—such as vigor mapping—where relative differences are crucial. For application-specific evaluations, such as Botrytis bunch rot risk mapping, both GAN-derived and index-based methods can be effective, though vegetation indices may sometimes offer comparable or even better usability. Generalization remains a significant challenge. Models trained on datasets closely resembling the evaluation conditions perform best. Shifts in sensor types, vineyard architecture, soil characteristics, or temporal factors dramatically influence performance, often overshadowing differences between modeling approaches. The can2023 dataset exemplifies these difficulties, showing that sensor selection alone cannot fully explain poor results. Ultimately, improving model performance and applicability requires not only refining algorithms and indices but also carefully considering the unique environmental and data properties of each vineyard setting.
This study presented an approach for testing a domain shift: predicting NDVI from RGB imagery. However, this approach also suffers from domain shift, as the can2023 dataset proved to be difficult to model by just the training dataset. For Deep Learning applications to take further hold, and address real-world issues, further domain shift research within UAV applications should also be addressed, starting points can be found in acquisition angle (image perspective), time of day (lighting conditions), seasonal variability, sensor variability and subject matter. UAV flights are often performed at a preset sensor angle. This preset image perspective has a large influence on the shape and size of objects, such as vines, vineyard rows, and root stock. Furthermore, the different angles of the sun or clouds can have a large influence on the color intensity of the objects in the image. Finally, generalization across subject matter could also be relevant, the presented evaluation was only performed for vineyards, however the a generalization test could also incorporate different field crops.
It is important for UAV studies using Deep Learning to take these generalization challenges seriously and create models that are usable outside of training and validation data. This is achieved through careful testing-set selection and experimental setup. It is up to future research to overcome these challenges using creative approaches, share their datasets, and properly evaluate such approaches.
Author Contributions
Conceptualization, J.D., Ö.B. and J.V.; methodology, J.D.; software, J.D.; formal analysis, J.D.; writing—original draft preparation, J.D.; writing—review and editing, Ö.B. and J.V.; supervision, Ö.B. and J.V.; project administration, Ö.B. and J.V.; funding acquisition, J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Horizon Europe program in the scope of the ICAERUS project (contract number 101060643).
Data Availability Statement
The original code, model weights and analysis presented in the study are openly available on GitHub at https://github.com/jurriandoornbos/RGBtoNDVIconversion, accessed on 15 December 2024. The employed datasets are available as mentioned in Table 1.
Acknowledgments
The authors acknowledge valuable help and contributions from all partners of the ICAERUS project.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| UAV | Uncrewed Aerial Vehicle |
| PA | Precision Agriculture |
| DL | Deep Learning |
| GAN | Generative Adversarial Network |
| NDVI | Normalized Difference Vegetation Index |
| RGBVI | Red Green Blue Vegetation Index |
| NIR | Near-Infrared |
| MS | Multispectral |
| SSIM | Structurual Similarity |
| RMSE | Root Mean Squared Error |
Appendix A. Visualizations of GAN Models for Artificial Generation of NDVI
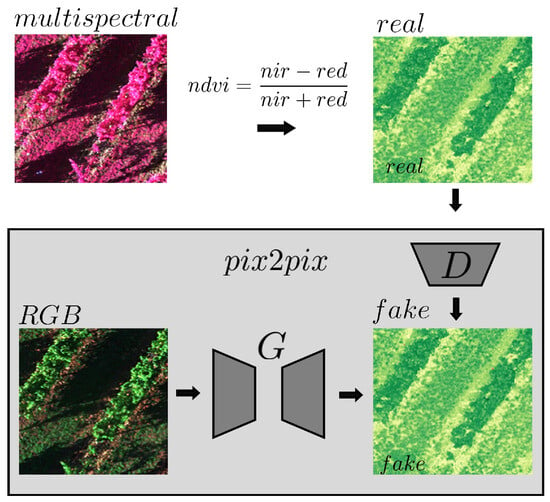
Figure A1.
Pix2Pix architecture abstraction for RGBtoNDVI conversion. The top left pink image is the multispectral source image, which is used to calculate the real NDVI, shown on the top right. The bottom left image shows the input RGB image, and the bottom right shows the generated artificial NDVI. They grey box contains the network architecture. The G and the D denote the Generator and Discriminator Networks in Pix2Pix.
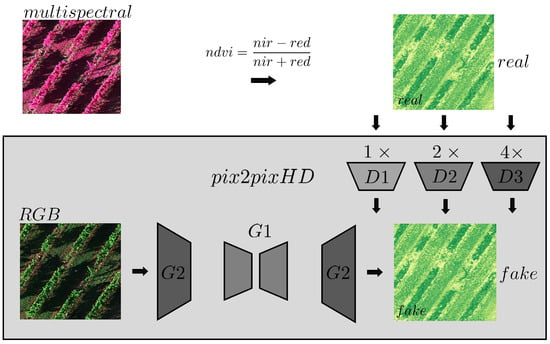
Figure A2.
Pix2PixHD architecture abstraction for RGBtoNDVI conversion. The top left pink image is the multispectral source image, which is used to calculate the real NDVI, shown on the top right. The bottom left image shows the input RGB image, and the bottom right shows the generated artificial NDVI. They grey box contains the network architecture. The Generator networks are shown as G1 for the original network and G2 for the upsampling network, and the three Discriminator Networks (D1, D2, D3), with their respective upsampling size of 1×, 2× and 4×.
Appendix B. NDVI Map Visualizations

Figure A3.
btg2021 NDVI map. VI color range is scaled (−1, 1). The NDVI and generated NDVI maps show a high similarity across both canopy and in-between rows. the RGBVI map has lower values for the bare soil.

Figure A4.
btg2022ms NDVI maps. VI color range is scaled (−1, 1). The NDVI map shows some in-between-row vegetation in the northern section. These are not visible in the Pix2Pix and Pix2PixHD maps. RGBVI has a lower estimation across the canopy and in-between-row. Pix2Pix has some hard boundaries between the edges of the reconstructed chips. vNDVI has lower values overall.

Figure A5.
btg2022rgb NDVI maps. VI color range is scaled (−1, 1). The NDVI map shows some in-between-row vegetation in the northern section. These are not visible in the Pix2Pix and Pix2PixHD maps. RGBVI has a lower estimation across the canopy and in-between rows. Pix2Pix has some hard boundaries between the edges of the reconstructed chips. vNDVI has lower values overall.
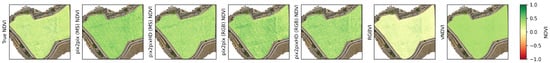
Figure A6.
can2023 NDVI map. VI color range is scaled (−1, 1). The NDVI map shows low values between the rows. In the middle section slightly higher intensity of the canopy is observed, which is low in the generated maps. RGBVI has lower values overall, but a highly similar structure across the vineyard compared to NDVI. Pix2Pix has some hard boundaries between the edges of the reconstructed chips.
Appendix C. BBR Difference Map Visualizations

Figure A7.
btg2021 Botrytis bunch rot risk (BBR) difference maps, compared to the true NDVI input.

Figure A8.
btg2022ms Botrytis bunch rot risk (BBR) difference maps, compared to the true NDVI input.

Figure A9.
btg2022rgb Botrytis bunch rot risk (BBR) difference maps, compared to the true NDVI input.
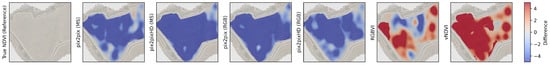
Figure A10.
can2023 Botrytis bunch rot risk (BBR) difference map compared to the true NDVI map.
Appendix D. Vigor Map Visualizations

Figure A11.
btg2021 Vigor maps using the different vegetation maps as input. Red indicates low vigor, yellow indicates medium vigor and green indicates high vigor.

Figure A12.
btg2022ms Vigor maps using the different vegetation maps as input. Red indicates low vigor, yellow indicates medium vigor and green indicates high vigor.

Figure A13.
btg2022rgb Vigor maps using the different vegetation maps as input. Red indicates low vigor, yellow indicates medium vigor and green indicates high vigor.
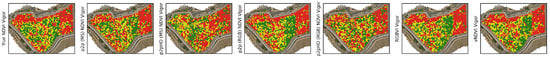
Figure A14.
can2023 Vigor maps using the different vegetation maps as input. Red indicates low vigor, yellow indicates medium vigor and green indicates high vigor. The true NDVI-based vigor map shows a medium to high vigor area in the middle. Wheres the top left and right side are convincingly low-vigor. The RGBVI-based vigor map most accurately models this pattern. Pix2Pix has a similar low vigor area in the top-left of the vineyard. Pix2PixHD models misses the top-left low vigor section.
Appendix E. Confusion Matrices of Vigor Estimation

Table A1.
btg2021 pix2pix (MS) vigor class confusion matrix, bold is the highest value.
Table A1.
btg2021 pix2pix (MS) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pix Low | 125 | 10 | 7 |
| pix2pix Medium | 15 | 88 | 18 |
| pix2pix High | 2 | 23 | 96 |

Table A2.
btg2021 pix2pixHD (MS) vigor class confusion matrix, bold is the highest value.
Table A2.
btg2021 pix2pixHD (MS) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pixHD Low | 120 | 15 | 7 |
| pix2pixHD Medium | 19 | 76 | 26 |
| pix2pixHD High | 3 | 30 | 88 |

Table A3.
btg2021 pix2pix (RGB) vigor class confusion matrix, bold is the highest value.
Table A3.
btg2021 pix2pix (RGB) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pix Low | 97 | 27 | 18 |
| pix2pix Medium | 30 | 50 | 41 |
| pix2pix High | 15 | 44 | 62 |

Table A4.
btg2021 pix2pixHD (RGB) vigor class confusion matrix, bold is the highest value.
Table A4.
btg2021 pix2pixHD (RGB) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pixHD Low | 99 | 34 | 9 |
| pix2pixHD Medium | 27 | 48 | 46 |
| pix2pixHD High | 16 | 39 | 66 |

Table A5.
btg2021 RGBVI vigor class confusion matrix, bold is the highest value.
Table A5.
btg2021 RGBVI vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| RGBVI Low | 127 | 9 | 6 |
| RGBVI Medium | 14 | 95 | 12 |
| RGBVI High | 0 | 18 | 103 |

Table A6.
btg2021 vNDVI vigor class confusion matrix, bold is the highest value.
Table A6.
btg2021 vNDVI vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| vNDVI Low | 122 | 14 | 6 |
| vNDVI Medium | 20 | 83 | 18 |
| vNDVI High | 0 | 24 | 97 |

Table A7.
btg2022ms pix2pix (MS) vigor class confusion matrix, bold is the highest value.
Table A7.
btg2022ms pix2pix (MS) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pix Low | 40 | 2 | 0 |
| pix2pix Medium | 2 | 24 | 10 |
| pix2pix High | 0 | 10 | 26 |

Table A8.
btg2022ms pix2pixHD (MS) vigor class confusion matrix, bold is the highest value.
Table A8.
btg2022ms pix2pixHD (MS) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pixHD Low | 39 | 3 | 0 |
| pix2pixHD Medium | 3 | 25 | 8 |
| pix2pixHD High | 0 | 8 | 28 |

Table A9.
btg2022ms pix2pix (RGB) vigor class confusion matrix, bold is the highest value.
Table A9.
btg2022ms pix2pix (RGB) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pix Low | 32 | 5 | 5 |
| pix2pix Medium | 8 | 15 | 13 |
| pix2pix High | 2 | 16 | 18 |

Table A10.
btg2022ms pix2pixHD (RGB) vigor class confusion matrix, bold is the highest value.
Table A10.
btg2022ms pix2pixHD (RGB) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pixHD Low | 32 | 7 | 3 |
| pix2pixHD Medium | 8 | 16 | 12 |
| pix2pixHD High | 2 | 13 | 21 |

Table A11.
btg2021ms RGBVI vigor class confusion matrix, bold is the highest value.
Table A11.
btg2021ms RGBVI vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| RGBVI Low | 41 | 1 | 0 |
| RGBVI Medium | 0 | 31 | 5 |
| RGBVI High | 0 | 5 | 31 |

Table A12.
btg2022ms vNDVI vigor class confusion matrix, bold is the highest value.
Table A12.
btg2022ms vNDVI vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| vNDVI Low | 41 | 1 | 0 |
| vNDVI Medium | 1 | 28 | 7 |
| vNDVI High | 0 | 7 | 29 |

Table A13.
can2023 pix2pix (MS) vigor class confusion matrix, bold is the highest value.
Table A13.
can2023 pix2pix (MS) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pix Low | 551 | 135 | 31 |
| pix2pix Medium | 141 | 228 | 119 |
| pix2pix High | 25 | 125 | 338 |

Table A14.
can2023 pix2pixHD (MS) vigor class confusion matrix, bold is the highest value.
Table A14.
can2023 pix2pixHD (MS) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pixHD Low | 439 | 148 | 130 |
| pix2pixHD Medium | 171 | 169 | 148 |
| pix2pixHD High | 107 | 171 | 210 |

Table A15.
can2023 pix2pix (RGB) vigor class confusion matrix, bold is the highest value.
Table A15.
can2023 pix2pix (RGB) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pix Low | 569 | 119 | 29 |
| pix2pix Medium | 120 | 223 | 145 |
| pix2pix High | 28 | 146 | 314 |

Table A16.
can2023 pix2pixHD (RGB) vigor class confusion matrix, bold is the highest value.
Table A16.
can2023 pix2pixHD (RGB) vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| pix2pixHD Low | 510 | 128 | 79 |
| pix2pixHD Medium | 150 | 203 | 135 |
| pix2pixHD High | 57 | 157 | 274 |

Table A17.
can2023 RGBVI vigor class confusion matrix, bold is the highest value.
Table A17.
can2023 RGBVI vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| RGBVI Low | 572 | 109 | 36 |
| RGBVI Medium | 127 | 228 | 133 |
| RGBVI High | 18 | 151 | 319 |

Table A18.
can2023 vNDVI vigor class confusion matrix, bold is the highest value.
Table A18.
can2023 vNDVI vigor class confusion matrix, bold is the highest value.
| True Low | True Medium | True High | |
|---|---|---|---|
| vNDVI Low | 570 | 113 | 34 |
| vNDVI Medium | 122 | 230 | 136 |
| vNDVI High | 25 | 144 | 319 |
References
- Fraser, E.D.; Campbell, M. Agriculture 5.0: Reconciling Production with Planetary Health. One Earth 2019, 1, 278–280. [Google Scholar] [CrossRef]
- Tsouros, D.; Bibi, S.; Sarigiannidis, P. A Review on UAV-based Applications for Precision Agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef]
- Doornbos, J.; Bennin, K.E.; Babur, Ç.Ö.; ao Valente, J. Drone Technologies: A Tertiary Systematic Literature Review on a Decade of Improvements. IEEE Access 2024, 12, 23220–23240. [Google Scholar] [CrossRef]
- Matese, A.; Baraldi, R.; Berton, A.; Cesaraccio, C.; Gennaro, S.F.D.; Duce, P.; Facini, O.; Mameli, M.G.; Piga, A.; Zaldei, A. Estimation of Water Stress in Grapevines Using Proximal and Remote Sensing Methods. Remote Sens. 2018, 10, 114. [Google Scholar] [CrossRef]
- Caruso, G.; Tozzini, L.; Rallo, G.; Primicerio, J.; Moriondo, M.; Palai, G.; Gucci, R. Estimating Biophysical and Geometrical Parameters of Grapevine Canopies (‘Sangiovese’) by an Unmanned Aerial Vehicle (UAV) and VIS-NIR Cameras. Vitis-J. Grapevine Res. 2017, 56, 63–70. [Google Scholar] [CrossRef]
- Primicerio, J.; Caruso, G.; Comba, L.; Crisci, A.; Gay, P.; Guidoni, S.; Genesio, L.; Aimonino, D.R.; Vaccari, F.P. Individual Plant Definition and Missing Plant Characterization in Vineyards from High-Resolution UAV Imagery. Eur. J. Remote Sens. 2017, 50, 179–186. [Google Scholar] [CrossRef]
- Vélez, S.; Ariza-Sentís, M.; Valente, J. Mapping the Spatial Variability of Botrytis Bunch Rot Risk in Vineyards Using UAV Multispectral Imagery. Eur. J. Agron. 2023, 142, 126691. [Google Scholar] [CrossRef]
- Kerkech, M.; Hafiane, A.; Canals, R. Vine Disease Detection in UAV Multispectral Images Using Optimized Image Registration and Deep Learning Segmentation Approach. Comput. Electron. Agric. 2020, 174, 105446. [Google Scholar] [CrossRef]
- Pádua, L.; Marques, P.; Adão, T.; Guimarães, N.; Sousa, A.; Peres, E.; Sousa, J.J. Vineyard Variability Analysis through UAV-Based Vigour Maps to Assess Climate Change Impacts. Agronomy 2019, 9, 581. [Google Scholar] [CrossRef]
- Deng, L.; Mao, Z.; Li, X.; Hu, Z.; Duan, F.; Yan, Y. UAV-Based Multispectral Remote Sensing for Precision Agriculture: A Comparison between Different Cameras. ISPRS J. Photogramm. Remote Sens. 2018, 146, 124–136. [Google Scholar] [CrossRef]
- Rouse, J.W.J.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS; NASA Goddard Space Flight Center, Third ERTS-1 Symposium: Washington, DC, USA, 1974; Volume 1, pp. 309–317. [Google Scholar]
- Caruso, G.; Palai, G.; Tozzini, L.; D’Onofrio, C.; Gucci, R. The Role of LAI and Leaf Chlorophyll on NDVI Estimated by UAV in Grapevine Canopies. Sci. Hortic. 2023, 322, 112398. [Google Scholar] [CrossRef]
- Matese, A.; Gennaro, S.F.D. Beyond the Traditional NDVI Index as a Key Factor to Mainstream the Use of UAV in Precision Viticulture. Sci. Rep. 2021, 11, 2721. [Google Scholar] [CrossRef] [PubMed]
- Kasimati, A.; Psiroukis, V.; Darra, N.; Kalogrias, A.; Kalivas, D.; Taylor, J.; Fountas, S. Investigation of the Similarities between NDVI Maps from Different Proximal and Remote Sensing Platforms in Explaining Vineyard Variability. Precis. Agric. 2023, 24, 1220–1240. [Google Scholar] [CrossRef]
- Aasen, H.; Honkavaara, E.; Lucieer, A.; Zarco-Tejada, P.J. Quantitative Remote Sensing at Ultra-High Resolution with UAV Spectroscopy: A Review of Sensor Technology, Measurement Procedures, and Data Correction Workflows. Remote Sens. 2018, 10, 1091. [Google Scholar] [CrossRef]
- Ashapure, A.; Jung, J.; Chang, A.; Oh, S.; Maeda, M.; Landivar, J. A Comparative Study of RGB and Multispectral Sensor-Based Cotton Canopy Cover Modelling Using Multi-Temporal UAS Data. Remote Sens. 2019, 11, 2757. [Google Scholar] [CrossRef]
- Agapiou, A. Vegetation Extraction Using Visible-Bands from Openly Licensed Unmanned Aerial Vehicle Imagery. Drones 2020, 4, 27. [Google Scholar] [CrossRef]
- Costa, L.; Nunes, L.; Ampatzidis, Y. A new visible band index (vNDVI) for estimating NDVI values on RGB images utilizing genetic algorithms. Comput. Electron. Agric. 2020, 172, 105334. [Google Scholar] [CrossRef]
- Suárez, P.L.; Sappa, A.D.; Vintimilla, B.X. Cycle Generative Adversarial Network: Towards a Low Cost Vegetation Index Estimation. In Proceedings of the International Conference on Image Processing, Anchorage, AK, USA, 19–22 September 2021; pp. 2783–2787. [Google Scholar] [CrossRef]
- Farooque, A.A.; Afzaal, H.; Benlamri, R.; Al-Naemi, S.; MacDonald, E.; Abbas, F.; MacLeod, K.; Ali, H. Red-Green-Blue to Normalized Difference Vegetation Index Translation: A Robust and Inexpensive Approach for Vegetation Monitoring Using Machine Vision and Generative Adversarial Networks. Precis. Agric. 2023, 24, 1097–1115. [Google Scholar] [CrossRef]
- Boroujeni, S.P.H.; Razi, A. IC-GAN: An Improved Conditional Generative Adversarial Network for RGB-to-IR Image Translation with Applications to Forest Fire Monitoring. Expert Syst. Appl. 2024, 238, 121962. [Google Scholar] [CrossRef]
- Davidson, C.; Jaganathan, V.; Sivakumar, A.N.; Czarnecki, J.M.P.; Chowdhary, G. NDVI/NDRE Prediction from Standard RGB Aerial Imagery Using Deep Learning. Comput. Electron. Agric. 2022, 203, 107396. [Google Scholar] [CrossRef]
- Safonova, A.; Ghazaryan, G.; Stiller, S.; Main-Knorn, M.; Nendel, C.; Ryo, M. Ten Deep Learning Techniques to Address Small Data Problems with Remote Sensing. Int. J. Appl. Earth Obs. Geoinf. 2023, 125, 103569. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Chen, W. UAV Aerial Image Generation of Crucial Components of High-Voltage Transmission Lines Based on Multi-Level Generative Adversarial Network. Remote Sens. 2023, 15, 1412. [Google Scholar] [CrossRef]
- Mahara, A.; Rishe, N. Multispectral Band-Aware Generation of Satellite Images across Domains Using Generative Adversarial Networks and Contrastive Learning. Remote Sens. 2024, 16, 1154. [Google Scholar] [CrossRef]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning Transferable Visual Models From Natural Language Supervision. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; ML Research Press: Cambridge, MA, USA, 2021; Volume 139, pp. 8748–8763. [Google Scholar]
- Ahmedt-Aristizabal, D.; Smith, D.; Khokher, M.R.; Li, X.; Smith, A.L.; Petersson, L.; Rolland, V.; Edwards, E.J. An In-Field Dynamic Vision-Based Analysis for Vineyard Yield Estimation. IEEE Access 2024, 12, 102146–102166. [Google Scholar] [CrossRef]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-Resolution Image Synthesis and Semantic Manipulation with Conditional GANs. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8798–8807. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar] [CrossRef]
- Jozdani, S.; Chen, D.; Pouliot, D.; Johnson, B.A. A Review and Meta-analysis of Generative Adversarial Networks and Their Applications in Remote Sensing. Int. J. Appl. Earth Obs. Geoinf. 2022, 108, 102734. [Google Scholar] [CrossRef]
- Tuia, D.; Kellenberger, B.; Beery, S.; Costelloe, B.R.; Zuffi, S.; Risse, B.; Mathis, A.; Mathis, M.W.; van Langevelde, F.; Burghardt, T.; et al. Perspectives in Machine Learning for Wildlife Conservation. Nat. Commun. 2022, 13, 792. [Google Scholar] [CrossRef]
- Diez, Y.; Kentsch, S.; Fukuda, M.; Caceres, M.; Moritake, K.; Cabezas, M. Deep Learning in Forestry Using UAV-acquired RGB Data: A Practical Review. Remote Sens. 2021, 13, 2837. [Google Scholar] [CrossRef]
- Ariza-Sentís, M.; Vélez, S.; Valente, J. BBR: An Open-Source Standard Workflow Based on Biophysical Crop Parameters for Automatic Botrytis Cinerea Assessment in Vineyards. SoftwareX 2023, 24, 101542. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Pádua, L.; Marques, P.; Hruška, J.; Adão, T.; Bessa, J.; Sousa, A.; Peres, E.; Morais, R.; Sousa, J.J. Vineyard properties extraction combining UAS-based RGB imagery with elevation data. Int. J. Remote. Sens. 2018, 39, 5377–5401. [Google Scholar] [CrossRef]
- Vera, E.; Arroyo, O.; Sollazzo, A.; Rangholia, C.; Osés, P. UAV Canyelles Vineyard Dataset 2023-06-09. 2023. Available online: https://zenodo.org/records/10171243 (accessed on 15 December 2024). [CrossRef]
- Agisoft LLC. Agisoft Metashape Professional (Version 1.7.1). Available online: https://www.agisoft.com/ (accessed on 29 January 2025).
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation. 2009. Available online: http://qgis.osgeo.org (accessed on 29 January 2025).
- Vélez, S.; Ariza-Sentís, M.; Valente, J. UAV Multispectral Imagery Dataset over a Vineyard Affected by Botrytis in ‘Tomiño’, Pontevedra, Spain. It Includes GPS Location of Vine Trunks, Diseases and GCP Points. 2022. Available online: https://zenodo.org/records/7383601 (accessed on 15 December 2024). [CrossRef]
- Ho, J.; Jain, A.; Abbeel, P. Denoising Diffusion Probabilistic Models. arXiv 2020, arXiv:2006.11239. [Google Scholar]
- Su, X.; Song, J.; Meng, C.; Ermon, S. Dual Diffusion Implicit Bridges for Image-to-Image Translation. arXiv 2022, arXiv:2203.08382. [Google Scholar]
- Liu, G.H.; Vahdat, A.; Song, J.; Huang, D.A.; Theodorou, E.A.; Nie, W.; Anandkumar, A. I2SB: Image-to-Image Schrödinger Bridge. arXiv 2023, arXiv:2302.05872. [Google Scholar]
- Müller-Franzes, G.; Niehues, J.M.; Khader, F.; Arasteh, S.T.; Haarburger, C.; Kuhl, C.; Wang, T.; Han, T.; Nebelung, S.; Kather, J.N.; et al. Diffusion Probabilistic Models Beat GANs on Medical Images. Sci. Rep. 2022, 13, 12098. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Song, J.; Liu, G.; Kautz, J.; Vahdat, A. DiffiT: Diffusion Vision Transformers for Image Generation. arXiv 2023, arXiv:2312.02139. [Google Scholar]
- Wei, J.; Tay, Y.; Bommasani, R.; Raffel, C.; Zoph, B.; Borgeaud, S.; Yogatama, D.; Bosma, M.; Zhou, D.; Metzler, D.; et al. Emergent Abilities of Large Language Models. arXiv 2022, arXiv:2206.07682. [Google Scholar]
- Schuhmann, C.; Beaumont, R.; Vencu, R.; Gordon, C.; Wightman, R.; Cherti, M.; Coombes, T.; Katta, A.; Mullis, C.; Wortsman, M.; et al. LAION-5B: An Open Large-scale Dataset for Training Next Generation Image-text Models. In Proceedings of the 36th Conference on Neural Information Processing Systems (NeurIPS 2022), New Orleans, LA, USA, 28 November–9 December 2022; Volume 35, pp. 25278–25294. [Google Scholar]
- Feyisa, G.L.; Meilby, H.; Fensholt, R.; Proud, S.R. Automated Water Extraction Index: A New Technique for Surface Water Mapping Using Landsat Imagery. Remote Sens. Environ. 2014, 140, 23–35. [Google Scholar] [CrossRef]
- Babaeian, E.; Sadeghi, M.; Jones, S.B.; Montzka, C.; Vereecken, H.; Tuller, M. Ground, Proximal, and Satellite Remote Sensing of Soil Moisture. Rev. Geophys. 2019, 57, 530–616. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).