Hyperspectral Data Can Differentiate Species and Cultivars of C3 and C4 Turf Despite Measurable Diurnal Variation
Abstract
1. Introduction
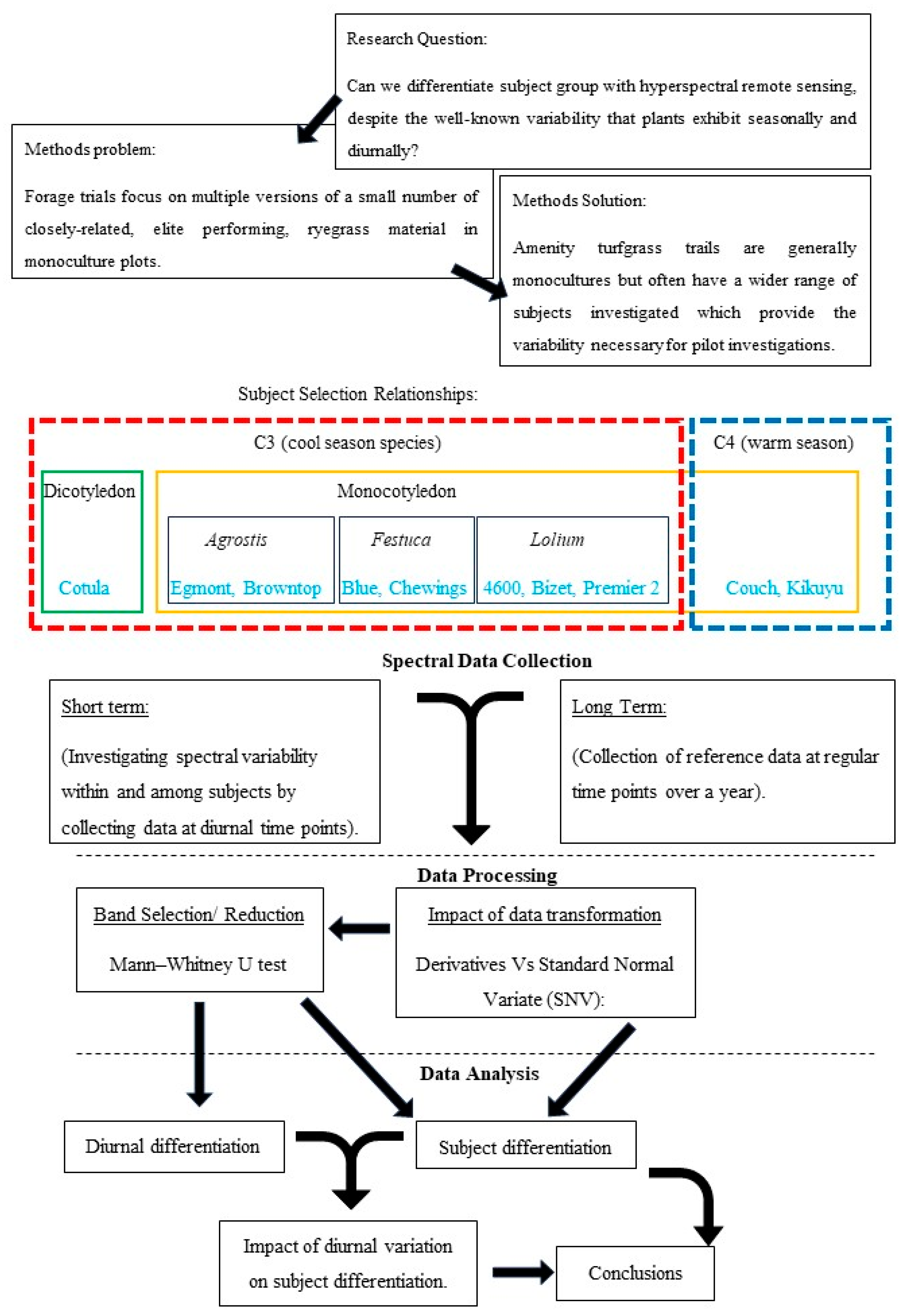
2. Materials and Methods
2.1. Site Description
2.2. Reflectance Data Collection
2.3. Collection of Data for Diurnal Analysis
2.4. Continued Data Collection for Subject Differentiation
2.5. Data Handling
2.6. Statistics and Analysis
2.6.1. Pre-Processing
2.6.2. Band Ranking and Band Reduction, U-Test
2.6.3. Subject Differentiation with Linear Discriminate Analysis (LDA)
2.6.4. Stepwise LDA
3. Results
3.1. Datasets
- Diurnal dataset: This dataset (n = 2000) was examined for fluctuations in diurnal reflectance for each subject. Forty spectral samples per subject, at each of the five time periods, were collected over two days. Each spectral sample represents the average of ten automatically collected and averaged readings.
- Differentiation dataset: The total dataset (n = 2800) collated data from all subjects over the 12 months, including the data used for diurnal analysis, to investigate subject separability and explore dimensionality reduction through band selection.
3.2. Subject Differentiation with LDA
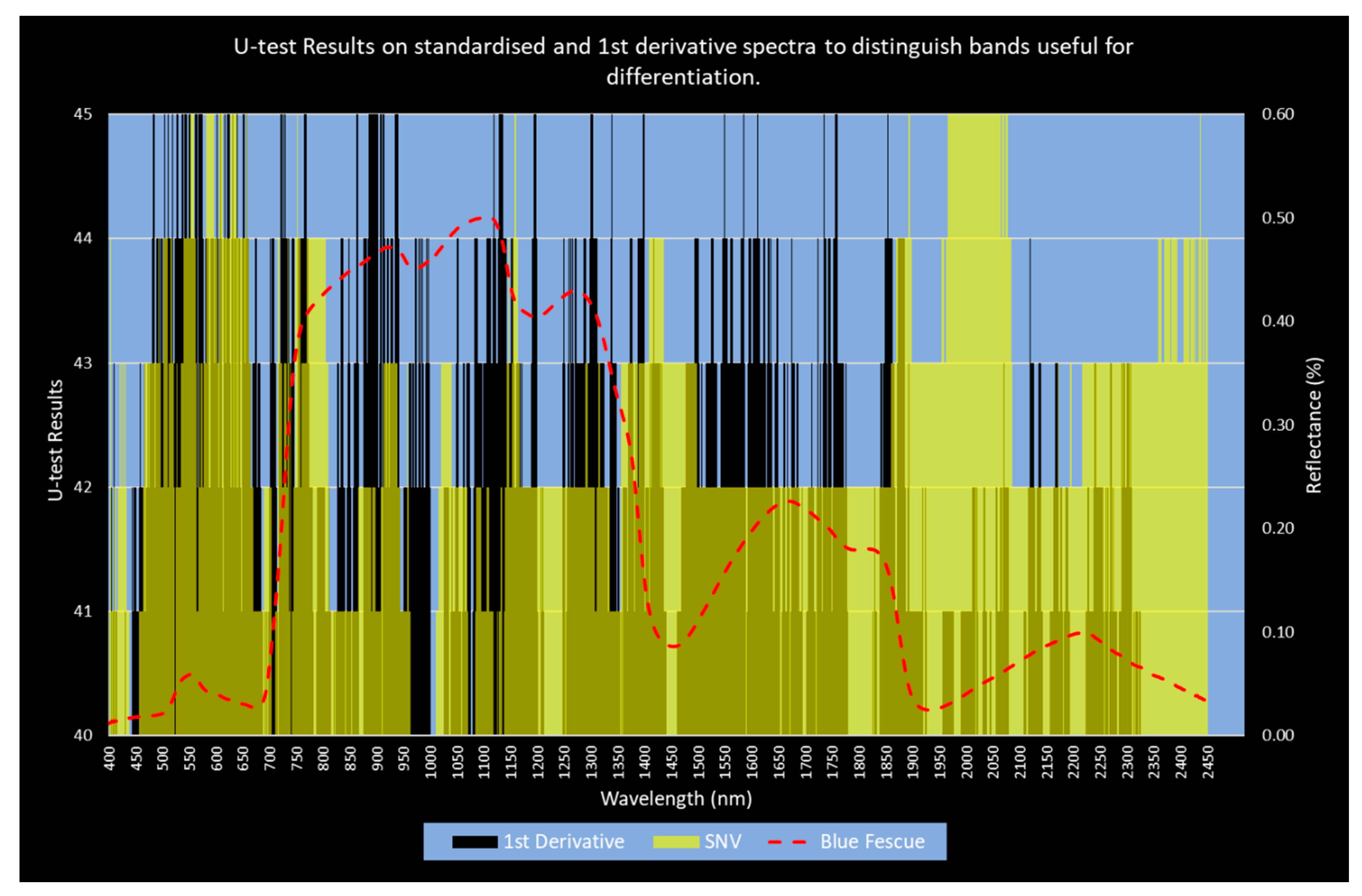
3.3. Band Reduction
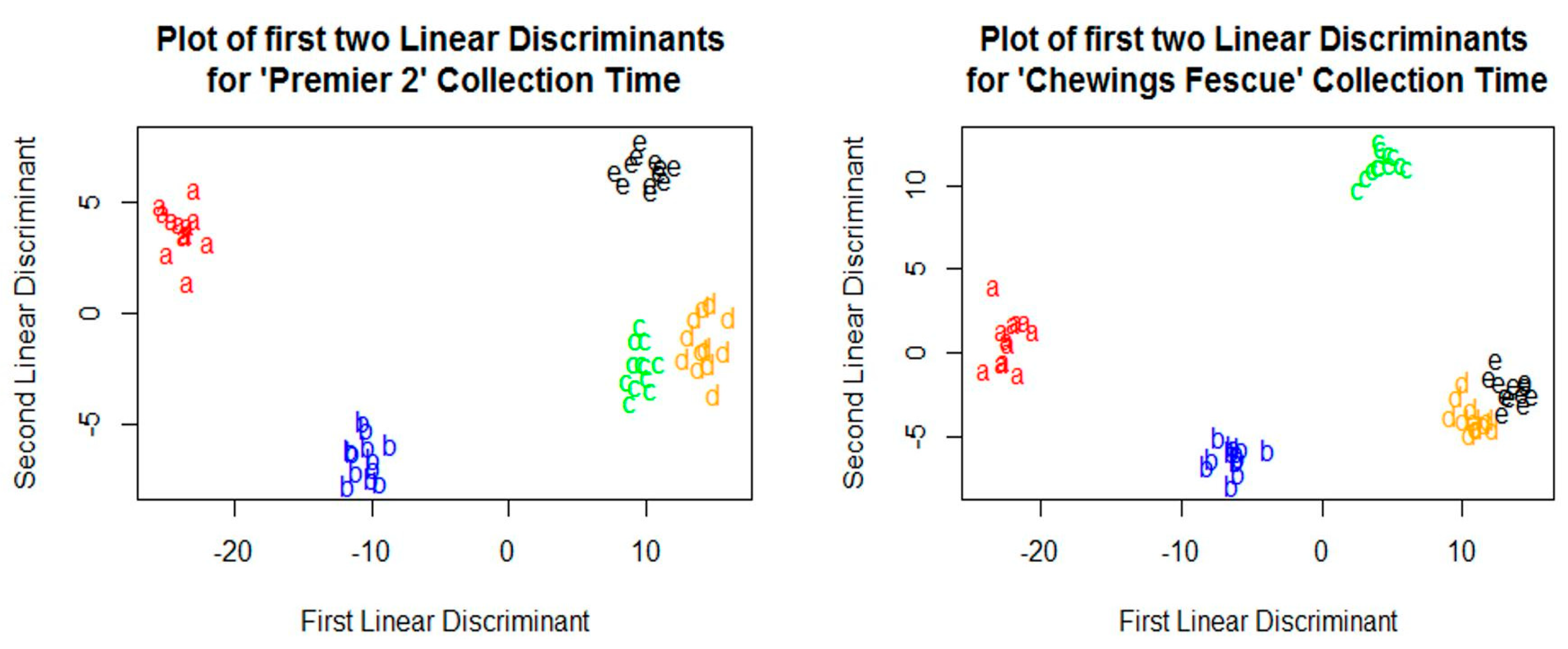
3.4. Prediction of Sample Collection Time
3.5. Analysis of Collection Time Predictions for Colated Samples
4. Discussions
4.1. Subject Differentiation
4.2. Band Selection
4.3. Band Reduction and Selection
4.4. Diurnal Identification
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dodd, M.B.; Chapman, D.F.; Ludemann, C.I.; Griffiths, W.; Tozer, K.N.; Donnelly, L. The measurement of perennial ryegrass persistence. J. N. Z. Grassl. 2018, 80, 161–168. [Google Scholar] [CrossRef]
- Morrison, L.W. Observer error in vegetation surveys: A review. J. Plant Ecol. 2015, 9, 367–379. [Google Scholar] [CrossRef]
- Schaaf, C.B.; Strahler, A.H. Solar zenith angle effects on forest canopy hemispherical reflectances calculated with a geometric-optical bidirectional reflectance model. IEEE Trans. Geosci. Remote Sens. 1993, 31, 921–927. [Google Scholar] [CrossRef]
- Middleton, E.M. Solar zenith angle effects on vegetation indices in tallgrass prairie. Remote Sens. Environ. 1991, 38, 45–62. [Google Scholar] [CrossRef]
- Huete, A.R. Soil and sun angle interactions on partial canopy spectra. Int. J. Remote Sens. 1987, 8, 1307–1317. [Google Scholar] [CrossRef]
- Ma, X.; Huete, A.; Tran, N.N.; Bi, J.; Gao, S.; Zeng, Y. Sun-Angle Effects on Remote-Sensing Phenology Observed and Modelled Using Himawari-8. Remote Sens. 2020, 12, 1339. [Google Scholar] [CrossRef]
- Jafarbiglu, H.; Pourreza, A. Impact of sun-view geometry on canopy spectral reflectance variability. ISPRS J. Photogramm. Remote Sens. 2023, 196, 270–286. [Google Scholar] [CrossRef]
- Pinter, P.J.; Jackson, R.D.; Idso, S.B.; Reginato, R.J. Diurnal patterns of wheat spectral reflectances. IEEE Trans. Geosci. Remote Sens. 1983, 2, 156–163. [Google Scholar] [CrossRef]
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef]
- Horton, P.; Ruban, A.V.; Walters, R.G. Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 655–684. [Google Scholar] [CrossRef]
- Grime, J.P.; Thompson, K.; Hunt, R.; Hodgson, J.G.; Cornelissen, J.H.C.; Rorison, I.H.; Hendry, G.A.F.; Ashenden, T.W.; Askew, A.P.; Band, S.R.; et al. Integrated screening validates primary axes of specialisation in plants. Oikos 1997, 79, 259–281. [Google Scholar] [CrossRef]
- Ollinger, S. Sources of variability in canopy reflectance and the convergent properties of plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Girolamo-Neto, C.D.; Sanches, I.D.A.; Neves, A.K.; Prudente, V.H.R.; Körting, T.S.; Picoli, M.C.A.; Aragão, L.E.O.e.C.d. Assessment of texture features for bermudagrass (cynodon dactylon) detection in sugarcane plantations. Drones 2019, 3, 36. [Google Scholar] [CrossRef]
- Wang, C.; Hunt, E.R.; Zhang, L.; Guo, H. Phenology-assisted classification of C3 and C4 grasses in the U.S. Great Plains and their climate dependency with MODIS time series. Remote Sens. Environ. 2013, 138, 90–101. [Google Scholar] [CrossRef]
- Lilienthal, H.; Wilde, P.; Schnug, E. Proximal hyperspectral sensing in plant breeding. In Proceedings of the 13th International Conference on Precision Agriculture, St. Louis, MI, USA, 31 July–3 August 2016. [Google Scholar]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Walter, A.; Studer, B.; Kölliker, R. Advanced phenotyping offers opportunities for improved breeding of forage and turf species. Ann. Bot. 2012, 110, 1271–1279. [Google Scholar] [CrossRef]
- Ghamkhar, K. Phenomics for the Improvement of Crop Adaptation. In Plant Genetic Resources for the 21st Century, 1st ed.; Apple Academic Press: Palm Bay, FL, USA, 2023; pp. 115–129. ISBN 9781003302957. [Google Scholar]
- Caturegli, L.; Lulli, F.; Foschi, L.; Guglielminetti, L.; Bonari, E.; Volterrani, M. Turfgrass spectral reflectance: Simulating satellite monitoring of spectral signatures of main C3 and C4 species. Precis. Agric 2015, 16, 297–310. [Google Scholar] [CrossRef]
- Irisarri, J.; Oesterheld, M.; Verón, S.; Paruelo, J. Grass species differentiation through canopy hyperspectral reflectance. Int. J. Remote Sens. 2009, 30, 5959–5975. [Google Scholar] [CrossRef]
- Caturegli, L.; Corniglia, M.; Gaetani, M.; Grossi, N.; Magni, S.; Migliazzi, M.; Angelini, L.; Mazzoncini, M.; Silvestri, N.; Fontanelli, M. Unmanned aerial vehicle to estimate nitrogen status of turfgrasses. PLoS ONE 2016, 11, e0158268. [Google Scholar] [CrossRef]
- Caturegli, L.; Gaetani, M.; Volterrani, M.; Magni, S.; Minelli, A.; Baldi, A.; Brandani, G.; Mancini, M.; Lenzi, A.; Orlandini, S. Normalized Difference Vegetation Index versus Dark Green Colour Index to estimate nitrogen status on bermudagrass hybrid and tall fescue. Int. J. Remote Sens. 2020, 41, 455–470. [Google Scholar] [CrossRef]
- Caturegli, L.; Matteoli, S.; Gaetani, M.; Grossi, N.; Magni, S.; Minelli, A.; Corsini, G.; Remorini, D.; Volterrani, M. Effects of water stress on spectral reflectance of bermudagrass. Sci. Rep. 2020, 10, 15055. [Google Scholar] [CrossRef]
- Marín, J.; Yousfi, S.; Mauri, P.V.; Parra, L.; Lloret, J.; Masaguer, A. RGB vegetation indices, NDVI, and biomass as indicators to evaluate C3 and C4 turfgrass under different water conditions. Sustainability 2020, 12, 2160. [Google Scholar] [CrossRef]
- Fotia, K.; Ntoulas, N.; Koliopanos, C.; Tsirogiannis, I.; Nektarios, P. Utilization of reflectance indices to evaluate the impact of grey or recycled irrigation water on Festuca arundinacea turfgrass. In Proceedings of the International Symposium on Sensing Plant Water Status-Methods and Applications in Horticultural Science 1197, Potsdam, Germany, 5−7 October 2016; pp. 103–108. [Google Scholar]
- Schmidt, K.S.; Skidmore, A.K. Spectral discrimination of vegetation types in a coastal wetland. Remote Sens. Environ. 2003, 85, 92–108. [Google Scholar] [CrossRef]
- Möckel, T.; Dalmayne, J.; Schmid, B.; Prentice, H.; Hall, K. Airborne Hyperspectral Data Predict Fine-Scale Plant Species Diversity in Grazed Dry Grasslands. Remote Sens. 2016, 8, 133. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K.; Anderson, J.E.; Hoch, G.A.; Smith, M.D. Indicators of plant species richness in AVIRIS spectra of a mesic grassland. Remote Sens. Environ. 2005, 98, 304–316. [Google Scholar] [CrossRef]
- Davidson, A.; Csillag, F. A comparison of three approaches for predicting C4 species cover of northern mixed grass prairie. Remote Sens. Environ. 2003, 86, 70–82. [Google Scholar] [CrossRef]
- Liu, L.; Cheng, Z. Mapping C3 and C4 plant functional types using separated solar-induced chlorophyll fluorescence from hyperspectral data. Int. J. Remote Sens. 2011, 32, 9171–9183. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and de-velopmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Hull, R.J. Energy relations and carbohydrate partitioning in turfgrasses. Turfgrass 1992, 32, 175–205. [Google Scholar]
- Sanches, I.D.A. Hyperspectral Proximal Sensing of the Botanical Composition and Nutrient Content of New Zealand Pastures: A Thesis Presented in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy in Earth Science. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2009. [Google Scholar]
- Sanches, I.; Tuohy, M.; Hedley, M.; Bretherton, M. Large, durable and low-cost reflectance standard for field remote sensing applications. Int. J. Remote Sens. 2009, 30, 2309–2319. [Google Scholar] [CrossRef]
- Pu, R. Broadleaf species recognition with in situ hyperspectral data. Int. J. Remote Sens. 2009, 30, 2759–2779. [Google Scholar] [CrossRef]
- Lu, Y.; Saeys, W.; Kim, M.; Peng, Y.; Lu, R. Hyperspectral imaging technology for quality and safety evaluation of horticultural products: A review and celebration of the past 20-year progress. Postharvest Biol. Technol. 2020, 170, 111318. [Google Scholar] [CrossRef]
- Barnes, R.; Dhanoa, M.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Penuelas, J.; Gamon, J.A.; Fredeen, A.L.; Merino, J.; Field, C.B. Reflectance Indexes Associated with Physiological-Changes in Nitrogen-Limited and Water-Limited Sunflower Leaves. Remote Sens. Environ. 1994, 48, 135–146. [Google Scholar] [CrossRef]
- Rubo, S.; Zinkernagel, J. Exploring hyperspectral reflectance indices for the estimation of water and nitrogen status of spinach. Biosyst. Eng. 2022, 214, 58–71. [Google Scholar] [CrossRef]
- Tsai, F.; Philpot, W. Derivative Analysis of Hyperspectral Data. Remote Sens. Environ. 1998, 66, 41–51. [Google Scholar] [CrossRef]
- Stevens, A.; Ramirez–Lopez, L. An Introduction to the Prospectr Package. R Package Vignette. 2014, Version 0.1.3. Available online: https://cran.r-project.org/web/packages/prospectr/index.html (accessed on 19 August 2024).
- Welling, M. Fisher Linear Discriminant Analysis; Department of Computer Science, University of Toronto: Toronto, STC, Canada, 2006; p. 3. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer series in statistics; Springer: New York, NY, USA, 2009. [Google Scholar]
- Bhardwaj, A.; Verma, P. A Textbook on Pattern Recognition; Alpha Science International: Oxford, UK, 2015. [Google Scholar]
- Lu, J.; Plataniotis, K.N.; Venetsanopoulos, A.N. Face recognition using LDA-based algorithms. IEEE Trans. Neural Netw. 2003, 14, 195–200. [Google Scholar] [PubMed]
- Dudoit, S.; Fridlyand, J.; Speed, T.P. Comparison of discrimination methods for the classification of tumors using gene expression data. J. Am. Stat. Assoc. 2002, 97, 77–87. [Google Scholar] [CrossRef]
- Acquah, G.E.; Via, B.K.; Billor, N.; Fasina, O.O.; Eckhardt, L.G. Identifying plant part composition of forest logging residue using infrared spectral data and linear discriminant analysis. Sensors 2016, 16, 1375. [Google Scholar] [CrossRef]
- Wu, W.; Mallet, Y.; Walczak, B.; Penninckx, W.; Massart, D.; Heuerding, S.; Erni, F. Comparison of regularized discriminant analysis linear discriminant analysis and quadratic discriminant analysis applied to NIR data. Anal. Chim. Acta 1996, 329, 257–265. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef]
- Bandos, T.V.; Bruzzone, L.; Camps-Valls, G. Classification of hyperspectral images with regularized linear discriminant analysis. IEEE Trans. Geosci. Remote Sens. 2009, 47, 862–873. [Google Scholar] [CrossRef]
- Weihs, C.; Ligges, U.; Luebke, K.; Raabe, N. klaR Analyzing German Business Cycles. In Data Analysis and Decision Support; Baier, D., Decker, R., Schmidt-Thieme, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 335–343. [Google Scholar]
- Prospere, K.; McLaren, K.; Wilson, B. Plant Species Discrimination in a Tropical Wetland Using In Situ Hyperspectral Data. Remote Sens. 2014, 6, 8494–8523. [Google Scholar] [CrossRef]
- Vaiphasa, C.; Skidmore, A.K.; de Boer, W.F.; Vaiphasa, T. A hyperspectral band selector for plant species discrimination. ISPRS J. Photogramm. Remote Sens. 2007, 62, 225–235. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.K. A new technique for extracting the red edge position from hyperspectral data: The linear extrapolation method. Remote Sens. Environ. 2006, 101, 181–193. [Google Scholar] [CrossRef]
- Zhang, J.; Rivard, B.; Sánchez-Azofeifa, A.; Castro-Esau, K. Intra- and inter-class spectral variability of tropical tree species at La Selva, Costa Rica: Implications for species identification using HYDICE imagery. Remote Sens. Environ. 2006, 105, 129–141. [Google Scholar] [CrossRef]
- Huberty, C.J. Issues in the use and interpretation of discriminant analysis. Psychol. Bull. 1984, 95, 156–171. [Google Scholar] [CrossRef]
- Asner, G.P. Biophysical and Biochemical Sources of Variability in Canopy Reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Brown, D.J.; Shepherd, K.D.; Walsh, M.G.; Mays, M.D.; Reinsch, T.G. Global soil characterization with VNIR diffuse reflectance spectroscopy. Geoderma 2006, 132, 273–290. [Google Scholar] [CrossRef]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. Inf. Theory IEEE Trans. 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Cho, M.A.; Debba, P.; Mathieu, R.; Naidoo, L.; van Aardt, J.; Asner, G.P. Improving discrimination of savanna tree species through a multiple-endmember spectral angle mapper approach: Canopy-level analysis. Geosci. Remote Sens. IEEE Trans. 2010, 48, 4133–4142. [Google Scholar] [CrossRef]
- Sobhan, I.; Vaiphasa, C.; Skidmore, A. Spectral regions for maximizing species discrimination. Species Discrimination Hyperspectral Perspective. 2007, pp. 35–37. Available online: https://www.google.com/search?q=Species+discrimination+from+a+hyperspectral+perspective&oq=Species+discrimination+from+a+hyperspectral+perspective&gs_lcrp=EgZjaHJvbWUyBggAEEUYOTIGCAEQRRg8MgYIAhBFGDzSAQczMjNqMGo0qAIAsAIB&sourceid=chrome&ie=UTF-8 (accessed on 19 August 2024).
- Demetriades-Shah, T.H.; Steven, M.D.; Clark, J.A. High resolution derivative spectra in remote sensing. Remote Sens. Environ. 1990, 33, 55–64. [Google Scholar] [CrossRef]
- Betteridge, K.; Haynes, D. Altering the growth pattern of kikuyu pastures with temperate grasses. In Proceedings of the Proceedings of the New Zealand Grassland Association, Whangarei, New Zealand, 1 January 1986; pp. 149–156. [Google Scholar]
- Axelsson, C.; Skidmore, A.K.; Schlerf, M.; Fauzi, A.; Verhoef, W. Hyperspectral analysis of mangrove foliar chemistry using PLSR and support vector regression. Int. J. Remote Sens. 2013, 34, 1724–1743. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Susič, N.; Žibrat, U.; Širca, S.; Strajnar, P.; Razinger, J.; Knapič, M.; Vončina, A.; Urek, G.; Gerič Stare, B. Discrimination between abiotic and biotic drought stress in tomatoes using hyperspectral imaging. Sens. Actuators B Chem. 2018, 273, 842–852. [Google Scholar] [CrossRef]
- Price, J.C. How unique are spectral signatures? Remote Sens. Environ. 1994, 49, 181–186. [Google Scholar] [CrossRef]
- Sobhan, M.I. Species Discrimination from a Hyperspectral Perspective; Wageningen University and Research: Wageningen, The Netherlands, 2007. [Google Scholar]
- Cole, B.; McMorrow, J.; Evans, M. Spectral monitoring of moorland plant phenology to identify a temporal window for hyperspectral remote sensing of peatland. ISPRS J. Photogramm. Remote Sens. 2014, 90, 49–58. [Google Scholar] [CrossRef]

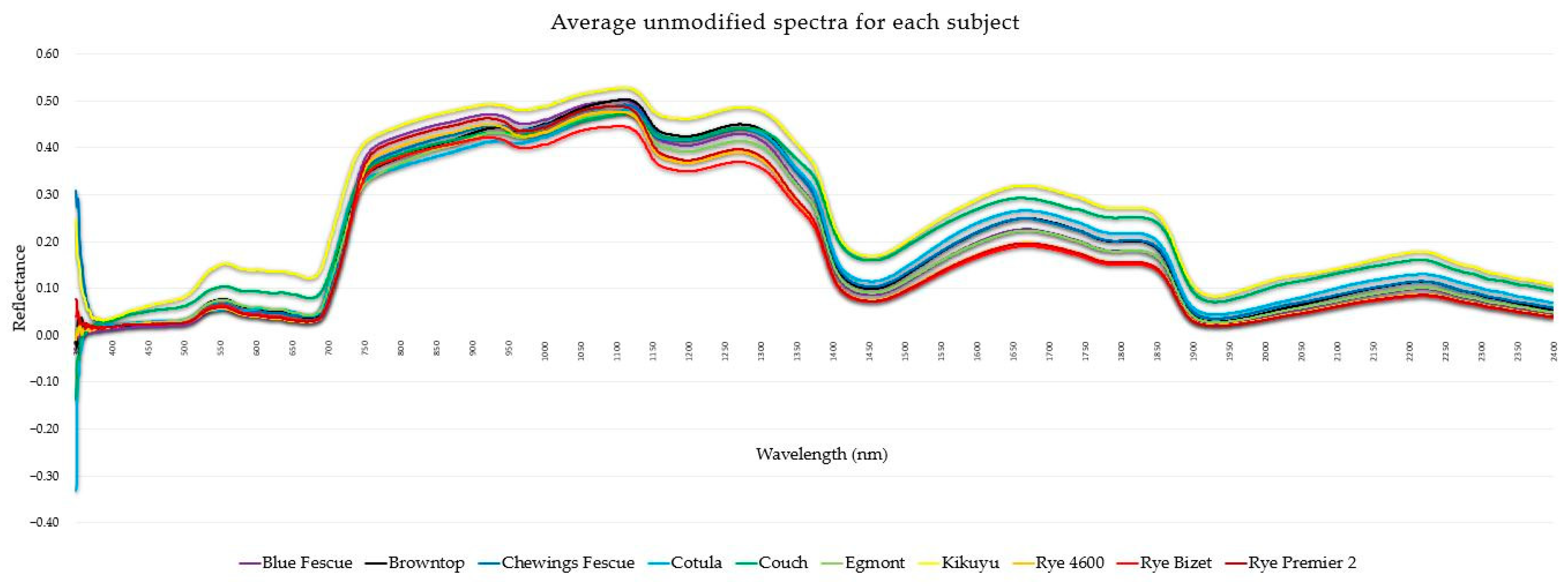
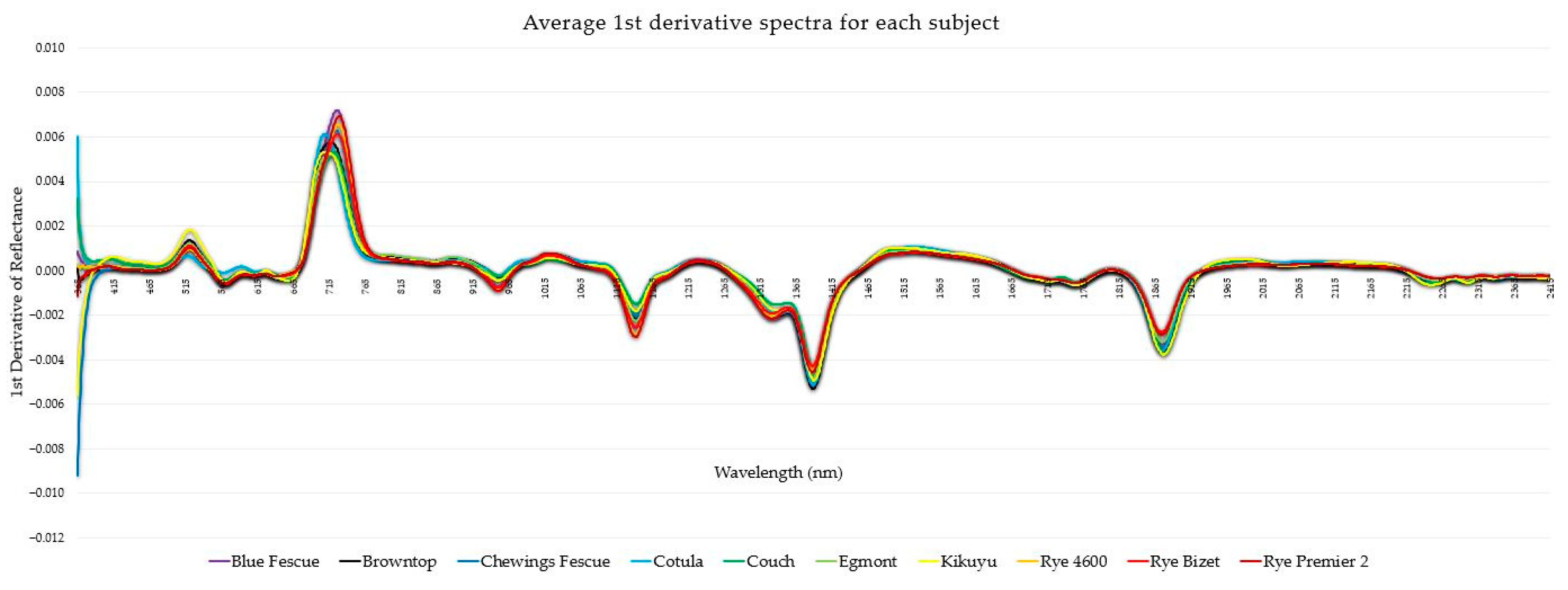

| Common Name | Latin Name |
|---|---|
| Cotula | Leptinella dioica cv. Pahia |
| Couch | Cynodon dactylon cv. Agridark |
| Kikuyu | Pennisetum clandestinum cv. Regal Stay Green |
| Egmont | Agrostis capillaris var. Egmont |
| Browntop | Agrostis capillaris cv. Arrowtown |
| Ryegrass ‘4600’ | Lolium perenne cv. 4600 |
| Ryegrass ‘Bizet’ | Lolium perenne cv. Bizet |
| Ryegrass ‘Premier 2’ | Lolium perenne cv. Premier 2 |
| Blue Fescue | Festuca sp. |
| Chewing’s Fescue | Festuca rubra subsp. Commutata |
| Reference Sample | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Blue Fescue | Browntop | Chewings Fescue | Cotula | Couch | Egmont | Kikuyu | Ryegrass—4600 | Ryegrass—Bizet | Ryegrass—Premier 2 | ||
| LDA Prediction | Blue Fescue | 50 | 2 | 7 | 4 | 0 | 2 | 3 | 0 | 0 | 0 |
| Browntop | 0 | 44 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Chewings Fescue | 0 | 3 | 34 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | |
| Cotula | 0 | 1 | 2 | 46 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Couch | 0 | 0 | 0 | 1 | 52 | 0 | 0 | 0 | 0 | 0 | |
| Egmont | 0 | 5 | 2 | 0 | 1 | 50 | 0 | 5 | 0 | 2 | |
| Kikuyu | 0 | 0 | 1 | 2 | 1 | 0 | 49 | 0 | 0 | 0 | |
| Ryegrass—4600 | 1 | 0 | 0 | 0 | 2 | 1 | 0 | 24 | 4 | 6 | |
| Ryegrass—Bizet | 5 | 0 | 8 | 2 | 0 | 0 | 4 | 7 | 50 | 9 | |
| Ryegrass—Premier 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 20 | 1 | 39 | |
| Overall Statistics | |||||||||||
| Accuracy: | 0.7821 | ||||||||||
| 95% Confidence Interval: | (0.7456, 0.8157) | ||||||||||
| No Information Rate: | 0.1 | ||||||||||
| p-Value [Acc > NIR]: | <2.2 × 10−16 | ||||||||||
| Kappa: | 0.7579 | ||||||||||
| Reference Sample | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Blue Fescue | Browntop | Chewings Fescue | Cotula | Couch | Egmont | Kikuyu | Ryegrass—4600 | Ryegrass—Bizet | Ryegrass—Premier 2 | ||
| LDA Prediction | Blue Fescue | 34 | 3 | 2 | 0 | 5 | 2 | 1 | 2 | 2 | 1 |
| Browntop | 0 | 50 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | |
| Chewings Fescue | 6 | 2 | 48 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | |
| Cotula | 0 | 0 | 1 | 54 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Couch | 5 | 0 | 2 | 0 | 46 | 0 | 0 | 2 | 6 | 2 | |
| Egmont | 0 | 0 | 0 | 0 | 1 | 47 | 0 | 0 | 0 | 0 | |
| Kikuyu | 0 | 0 | 0 | 0 | 0 | 0 | 54 | 0 | 0 | 0 | |
| Ryegrass—4600 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 43 | 1 | 3 | |
| Ryegrass—Bizet | 5 | 0 | 0 | 0 | 2 | 0 | 1 | 6 | 43 | 13 | |
| Ryegrass—Premier 2 | 6 | 0 | 2 | 2 | 0 | 0 | 0 | 3 | 4 | 36 | |
| Overall Statistics | |||||||||||
| Accuracy: | 0.8125 | ||||||||||
| 95% Confidence Interval: | (0.7777, 0.844) | ||||||||||
| No Information Rate: | 0.1 | ||||||||||
| p-Value [Acc > NIR]: | <2.2 × 10−16 | ||||||||||
| Kappa: | 0.7917 | ||||||||||
| Bands Used | 12 | 76 | 150 | 200 | 404 | 800 | 914 | 1740 | 2050 |
| Accuracy | 78.21% | 89.82% | 91.61% | 93.75% | 96.96% | 96.07% | 94.20% | 92.14% | 93.04% |
| kappa | 75.79% | 88.69% | 90.67% | 93.06% | 96.63% | 95.63% | 94.25% | 91.27% | 92.26% |
| Bands Used | 11 | 50 | 110 | 266 | 422 | 814 | 1177 | 1665 |
| Accuracy | 81.25% | 95.54% | 96.61% | 98.04% | 97.68% | 96.96% | 96.07% | 94.82% |
| kappa | 79.17% | 95.04% | 96.23% | 97.82% | 97.42% | 96.63% | 95.63% | 94.25% |
| Group Details | Important Bands | Bands in Range | Importance as a Proportion of the Group |
|---|---|---|---|
| 1st derivative (total) | 110 | 2020 bands | 5.4% |
| SNV (total) | 148 | 2051 bands | 7.3% |
| 1st derivative (VNIR only) | 98 | 1000 bands | 9.8% |
| SNV (VNIR only) | 41 | 1000 bands | 4.1% |
| 1st derivative (SWIR only) | 13 | 1020 bands | 1.3% |
| SNV (SWIR only) | 107 | 1051 bands | 10.2% |
| Target Subject | Prediction Accuracy | Kappa |
|---|---|---|
| Blue Fescue | 97.86% | 97.86% |
| Browntop | 95.71% | 95.71% |
| Chewings | 92.14% | 92.14% |
| Cotula | 98.57% | 98.57% |
| Couch | 94.29% | 94.29% |
| Egmont | 96.43% | 96.43% |
| Kikuyu | 98.57% | 98.57% |
| Ryegrass—‘4600’ | 97.86% | 97.86% |
| Ryegrass—‘Bizet’ | 94.29% | 94.29% |
| Ryegrass—‘Premier 2’ | 100.00% | 100.00% |
| Reference | |||||
|---|---|---|---|---|---|
| Prediction | a | b | c | d | e |
| a | 257 | 71 | 0 | 2 | 6 |
| b | 8 | 158 | 28 | 18 | 2 |
| c | 0 | 24 | 225 | 9 | 8 |
| d | 1 | 20 | 25 | 172 | 119 |
| e | 14 | 7 | 2 | 79 | 145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cushnahan, T.A.; Grafton, M.C.E.; Pearson, D.; Ramilan, T. Hyperspectral Data Can Differentiate Species and Cultivars of C3 and C4 Turf Despite Measurable Diurnal Variation. Remote Sens. 2024, 16, 3142. https://doi.org/10.3390/rs16173142
Cushnahan TA, Grafton MCE, Pearson D, Ramilan T. Hyperspectral Data Can Differentiate Species and Cultivars of C3 and C4 Turf Despite Measurable Diurnal Variation. Remote Sensing. 2024; 16(17):3142. https://doi.org/10.3390/rs16173142
Chicago/Turabian StyleCushnahan, Thomas A., Miles C. E. Grafton, Diane Pearson, and Thiagarajah Ramilan. 2024. "Hyperspectral Data Can Differentiate Species and Cultivars of C3 and C4 Turf Despite Measurable Diurnal Variation" Remote Sensing 16, no. 17: 3142. https://doi.org/10.3390/rs16173142
APA StyleCushnahan, T. A., Grafton, M. C. E., Pearson, D., & Ramilan, T. (2024). Hyperspectral Data Can Differentiate Species and Cultivars of C3 and C4 Turf Despite Measurable Diurnal Variation. Remote Sensing, 16(17), 3142. https://doi.org/10.3390/rs16173142







