Early-Season Crop Mapping by PRISMA Images Using Machine/Deep Learning Approaches: Italy and Iran Test Cases
Abstract
1. Introduction
2. Materials and Methods
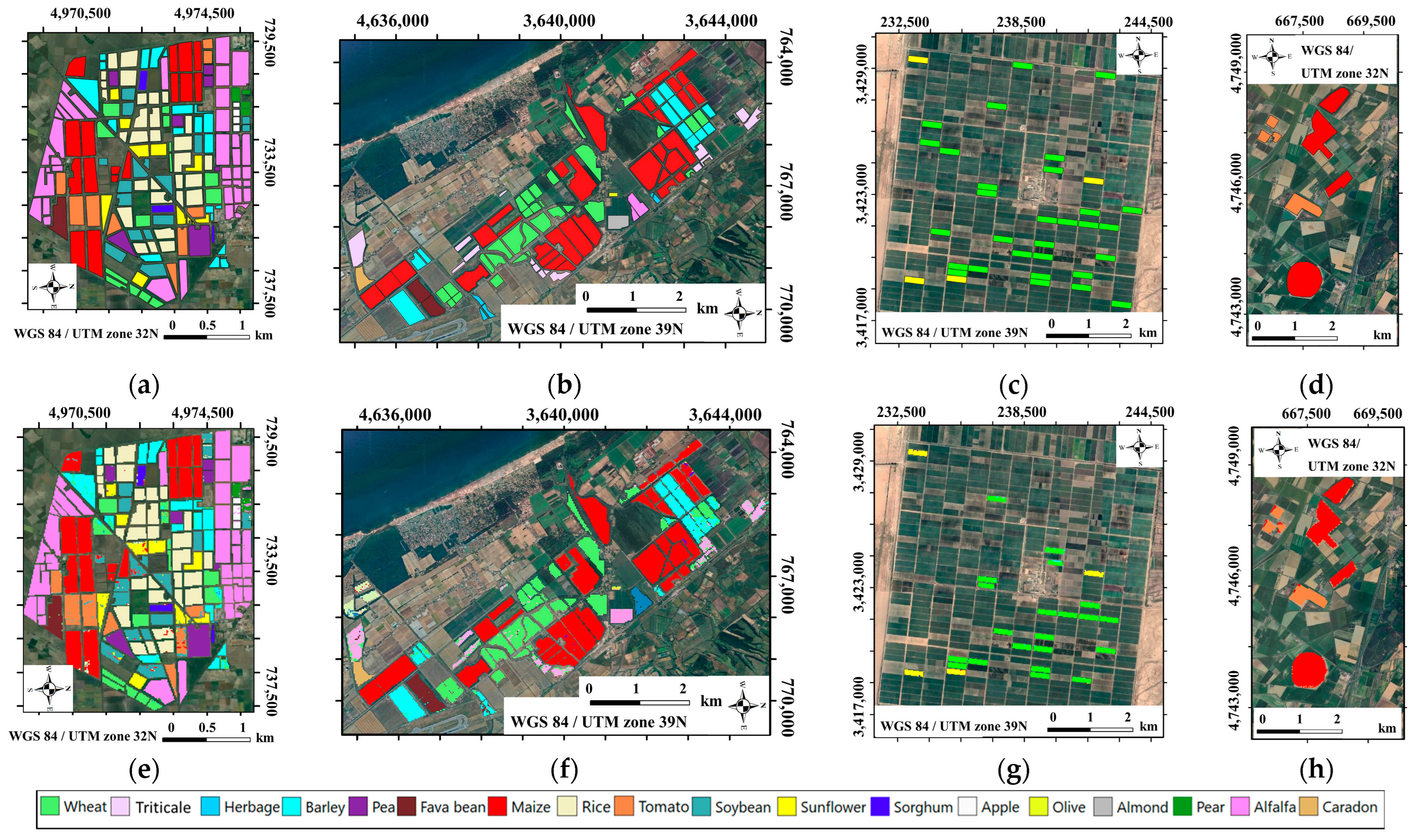
2.1. Study Areas
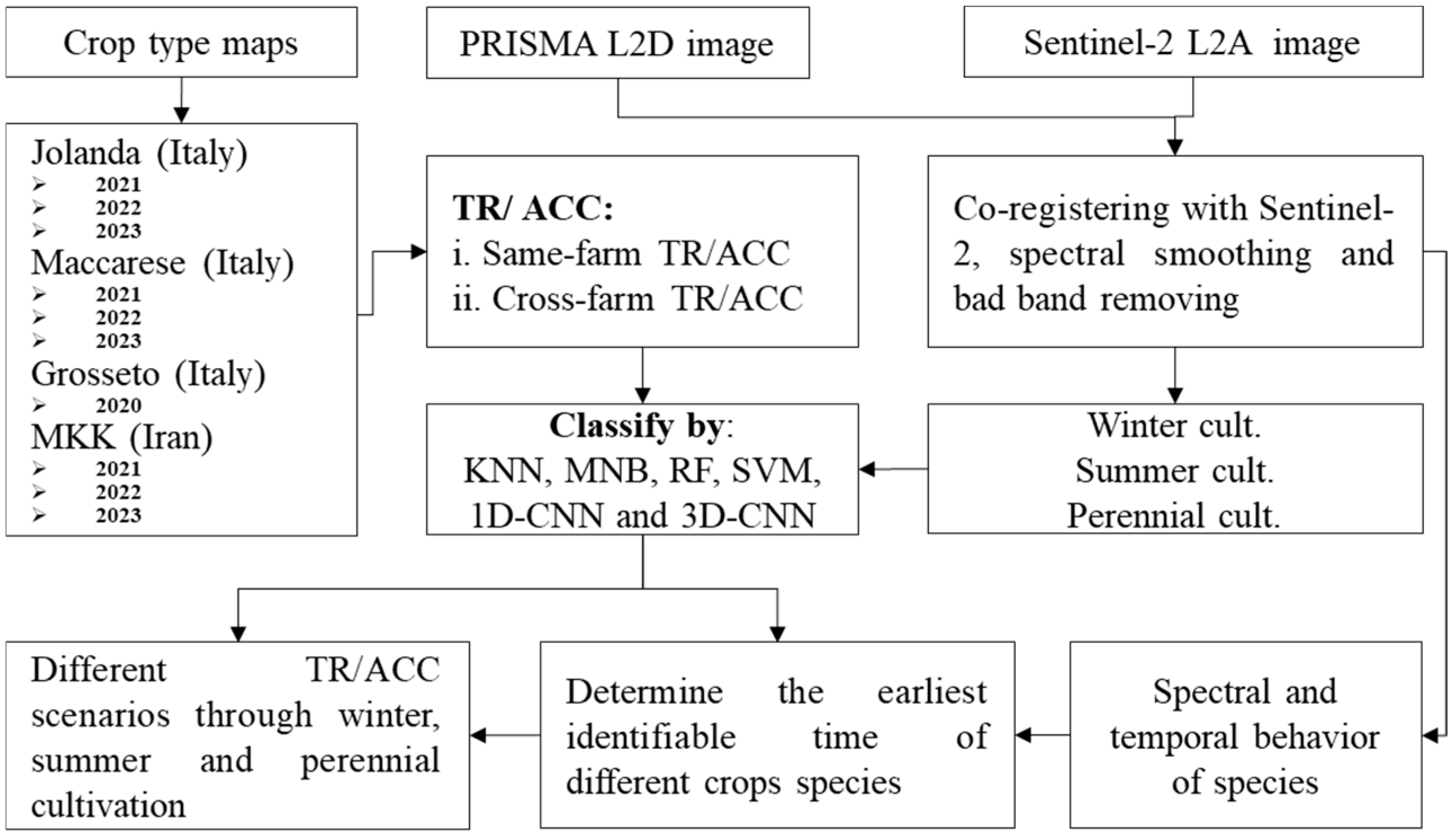
2.2. Overview of the Implemented Crop Mapping Procedure
2.3. Data Collection
2.3.1. Ground Reference Data
2.3.2. Satellite Imagery
2.4. Pre-Processing and Processing EO Data
2.4.1. Pre-Processing
2.4.2. Machine Learning Classification Algorithms
2.4.3. Deep Learning Classification Algorithms
2.5. Classification Scenarios
2.6. Accuracy Assessment Scenarios
3. Results
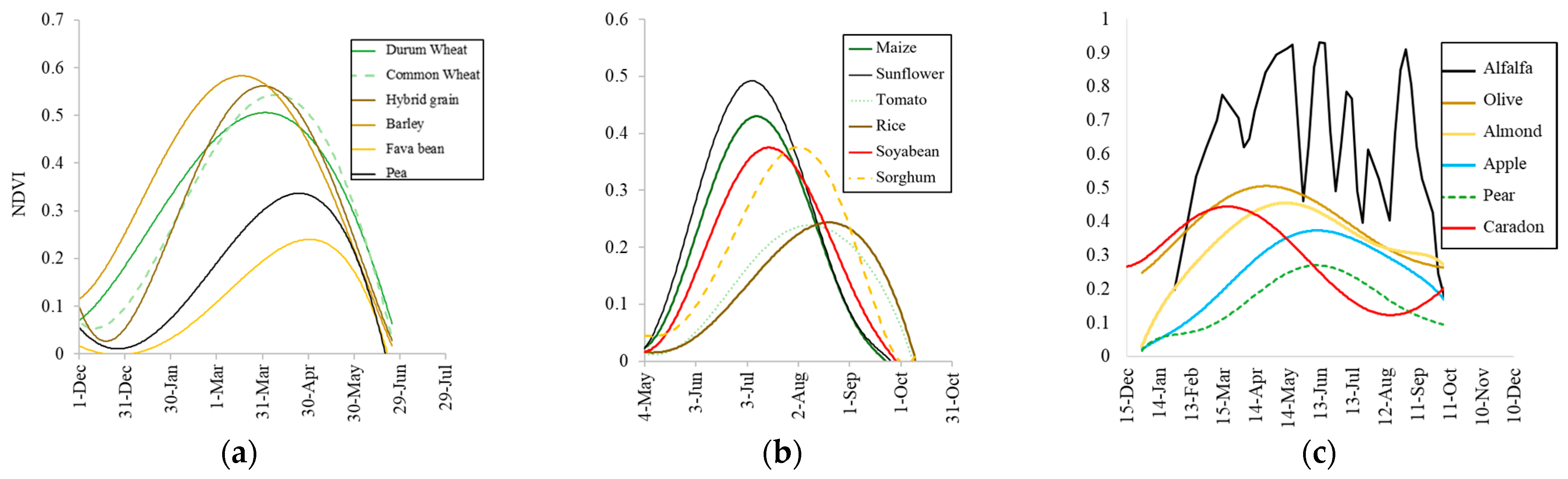
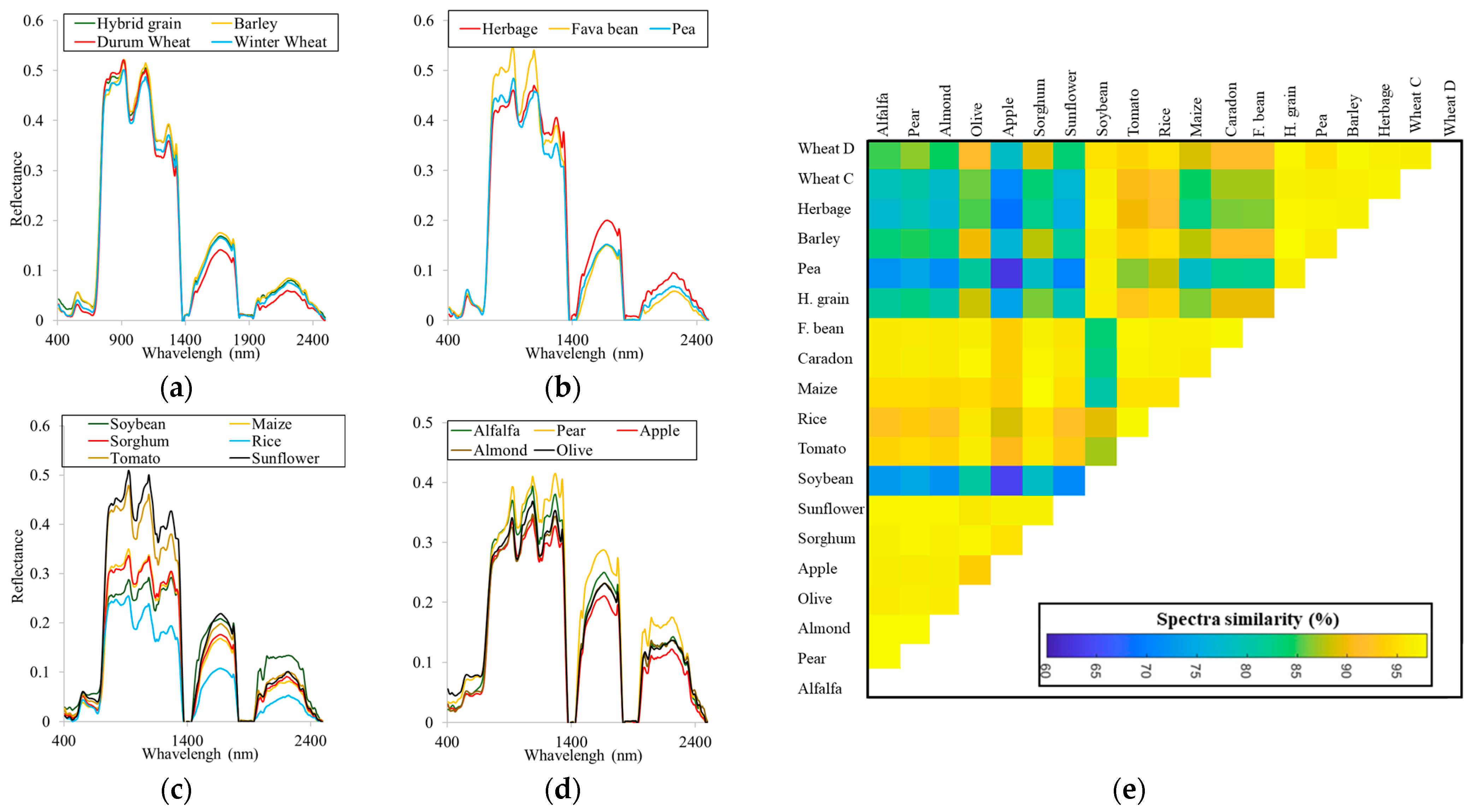
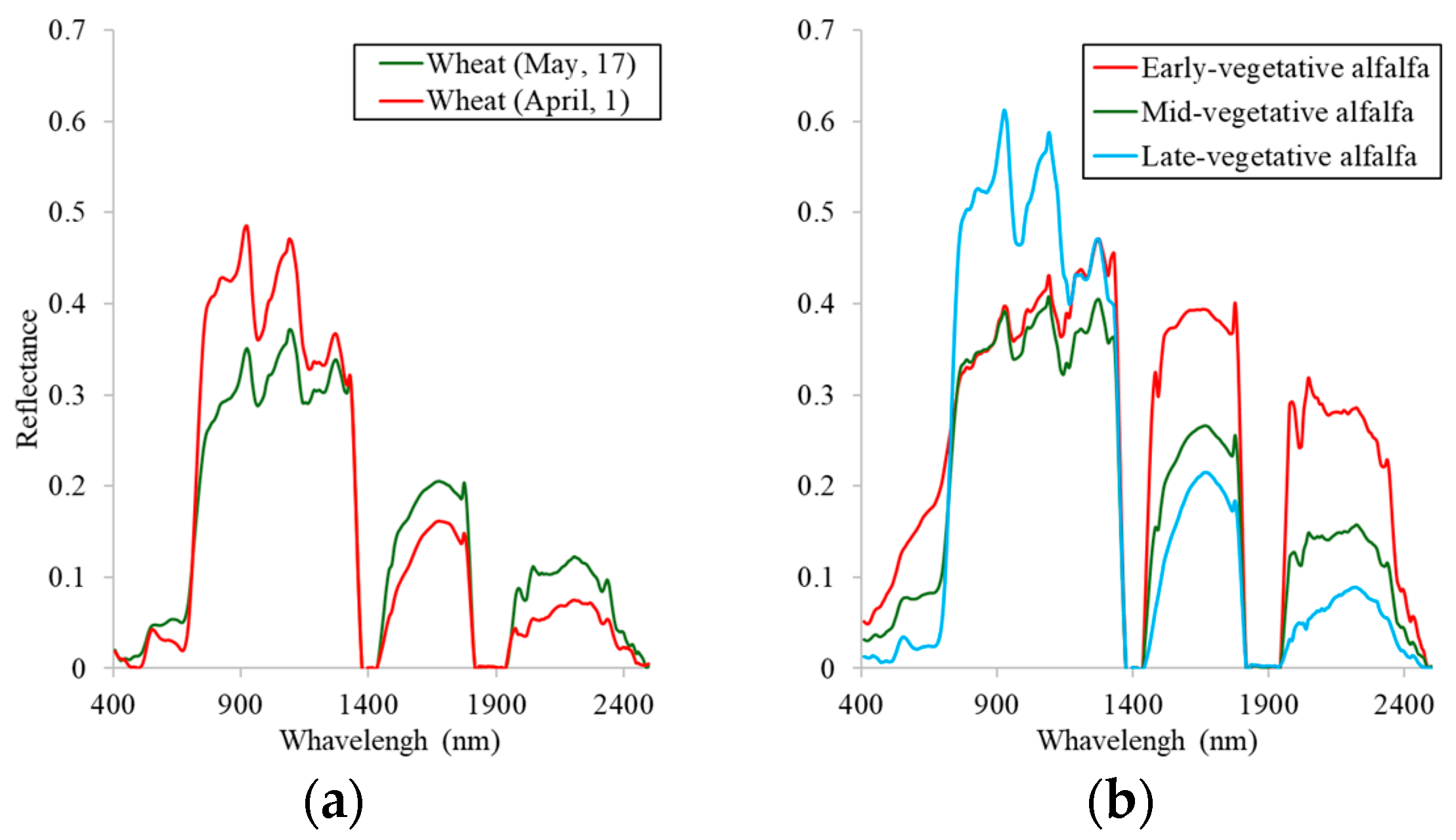
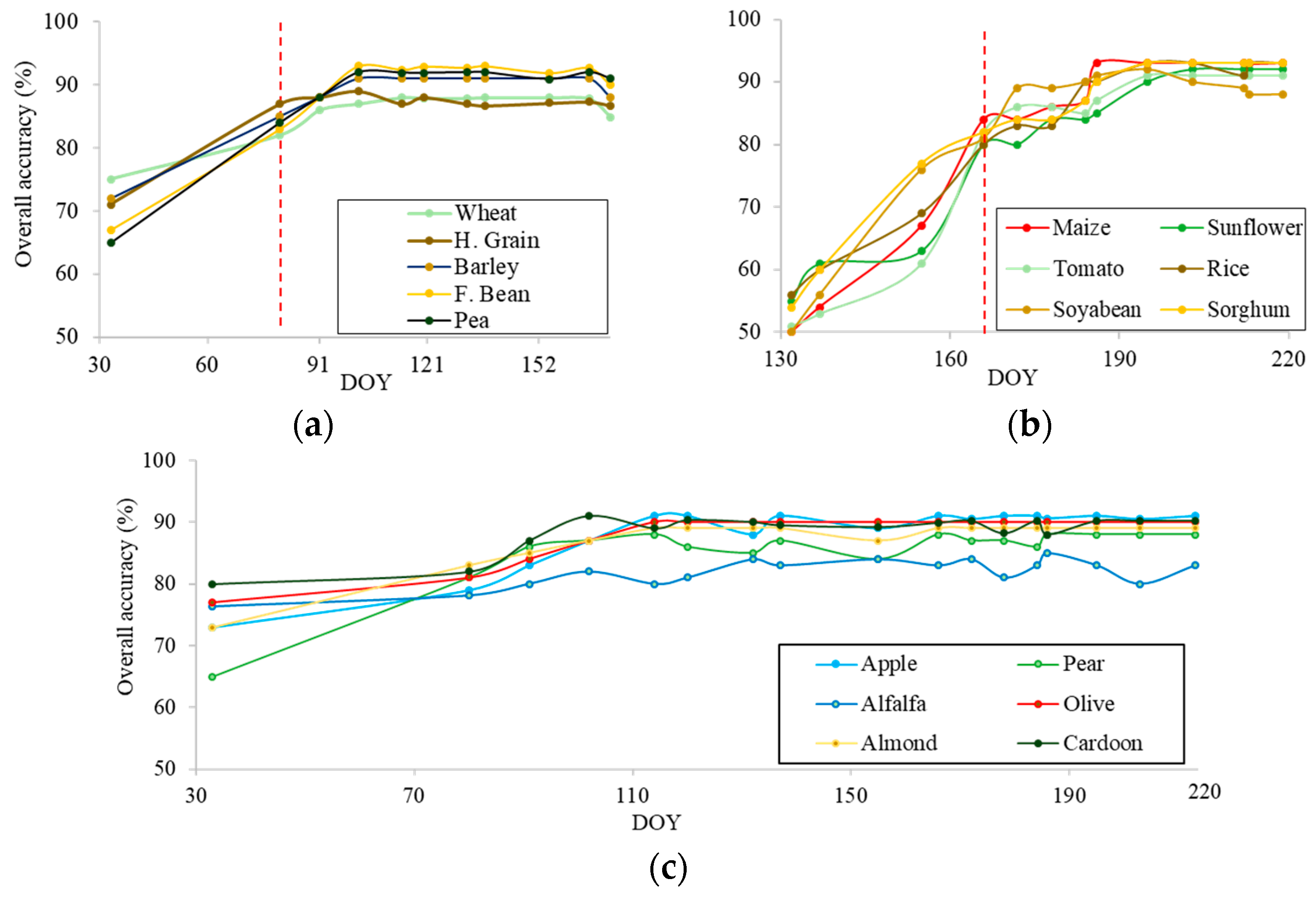
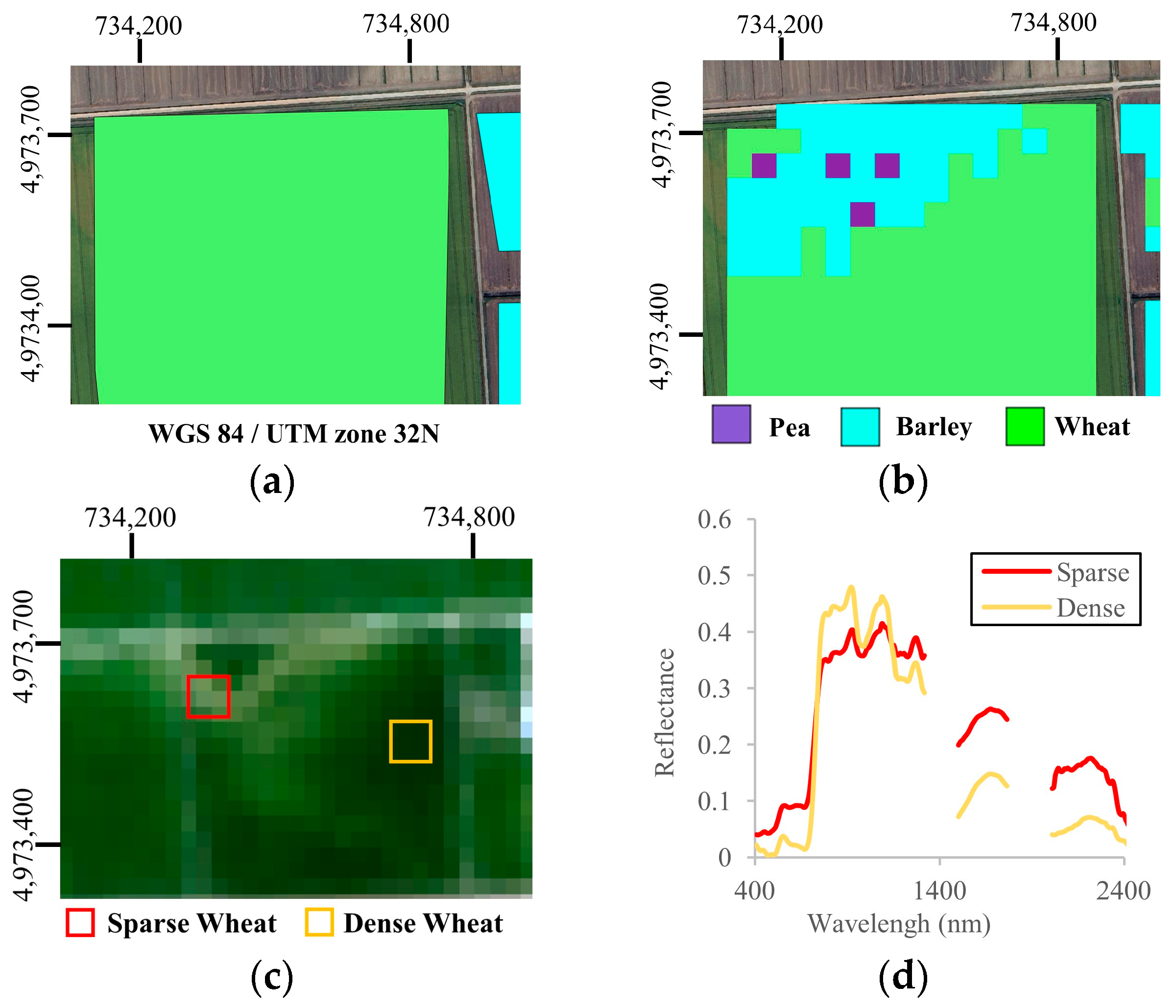
3.1. Spectral and Temporal Behavior of Species
3.2. Earliest Identifiable Time of Different Crops
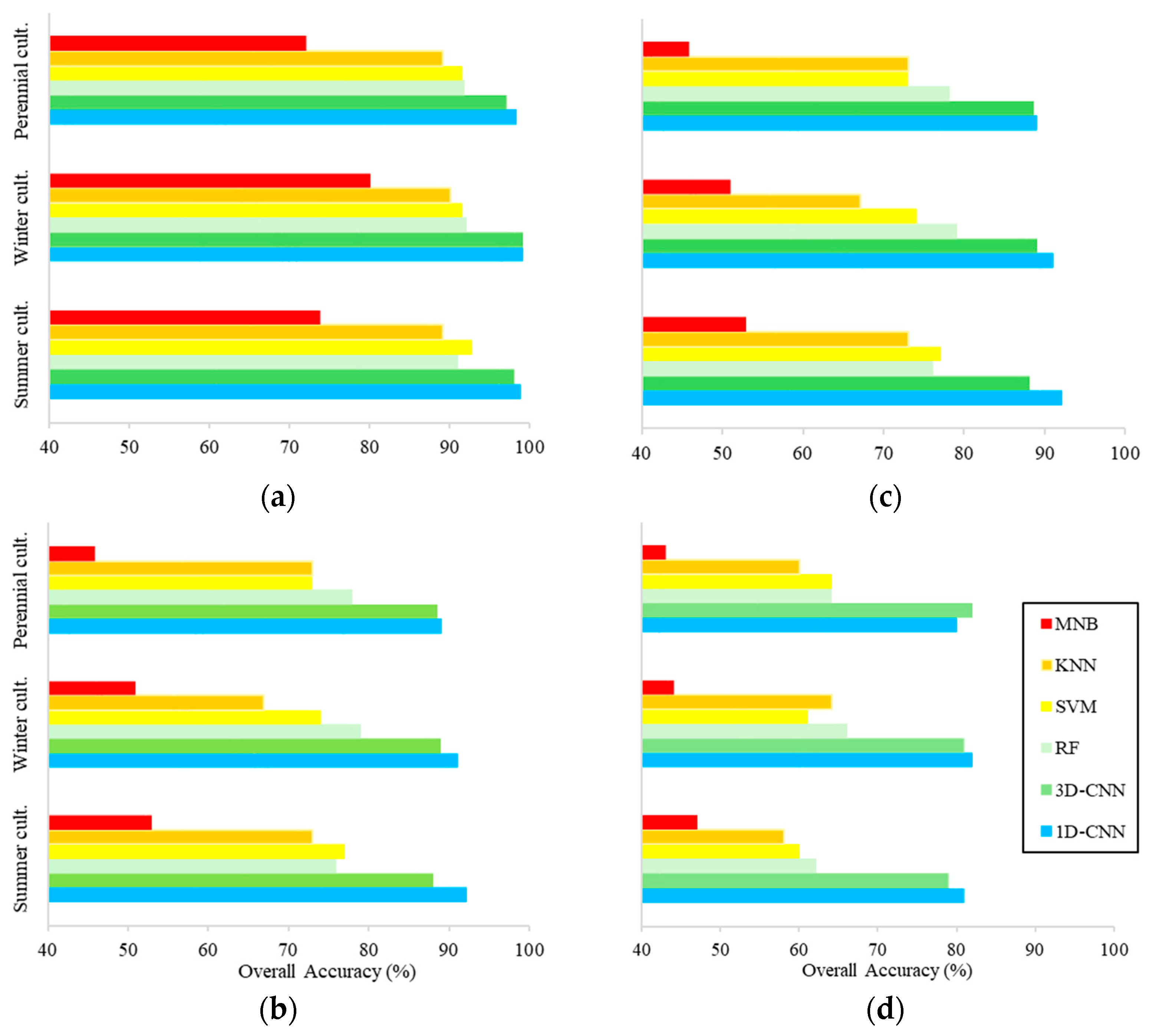
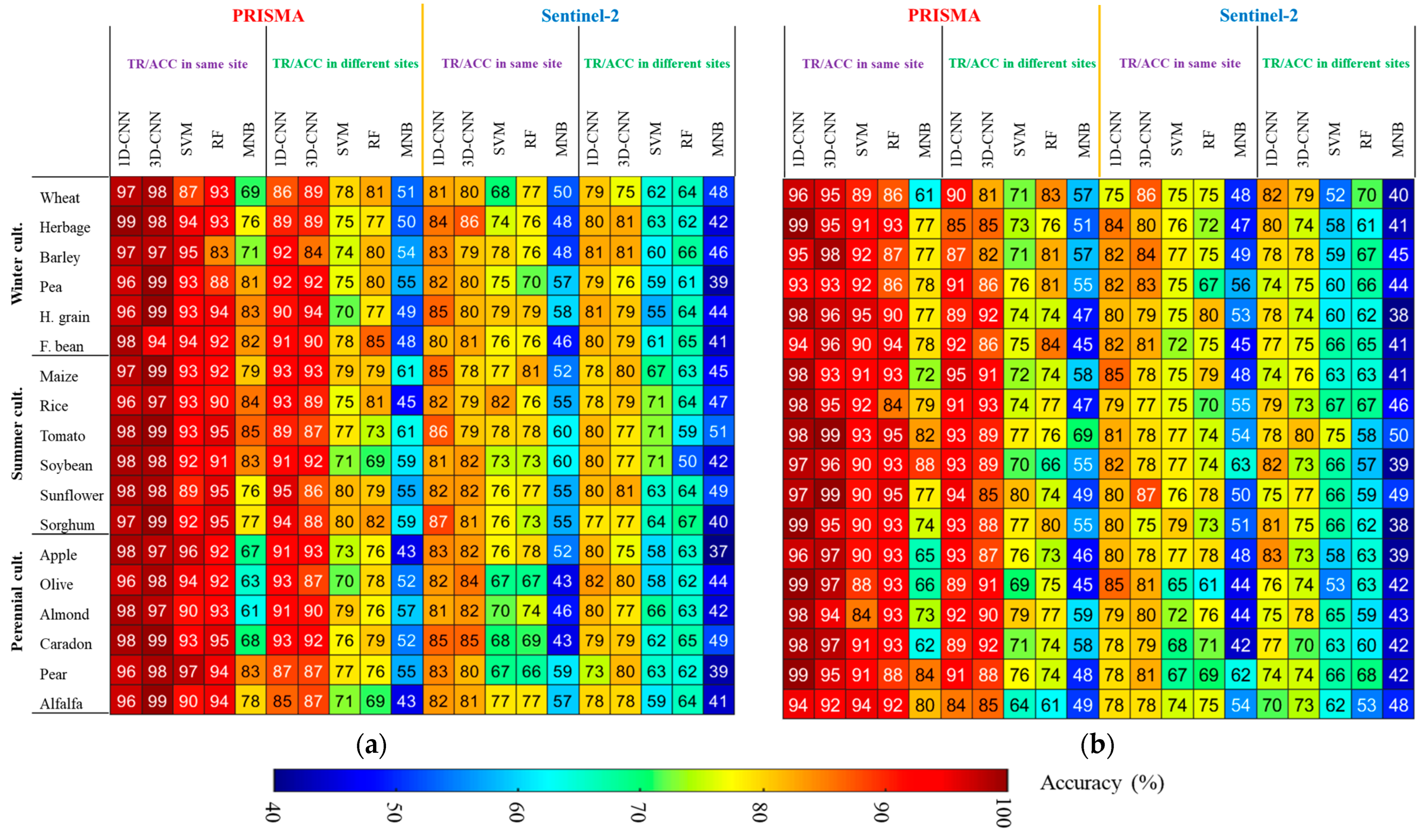
3.3. Classification Accuracy
4. Discussion
4.1. Effects of Reflectance Temporal Variation and Field Heterogeneity on Classifier Performances
4.2. Effects of Field Heterogeneity on Classifier Performances
4.3. Early-Stage Crop Mapping Using ML and DL Algorithms
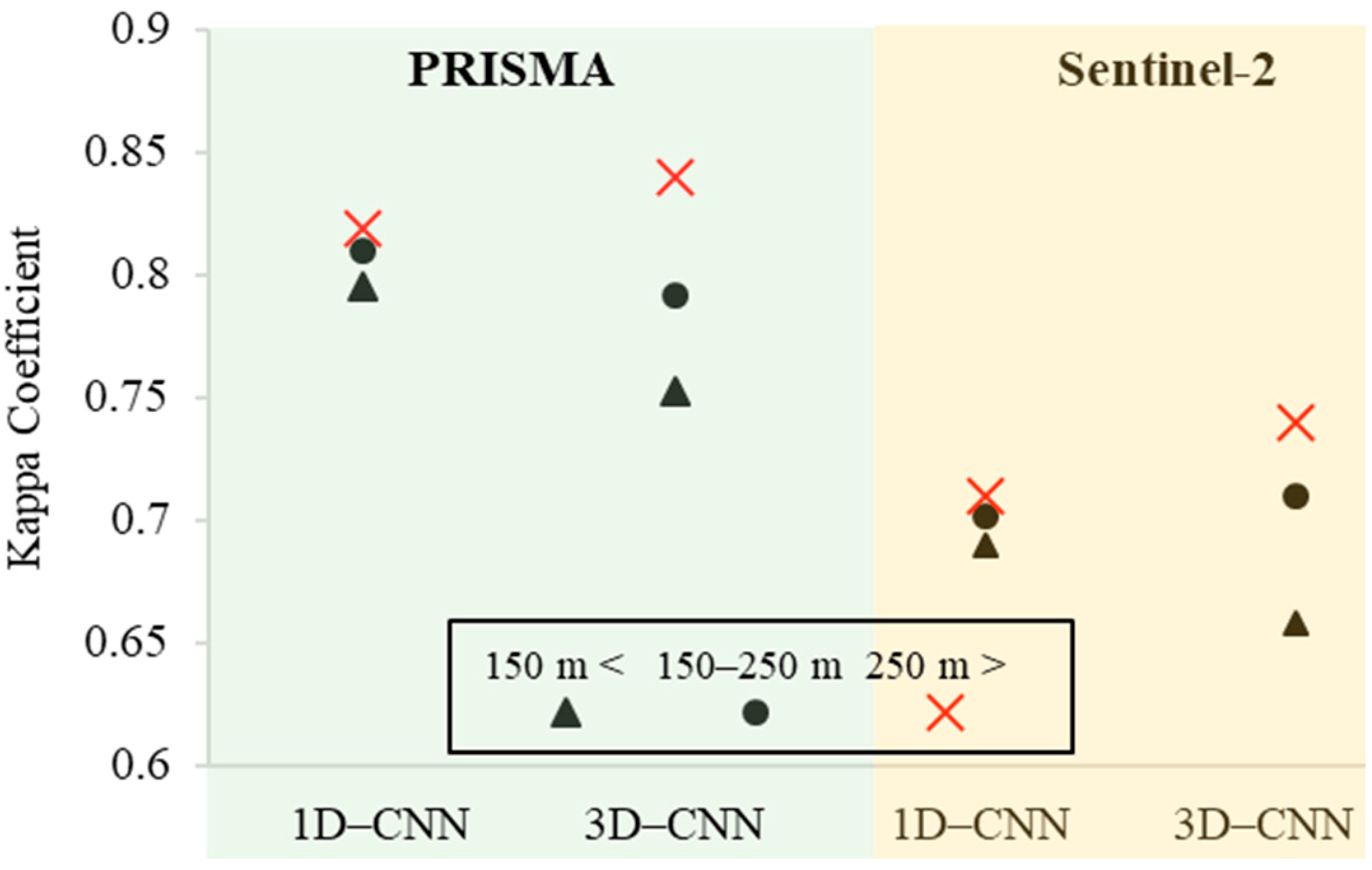
4.4. The Effects of Pixel/Field Size on the 3D-CNN
4.5. Cross-Farm TR/ACC for Unavailable Data Situation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, Y.N.; Luo, J.; Feng, L.; Zhou, X. DCN-based spatial features for improving parcel-based crop classification using high-resolution optical images and multi-temporal SAR data. Remote Sens 2019, 11, 1619. [Google Scholar] [CrossRef]
- Navidi, M.N.; Chatrenour, M.; Seyedmohammadi, J.; Delsous Khaki, B.; Moradi-Majd, N.; Mirzaei, S. Ecological potential assessment and land use area estimation of agricultural lands based on multi-time images of Sentinel-2 using ANP-WLC and GIS in Bastam, Iran. Environ. Monit. Assess 2023, 195, 36. [Google Scholar] [CrossRef] [PubMed]
- Turkoglu, M.O.; D’Aronco, S.; Perich, G.; Liebisch, F.; Streit, C.; Schindler, K.; Wegner, J.D. Crop mapping from image time series: Deep learning with multi-scale label hierarchies. Remote Sens. Environ 2021, 264, 112603. [Google Scholar] [CrossRef]
- Xie, Q.; Lai, K.; Wang, J.; Lopez-Sanchez, J.M.; Shang, J.; Liao, C.; Zhu, J.; Fu, H.; Peng, X. Crop monitoring and classification using polarimetric RADARSAT-2 time-series data across growing season: A case study in southwestern Ontario, Canada. Remote Sens. 2021, 13, 1394. [Google Scholar] [CrossRef]
- Wei, P.; Ye, H.; Qiao, S.; Liu, R.; Nie, C.; Zhang, B.; Lijuan, S.; Huang, S. Early crop mapping based on Sentinel-2 time-series data and the random forest algorithm. Remote Sens. 2023, 15, 3212. [Google Scholar] [CrossRef]
- Liu, X.; Li, X.; Gao, L.; Zhang, J.; Qin, D.; Wang, K.; Li, Z. Early-season and refined mapping of winter wheat based on phenology algorithms-a case of Shandong, China. Front. Plant Sci. 2023, 14, 1016890. [Google Scholar] [CrossRef]
- Hao, P.Y.; Tang, H.J.; Chen, Z.X.; Meng, Q.Y.; Kang, Y.P. Early-season crop type mapping using 30-m reference time series. J. Integr. Agric. 2020, 19, 1897–1911. [Google Scholar] [CrossRef]
- Liu, Y.; Kim, J.; Fleisher, D.H.; Kim, K.-S. Analogy-based crop yield forecasts based on temporal similarity of leaf area index. Remote Sens. 2021, 13, 3069. [Google Scholar] [CrossRef]
- Darvishi Boloorani, A.; Papi, R.; Soleimani, M.; Amiri, F.; Karami, L.; Neysani Samany, N.; Bakhtiari, M.; Mirzaei, S. Visual interpretation of satellite imagery for hotspot dust source identification. Remote Sens. Appl. Soc. Environ. 2023, 29, 100888. [Google Scholar] [CrossRef]
- Alajmi, M.; Mengash, H.A.; Eltahir, M.M.; Assiri, M.; Ibrahim, S.S.; Salama, A.S. Exploiting hyperspectral imaging and optimal deep learning for crop type detection and classification. IEEE Access 2023, 11, 124985–124995. [Google Scholar] [CrossRef]
- Liu, K.H.; Yang, M.H.; Huang, S.T.; Lin, C. Plant species classification based on hyperspectral imaging via a lightweight convolutional neural network model. Front. Plant Sci. 2022, 13, 855660. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Z.; Yin, C. Fine Crop Classification Based on UAV Hyperspectral Images and Random Forest. ISPRS Int. J. Geo-Inf. 2022, 11, 252. [Google Scholar] [CrossRef]
- Niu, B.; Feng, Q.; Chen, B.; Ou, C.; Liu, Y.; Yang, J. HSI-TransUNet: A transformer based semantic segmentation model for crop mapping from UAV hyperspectral imagery. Comput. Electron. Agric. 2022, 201, 107297. [Google Scholar] [CrossRef]
- Wei, L.; Wang, K.; Lu, Q.; Liang, Y.; Li, H.; Wang, Z.; Wang, R.; Cao, L. Crops fine classification in airborne hyperspectral imagery based on multi-feature fusion and deep learning. Remote Sens. 2021, 13, 2917. [Google Scholar] [CrossRef]
- Gimenez, R.; Lassalle, G.; Elger, A.; Dubucq, D.; Credoz, A.; Fabre, S. Mapping plant species in a former industrial site using airborne hyperspectral and time series of Sentinel-2 data sets. Remote Sens. 2022, 14, 3633. [Google Scholar] [CrossRef]
- Spiller, D.; Ansalone, L.; Carotenuto, F.; Mathieu, P.P. Crop type mapping using PRISMA hyperspectral images and one-dimensional convolutional neural network. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 8166–8169. [Google Scholar]
- Meng, S.; Wang, X.; Hu, X.; Luo, C.; Zhong, Y. Deep learning-based crop mapping in the cloudy season using one-shot hyperspectral satellite imagery. Comput. Electron. Agric 2021, 186, 106188. [Google Scholar] [CrossRef]
- Farmonov, N.; Amankulova, K.; Szatmári, J.; Sharifi, A.; Abbasi-Moghadam, D.; Mirhoseini Nejad, S.M.; Mucsi, L. Crop type classification by DESIS hyperspectral imagery and machine learning algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2023, 16, 1576–1588. [Google Scholar] [CrossRef]
- Ungar, S.G.; Pearlman, J.S.; Mendenhall, J.A.; Reuter, D. Overview of the earth observing one (EO-1) mission. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1149–1159. [Google Scholar] [CrossRef]
- Barnsley, M.J.; Settle, J.J.; Cutter, M.; Lobb, D.; Teston, F. The PROBA/CHRIS mission: A low-cost smallsat for hyperspectral, multi-angle, observations of the Earth surface and atmosphere. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1512–1520. [Google Scholar] [CrossRef]
- Pignatti, S.; Palombo, A.; Pascucci, S.; Romano, F.; Santini, F.; Simoniello, T.; Umberto, A.; Vincenzo, C.; Acito, N.; Diani, M.; et al. The PRISMA hyperspectral mission: Science activities and opportunities for agriculture and land monitoring. In Proceedings of the 2013 IEEE International Geoscience and Remote Sensing Symposium-IGARSS, Melbourne, VIC, Australia, 21–26 July 2013; pp. 4558–4561. [Google Scholar]
- Guanter, L.; Kaufmann, H.; Segl, K.; Foerster, S.; Rogass, C.; Chabrillat, S.; Kuester, T.; Hollstein, A.; Rossner, G.; Chlebek, C.; et al. The EnMAP spaceborne imaging spectroscopy mission for earth observation. Remote Sens. 2015, 7, 8830–8857. [Google Scholar] [CrossRef]
- Green, R.O.; Mahowald, N.; Ung, C.; Thompson, D.R.; Bator, L.; Bennet, M.; Bernas, M.; Blackway, N.; Bradley, C.; Cha, J.; et al. The Earth surface mineral dust source investigation: An Earth science imaging spectroscopy mission. In Proceedings of the 2020 IEEE Aerospace Conference, Big Sky, MT, USA, 7–14 March 2020; pp. 1–15. [Google Scholar]
- Müller, R.; Avbelj, J.; Carmona, E.; Eckardt, A.; Gerasch, B.; Graham, L.; Walter, I. The new hyperspectral sensor DESIS on the multi-payload platform muses installed on the ISS. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 461–467. [Google Scholar]
- Buschkamp, P.; Hofmann, J.; Rio-Fernandes, D.; Haberler, P.; Gerstmeier, M.; Bartscher, C.; Bianchi, S.; Delpet, P.; Weber, H.; Strese, H.; et al. CHIME’s hyperspectral imager (HSI): Status of instrument design and performance at PDR. Int. Conf. Space Opt. 2023, 12777, 1379–1398. [Google Scholar]
- Cawse-Nicholson, K.; Townsend, P.A.; Schimel, D.; Assiri, A.M.; Blake, P.L.; Buongiorno, M.F.; Campbell, P.; Carmon, N.; Casey, K.A.; Correa-Pabón, R.E.; et al. NASA’s surface biology and geology designated observable: A perspective on surface imaging algorithms. Remote Sens. Environ. 2021, 257, 112349. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera-Caicedo, J.P.; Reyes-Muñoz, P.; Morata, M.; Amin, E.; Tagliabue, G.; Panigada, C.; Hank, T.; Berger, K. Mapping landscape canopy nitrogen content from space using PRISMA data. ISPRS J. Photogramm. Remote Sens. 2021, 178, 382–395. [Google Scholar] [CrossRef]
- Asadi, B.; Shamsoddini, A. Crop mapping through a hybrid machine learning and deep learning method. Remote Sens. Appl. Soc. Environ. 2023, 33, 101090. [Google Scholar] [CrossRef]
- Aneece, I.; Foley, D.; Thenkabail, P.; Oliphant, A.; Teluguntla, P. New generation hyperspectral data from DESIS compared to high spatial resolution PlanetScope data for crop type classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 7846–7858. [Google Scholar]
- Patel, U.; Pathan, M.; Kathiria, P.; Patel, V. Crop type classification with hyperspectral images using deep learning: A transfer learning approach. Model. Earth Syst. Environ. 2023, 9, 1977–1987. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, H.; Wang, A.; Iwahori, Y. Precise crop classification of hyperspectral images using multi-branch feature fusion and dilation-based MLP. Remote Sens. 2022, 14, 2713. [Google Scholar] [CrossRef]
- Joshi, A.; Pradhan, B.; Gite, S.; Chakraborty, S. Remote-Sensing Data and Deep-Learning Techniques in Crop Mapping and Yield Prediction: A Systematic Review. Remote Sens. 2023, 15, 2014. [Google Scholar] [CrossRef]
- Alami Machichi, M.; Mansouri, L.E.; Imani, Y.; Bourja, O.; Lahlou, O.; Zennayi, Y.; Bourzeix, F.; Houmma, I.H.; Hadria, R. Crop mapping using supervised machine learning and deep learning: A systematic literature review. Int. J. Remote Sens. 2023, 44, 2717–2753. [Google Scholar] [CrossRef]
- Saha, D.; Manickavasagan, A. Machine learning techniques for analysis of hyperspectral images to determine quality of food products: A review. J. Curr. Res. Food Sci. 2021, 4, 28–44. [Google Scholar] [CrossRef]
- Zan, X.; Zhang, X.; Xing, Z.; Liu, W.; Zhang, X.; Su, W.; Liu, Z.; Zhao, Y.; Li, S. Automatic Detection of Maize Tassels from UAV Images by Combining Random Forest Classifier and VGG16. Remote Sens. 2020, 12, 3049. [Google Scholar] [CrossRef]
- Paoletti, M.E.; Haut, J.M.; Plaza, J.; Plaza, A. Deep learning classifiers for hyperspectral imaging: A review. ISPRS J. Photogramm. Remote Sens. 2019, 158, 279–317. [Google Scholar] [CrossRef]
- Kumar, B.; Dikshit, O.; Gupta, A.; Singh, M.K. Feature extraction for hyperspectral image classification: A review. Int. J. Remote Sens. 2020, 41, 6248–6287. [Google Scholar] [CrossRef]
- Cao, M.; Sun, Y.; Jiang, X.; Li, Z.; Xin, Q. Identifying leaf phenology of deciduous broadleaf forests from PhenoCam images using a convolutional neural network regression method. Remote Sens. 2021, 13, 2331. [Google Scholar] [CrossRef]
- Kun, X.; Wei, W.; Sun, Y.; Wang, Y.; Xin, Q. Mapping fine-spatial-resolution vegetation spring phenology from individual Landsat images using a convolutional neural network. Int. J. Remote Sens. 2023, 44, 3059–3081. [Google Scholar] [CrossRef]
- Yadav, D.P.; Kumar, D.; Jalal, A.S.; Kumar, A.; Khan, S.B.; Gadekallu, T.R.; Mashat, A.; Malibari, A.A. Spectral–spatial features exploitation using lightweight HResNeXt model for hyperspectral image classification. Can. J. Remote Sens. 2023, 49, 2248270. [Google Scholar] [CrossRef]
- Zhu, J.; Hu, J.; Jia, S.; Jia, X.; Li, Q. Multiple 3-D feature fusion framework for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 1873–1886. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Jiang, Y.; Wang, P.; Shen, Q.; Shen, C. Hyperspectral classification based on lightweight 3-D-CNN with transfer learning. IEEE Trans. Geosci. Remote Sens. 2019, 57, 5813–5828. [Google Scholar] [CrossRef]
- Ran, L.; Zhang, Y.; Wei, W.; Zhang, Q. A hyperspectral image classification framework with spatial pixel pair features. Sensors 2017, 17, 2421. [Google Scholar] [CrossRef]
- Kanthi, M.; Sarma, T.H.; Bindu, C.S. A 3d-Deep CNN based feature extraction and hyperspectral image classification. In Proceedings of the IEEE India Geoscience and Remote Sensing Symposium, Ahmedabad, India, 1–4 December 2020; pp. 229–232. [Google Scholar]
- Zhang, H.; Yu, H.; Xu, Z.; Zheng, K.; Gao, L. A novel classification framework for hyperspectral image classification based on multi-scale dense network. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 2238–2241. [Google Scholar]
- Lin, C.; Zhong, L.; Song, X.P.; Dong, J.; Lobell, D.B.; Jin, Z. Early- and in-season crop type mapping without current-year ground truth: Generating labels from historical information via a topology-based approach. Remote Sens. Environ. 2022, 274, 112994. [Google Scholar] [CrossRef]
- Moharrami, M.; Attarchi, S.; Gloaguen, R.; Alavipanah, S.K. Integration of Sentinel-1 and Sentinel-2 data for ground truth sample migration for multi-temporal land cover mapping. Remote Sens. 2024, 16, 1566. [Google Scholar] [CrossRef]
- Pham, V.D.; Tetteh, G.; Thiel, F.; Erasmi, S.; Schwieder, M.; Frantz, D.; van der Linden, S. Temporally transferable crop mapping with temporal encoding and deep learning augmentations. Int. J. Appl. Earth Obs. Geoinf. 2024, 129, 103867. [Google Scholar] [CrossRef]
- Priori, S.; Mzid, N.; Pascucci, S.; Pignatti, S.; Casa, R. Performance of a portable FT-NIR MEMS spectrometer to predict soil features. Soil Syst. 2022, 6, 66. [Google Scholar] [CrossRef]
- Ahmad, A.; Sakidin, H.; Abu Sari, M.Y.; Mat Amin, A.R.; Sufahani, S.F.; Rasib, A.W. Naïve Bayes classification of high-resolution aerial imagery. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 168–177. [Google Scholar] [CrossRef]
- Pacheco, A.d.P.; Junior, J.A.d.S.; Ruiz-Armenteros, A.M.; Henriques, R.F.F. Assessment of k-nearest neighbor and random forest classifiers for mapping forest fire areas in central Portugal using Landsat-8, Sentinel-2, and Terra imagery. Remote Sens. 2021, 13, 1345. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Pelletier, C.; Webb, G.I.; Petitjean, F. Temporal convolutional neural network for the classification of satellite image time series. Remote Sens. 2019, 11, 523. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, H.; Hsu, C.J.; Chen, W.H.; Wu, S. Credit rating analysis with support vector machines and neural networks: A market comparative study. Decis. Support Syst. 2004, 37, 543–558. [Google Scholar] [CrossRef]
- Feng, S.; Zhao, J.; Liu, T.; Zhang, H.; Zhang, Z.; Guo, X. Crop type identification and mapping using machine learning algorithms and sentinel-2 time series data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 3295–3306. [Google Scholar] [CrossRef]
- Shafaey, M.; ElBery, M.; Salem, M.; Moushier, H.; El-Dahshan, E.S.A.; Tolba, M. Hyperspectral image analysis using a custom spectral convolutional neural network. Int. J. Intell. Comput. Inf. Sci. 2020, 22, 146–158. [Google Scholar] [CrossRef]
- Song, J.; Gao, S.; Zhu, Y.; Ma, C. A survey of remote sensing image classification based on CNNs. Big Earth Data 2019, 3, 232–254. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, L.; Zhou, H. Deep learning based multi-temporal crop classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Bera, S.; Shrivastava, V.; Satapathy, S. Advances in Hyperspectral Image Classification Based on Convolutional Neural Networks: A Review. Comput. Model. Eng. Sci. 2022, 133, 219–250. [Google Scholar] [CrossRef]
- Acito, N.; Diani, M.; Corsini, G. PRISMA spatial resolution enhancement by fusion with sentinel-2 data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 15, 62–79. [Google Scholar] [CrossRef]
- Musto, R.; Tricomi, A.; Bruno, R.; Pasquali, G. Advancing PRISMA Pansharpening: A Deep Learning Approach with Synthetic Data Pretraining and Transfer Learning. In Proceedings of the 2023 13th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Athens, Greece, 31 October–2 November 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–7. [Google Scholar]
- Zini, S.; Barbato, M.P.; Piccoli, F.; Napoletano, P. Deep Learning Hyperspectral Pansharpening on Large-Scale PRISMA Dataset. Remote Sens. 2024, 16, 2079. [Google Scholar] [CrossRef]
- Wilson, J.H.; Zhang, C.; Kovacs, J.M. Separating crop species in northeastern Ontario using hyperspectral data. Remote Sens. 2014, 6, 925–945. [Google Scholar] [CrossRef]
- Buchhart, C.; Schmidhalter, U. Daytime and seasonal reflectance of maize grown in varying compass directions. Front. Plant Sci. 2022, 13, 1029612. [Google Scholar] [CrossRef]
- Darvishi Boloorani, A.; Ranjbareslamloo, S.; Mirzaie, S.; Bahrami, H.A.; Mirzapour, F.; Abbaszadeh Tehrani, N. Spectral behavior of Persian oak under compound stress of water deficit and dust storm. Int. J. Appl. Earth Obs. Geoinf. 2020, 88, 102082. [Google Scholar] [CrossRef]
- Yu, K.; Lenz-Wiedemann, V.; Chen, X.; Bareth, G. Estimating leaf chlorophyll of barley at different growth stages using spectral indices to reduce soil background and canopy structure effects. ISPRS J. Photogramm. Remote Sens 2014, 97, 58–77. [Google Scholar] [CrossRef]
- Liu, N.; Zhao, R.; Qiao, L.; Zhang, Y.; Li, M.; Sun, H.; Xing, Z.; Wang, X. Growth stages classification of potato crop based on analysis of spectral response and variables optimization. Sensors 2020, 20, 3995. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zheng, T.; Liu, N.; Cheng, M.; Li, M.; Zhang, Q. Vertical distribution of chlorophyll in potato plants based on hyperspectral imaging. Trans. Chin. Soc. Agric. Eng. 2018, 34, 149–156. [Google Scholar]
- Graeff, S.; Claupein, W. Identification and discrimination of water stress in wheat leaves (Triticum aestivum L.) by means of reflectance measurements. Irrig. Sci. 2007, 26, 61–70. [Google Scholar] [CrossRef]
- Yao, J.; Hong, D.; Li, C.; Chanussot, J. SpectralMamba: Efficient Mamba for Hyperspectral Image Classification. arXiv 2024, arXiv:2404.08489v1. [Google Scholar]
- Zsigmond, T.; Braun, P.; Mészáros, J.; Waltner, I.; Horel, Á. Investigating plant response to soil characteristics and slope positions in a small catchment. Land 2022, 11, 774. [Google Scholar] [CrossRef]
- Mzid, N.; Castaldi, F.; Tolomio, M.; Pascucci, S.; Casa, R.; Pignatti, S. Evaluation of agricultural bare soil properties retrieval from Landsat 8, Sentinel-2 and PRISMA satellite data. Remote Sens. 2022, 14, 714. [Google Scholar] [CrossRef]
- Defourny, P.; Bontemps, S.; Bellemans, N.; Cara, C.; Dedieu, G.; Guzzonato, E.; Hagolle, O.; Inglada, J.; Nicola, L.; Rabaute, T.; et al. Near real-time agriculture monitoring at national scale at parcel resolution: Performance assessment of the Sen2-Agri automated system in various cropping systems around the world. Remote Sens. Environ. 2019, 221, 551–568. [Google Scholar] [CrossRef]
- Cai, Y.P.; Guan, K.Y.; Peng, J.; Wang, S.W.; Seifert, C.; Wardlow, B.; Li, Z. A high-performance and in-season classification system of field-level crop types using time-series Landsat data and a machine learning approach. Remote Sens. Environ. 2018, 210, 35–47. [Google Scholar] [CrossRef]



| Site | Species | January | February | March | April | May | June | July | August | September | October | November | December |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maccarese, Jolanda and Grosseto | Wheat | ||||||||||||
| Herbage | |||||||||||||
| Barley | |||||||||||||
| Pea | |||||||||||||
| Triticale | |||||||||||||
| Fava bean | |||||||||||||
| Cardoon | |||||||||||||
| Maize | |||||||||||||
| Rice | |||||||||||||
| Tomato | |||||||||||||
| Soybean | |||||||||||||
| Sunflower | |||||||||||||
| Sorghum | |||||||||||||
| Apple | |||||||||||||
| Olive | |||||||||||||
| Almond | |||||||||||||
| Pear | |||||||||||||
| Cardoon | |||||||||||||
| Alfalfa | |||||||||||||
| MKK | Sunflower | ||||||||||||
| Wheat |
| Cult. | Species | Sites | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grosseto | MKK | Maccarese | Jolanda | |||||||||
| 2020 | 2021 | 2022 | 2023 | 2021 | 2022 | 2023 | 2021 | 2022 | 2023 | |||
 | ||||||||||||
| Site | Year | N. of Fields | Area (ha) | Sentinel-2 | PRISMA Image Date (Month/Day) |
|---|---|---|---|---|---|
| Maccarese | 2021 | 92 | 1600 | 6 | 04/01, 05/17, 06/27, 09/04 |
| 2022 | 165 | 1600 | 6 | 01/16, 04/12, 05/29, 06/15, 07/14 | |
| 2023 | 70 | 700 | 2 | 02/02, 03/21 | |
| Jolanda | 2021 | 176 | 3700 | 6 | 04/24, 06/04, 06/21 |
| 2022 | 210 | 4100 | 8 | 04/30, 05/12, 07/03, 08/01 | |
| 2023 | 105 | 2863 | 4 | 03/04, 04/07, 05/24, 07/03, 08/07 | |
| Grosseto | 2020 | 6 | 20 | 1 | 07/31 |
| MKK-Iran | 2021 | 24 | 120 | 5 | 01/23, 03/11, 04/09, 05/14, 05/19, 06/23, 07/22 |
| 2022 | 30 | 310 | 7 | 01/29, 02/27, 03/28, 05/08, 05/25, 07/05, 07/22 | |
| 2023 | 26 | 130 | 3 | 02/28, 03/17, 04/09 |
| MNB | Parameter | Distribution | Kernel type | ||
| Range | Gaussian, kernel | Box, Epanechnikov, Gaussian, Triangle | |||
| Tuned | Kernel | Gaussian | |||
| KNN | Parameter | Distance weights | n_neighbors | Distance metric | |
| Range | Equal, Inverse, Squared inverse | 1–21 | Euclidean, Minkowski, Spearman, Hamming, Jaccard | ||
| Tuned | Equal | 7 | Euclidean | ||
| RF | Parameter | NumLearningCycles | Method | MaxNumSplits | MinLeafSize |
| Range | 10–500 | Bag, LSBoost | 1–50 | 1–50 | |
| Tuned | 100 | LSBoost | 15 | 10 | |
| SVM | Parameter | Gamma | C | Kernel type | |
| Range | 0.001–100 | 0.001–100 | Gaussian, Linear, Quadratic, Cubic, RBF | ||
| Tuned | 10 | 1.27 | RBF | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirzaei, S.; Pascucci, S.; Carfora, M.F.; Casa, R.; Rossi, F.; Santini, F.; Palombo, A.; Laneve, G.; Pignatti, S. Early-Season Crop Mapping by PRISMA Images Using Machine/Deep Learning Approaches: Italy and Iran Test Cases. Remote Sens. 2024, 16, 2431. https://doi.org/10.3390/rs16132431
Mirzaei S, Pascucci S, Carfora MF, Casa R, Rossi F, Santini F, Palombo A, Laneve G, Pignatti S. Early-Season Crop Mapping by PRISMA Images Using Machine/Deep Learning Approaches: Italy and Iran Test Cases. Remote Sensing. 2024; 16(13):2431. https://doi.org/10.3390/rs16132431
Chicago/Turabian StyleMirzaei, Saham, Simone Pascucci, Maria Francesca Carfora, Raffaele Casa, Francesco Rossi, Federico Santini, Angelo Palombo, Giovanni Laneve, and Stefano Pignatti. 2024. "Early-Season Crop Mapping by PRISMA Images Using Machine/Deep Learning Approaches: Italy and Iran Test Cases" Remote Sensing 16, no. 13: 2431. https://doi.org/10.3390/rs16132431
APA StyleMirzaei, S., Pascucci, S., Carfora, M. F., Casa, R., Rossi, F., Santini, F., Palombo, A., Laneve, G., & Pignatti, S. (2024). Early-Season Crop Mapping by PRISMA Images Using Machine/Deep Learning Approaches: Italy and Iran Test Cases. Remote Sensing, 16(13), 2431. https://doi.org/10.3390/rs16132431












