A Machine-Learning Model Based on the Fusion of Spectral and Textural Features from UAV Multi-Sensors to Analyse the Total Nitrogen Content in Winter Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Area and Design
2.2. Acquisition and Processing of Spectral Data
2.3. Pre-Processing of UAV Images
2.4. Spectral and Textural Features
2.5. Model Framework
2.6. Parameters for Model Accuracy Evaluation
3. Results
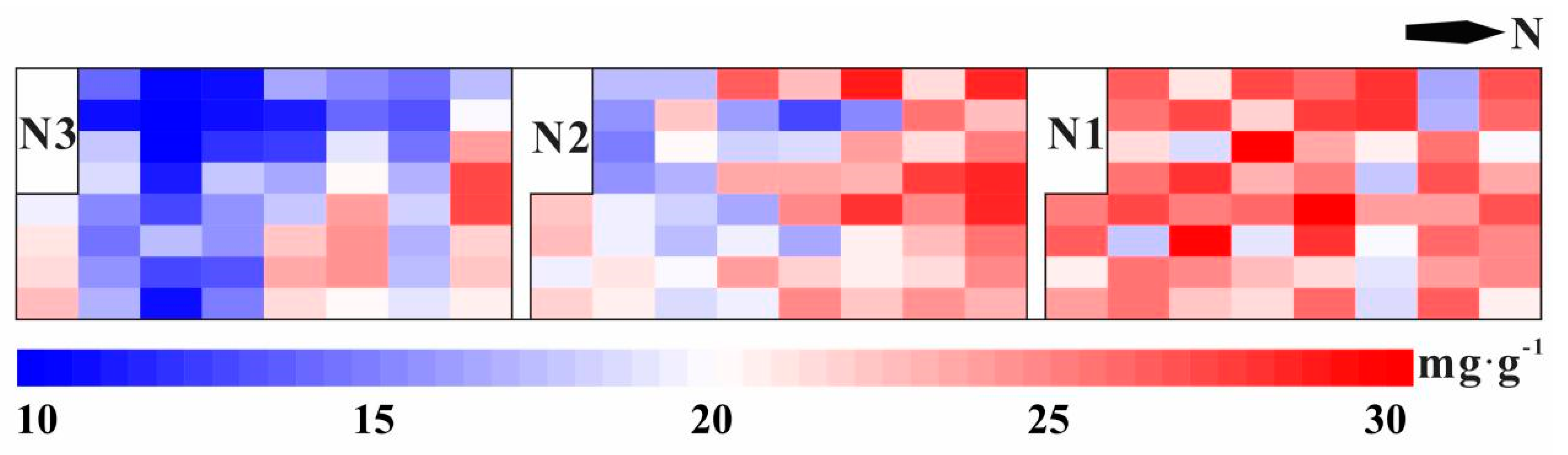
3.1. Sampling Statistics
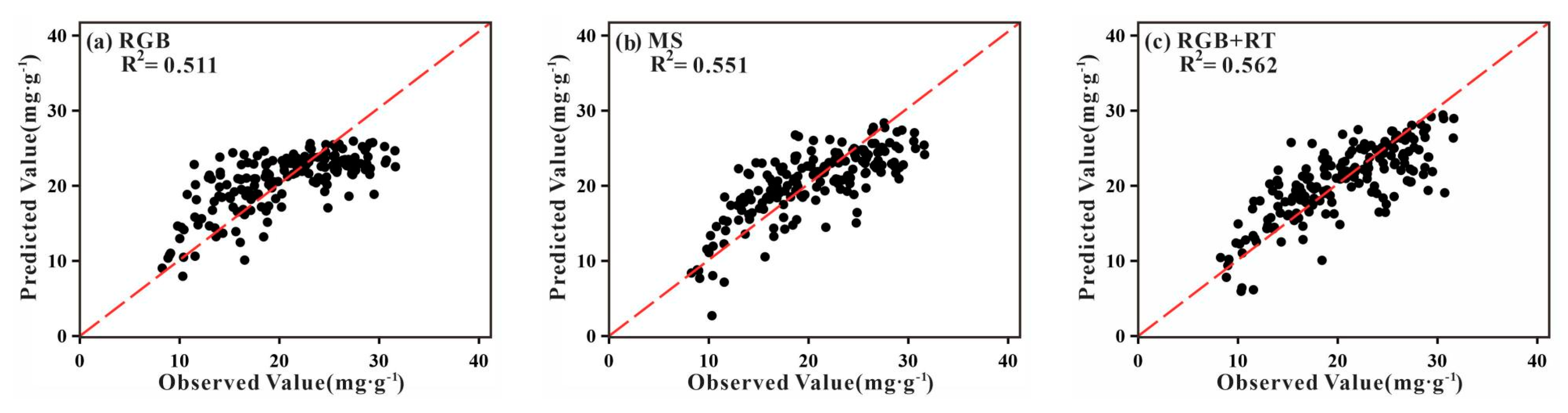
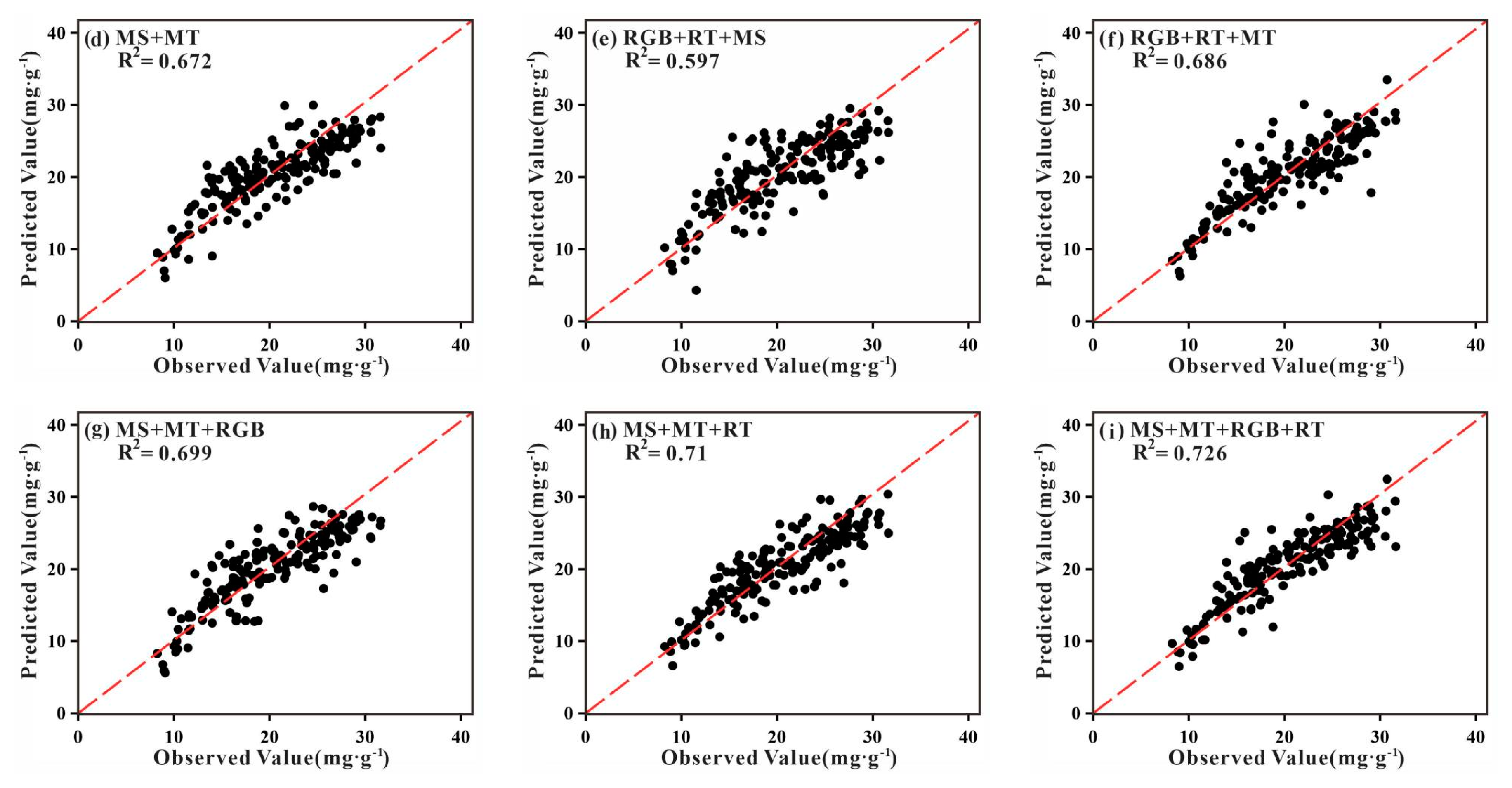
3.2. Analysis of TNC Prediction Accuracy
3.3. Analysis of TNC Observations and Predictions
4. Discussion
4.1. Analysis Based on Multi-Source Spectral Features and Texture Features
4.2. Potential for Ensemble Learning Models
4.3. Implications and Reflections
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Song, Y.; Wang, J. Soybean canopy nitrogen monitoring and prediction using ground based multispectral remote sensors. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 6389–6392. [Google Scholar]
- Tian, Z.; Zhang, Y.; Zhang, H.; Li, Z.; Li, M.; Wu, J.; Liu, K. Winter wheat and soil total nitrogen integrated monitoring based on canopy hyperspectral feature selection and fusion. Comput. Electron. Agric. 2022, 201, 107285. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, Q.; Xiong, S.; Shi, L.; Ma, X.; Du, P.; Guo, J. A spectral parameter for the estimation of soil total nitrogen and nitrate nitrogen of winter wheat growth period. Soil Use Manag. 2021, 37, 698–711. [Google Scholar] [CrossRef]
- Guo, B.; Zang, W.; Luo, W.; Yang, X. Detection model of soil salinization information in the Yellow River Delta based on feature space models with typical surface parameters derived from Landsat8 OLI image. Geomat. Nat. Hazards Risk 2020, 11, 288–300. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, B.; Lu, M.; Zang, W.; Yu, T.; Chen, D. Quantitative distinction of the relative actions of climate change and human activities on vegetation evolution in the Yellow River Basin of China during 1981–2019. J. Arid Land 2022, 15, 91–108. [Google Scholar] [CrossRef]
- Chen, S.T.; Guo, B.; Zhang, R.; Zang, W.Q.; Wei, C.X.; Wu, H.W.; Yang, X.; Zhen, X.Y.; Li, X.; Zhang, D.F.; et al. Quantitatively determine the dominant driving factors of the spatial-temporal changes of vegetation NPP in the Hengduan Mountain area during 2000–2015. J. Mt. Sci. 2021, 18, 427–445. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, J.; Du, Y. Combining multivariate method and spectral variable selection for soil total nitrogen estimation by Vis-NIR spectroscopy. Arch. Agron. Soil Sci. 2021, 67, 1665–1678. [Google Scholar] [CrossRef]
- Lopez-Calderon, M.J.; Estrada-Avalos, J.; Rodriguez-Moreno, V.M.; Mauricio-Ruvalcaba, J.E.; Martinez-Sifuentes, A.R.; Delgado-Ramirez, G.; Miguel-Valle, E. Estimation of Total Nitrogen Content in Forage Maize (Zea mays L.) Using Spectral Indices: Analysis by Random Forest. Agriculture 2020, 10, 451. [Google Scholar] [CrossRef]
- Qiu, Z.; Xiang, H.; Ma, F.; Du, C. Qualifications of Rice Growth Indicators Optimized at Different Growth Stages Using Unmanned Aerial Vehicle Digital Imagery. Remote Sens. 2020, 12, 3228. [Google Scholar] [CrossRef]
- Liu, S.; Yang, G.; Jing, H.; Feng, H.; Li, H.; Chen, P.; Yang, W. Retrieval of winter wheat nitrogen content based on UAV digital image. Trans. Chin. Soc. Agric. Eng. 2019, 35, 75–85. [Google Scholar]
- Possoch, M.; Bieker, S.; Hoffmeister, D.; Bolten, A.; Schellberg, J.; Bareth, G. Ulti-temporal crop surface models combined with the rgb vegetation index from uav-based images for forage monitoring in grassland. In Proceedings of the XXIII ISPRS Congress, Prague, Czech Republic, 12–19 July 2016; pp. 991–998. [Google Scholar]
- Lebourgeois, V.; Begue, A.; Labbe, S.; Houles, M.; Martine, J.F. A light-weight multi-spectral aerial imaging system for nitrogen crop monitoring. Precis. Agric. 2012, 13, 525–541. [Google Scholar] [CrossRef]
- Oscoa, L.P.; Marques Ramos, A.P.; Saito Moriya, E.A.; de Souza, M.; Marcato Junior, J.; Matsubara, E.T.; Imai, N.N.; Creste, J.E. Improvement of leaf nitrogen content inference in Valencia-orange trees applying spectral analysis algorithms in UAV mounted-sensor images. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101907. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Li, D.; Zhou, X.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Evaluation of RGB, Color-Infrared and Multispectral Images Acquired from Unmanned Aerial Systems for the Estimation of Nitrogen Accumulation in Rice. Remote Sens. 2018, 10, 824. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Liang, Y.; Cao, Q.; Tian, Y.; Zhu, Y.; Cao, W.; Liu, X. Using a Portable Active Sensor to Monitor Growth Parameters and Predict Grain Yield of Winter Wheat. Sensors 2019, 19, 1108. [Google Scholar] [CrossRef]
- Laliberte, A.S.; Rango, A. Texture and Scale in Object-Based Analysis of Subdecimeter Resolution Unmanned Aerial Vehicle (UAV) Imagery. IEEE Trans. Geosci. Remote Sens. 2009, 47, 761–770. [Google Scholar] [CrossRef]
- Murray, H.; Lucieer, A.; Williams, R. Texture-based classification of sub-Antarctic vegetation communities on Heard Island. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, 138–149. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, X.; Wu, Y.; Zhu, Y.; Cao, Q.; Liu, X.; Cao, W. Combining texture, color, and vegetation indices from fixed-wing UAS imagery to estimate wheat growth parameters using multivariate regression methods. Comput. Electron. Agric. 2021, 185, 106138. [Google Scholar] [CrossRef]
- Fu, Z.; Yu, S.; Zhang, J.; Xi, H.; Gao, Y.; Lu, R.; Zheng, H.; Zhu, Y.; Cao, W.; Liu, X. Combining UAV multispectral imagery and ecological factors to estimate leaf nitrogen and grain protein content of wheat. Eur. J. Agron. 2022, 132, 126405. [Google Scholar] [CrossRef]
- Geng, R.; Fu, B.; Jin, S.; Cai, J.; Geng, W.; Lou, P. Object-based Karst wetland vegetation classification using UAV images. Bull. Surv. Mapp. 2020, 13–18. [Google Scholar] [CrossRef]
- Jia, D.; Chen, P. Effect of Low-altitude UAV Image Resolution on Inversion of Winter Wheat Nitrogen Concentration. Trans. Chin. Soc. Agric. Mach. 2020, 51, 164–169. [Google Scholar]
- Zhang, H.; Huang, L.; Huang, W.; Dong, Y.; Weng, S.; Zhao, J.; Ma, H.; Liu, L. Detection of wheat Fusarium head blight using UAV-based spectral and image feature fusion. Front. Plant Sci. 2022, 13, 1004427. [Google Scholar] [CrossRef]
- Zha, H.; Miao, Y.; Wang, T.; Li, Y.; Zhang, J.; Sun, W.; Feng, Z.; Kusnierek, K. Improving Unmanned Aerial Vehicle Remote Sensing-Based Rice Nitrogen Nutrition Index Prediction with Machine Learning. Remote Sens. 2020, 12, 215. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, X.; Liu, M.; Wu, L. Random forest algorithm and regional applications of spectral inversion model for estimating canopy nitrogen concentration in rice. J. Remote Sens. 2014, 18, 923–945. [Google Scholar]
- Berger, K.; Verrelst, J.; Feret, J.; Hank, T.; Wocher, M.; Mauser, W.; Camps-Valls, G. Retrieval of aboveground crop nitrogen content with a hybrid machine learning method. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Su, C.; Duan, M. Experiment on sweet pepper nitrogen detection based on near-infrared reflectivity spectral ridge regression. J. Drain. Irrig. Mach. Eng. 2019, 37, 86–92. [Google Scholar]
- Mahajan, G.R.; Das, B.; Murgaokar, D.; Herrmann, I.; Berger, K.; Sahoo, R.N.; Patel, K.; Desai, A.; Morajkar, S.; Kulkarni, R.M. Monitoring the Foliar Nutrients Status of Mango Using Spectroscopy-Based Spectral Indices and PLSR-Combined Machine Learning Models. Remote Sens. 2021, 13, 641. [Google Scholar] [CrossRef]
- Comito, C.; Pizzuti, C. Artificial intelligence for forecasting and diagnosing COVID-19 pandemic: A focused review. Artif. Intell. Med. 2022, 128, 102286. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, Z.; Ma, Y.; Du, Q.; Williams, P.; Drewry, J.; Luck, B. Alfalfa Yield Prediction Using UAV-Based Hyperspectral Imagery and Ensemble Learning. Remote Sens. 2020, 12, 2028. [Google Scholar] [CrossRef]
- Fei, S.; Hassan, M.A.; Xiao, Y.; Su, X.; Chen, Z.; Cheng, Q.; Duan, F.; Chen, R.; Ma, Y. UAV-based multi-sensor data fusion and machine learning algorithm for yield prediction in wheat. Precis. Agric. 2022, 24, 187–212. [Google Scholar] [CrossRef]
- Yang, H.; Hu, Y.; Zheng, Z.; Qiao, Y.; Zhang, K.; Guo, T.; Chen, J. Estimation of Potato Chlorophyll Content from UAV Multispectral Images with Stacking Ensemble Algorithm. Agronomy 2022, 12, 2318. [Google Scholar] [CrossRef]
- Wu, T.; Li, Y.; Ge, Y.; Liu, L.; Xi, S.; Ren, M.; Yuan, X.; Zhuang, C. Estimation of nitrogen contents in citrus leaves using Stacking ensemble learning. Trans. Chin. Soc. Agric. Eng. 2021, 37, 163–171. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Schmidt, K.; Chakan, A.A.; Rentschler, T.; Scholten, T. Improving the Spatial Prediction of Soil Organic Carbon Content in Two Contrasting Climatic Regions by Stacking Machine Learning Models and Rescanning Covariate Space. Remote Sens. 2020, 12, 1095. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Louhaichi, M.; Borman, M.M.; Johnson, D.E. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 56–70. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Vonbargen, K.; Mortensen, D.A. Color indexes for weed identification under various soil, residue, and lighting conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Niu, Q.; Feng, H.; Yang, G.; Li, C.; Yang, H.; Xu, B.; Zhao, Y. Monitoring plant height and leaf area index of maize breeding material based on UAV digital images. Trans. Chin. Soc. Agric. Eng. 2018, 34, 73–82. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.S. Combined spectral and spatial processing of ERTS imagery data. Remote Sens. Environ. 1974, 3, 3–13. [Google Scholar] [CrossRef]
- Datt, B.; McVicar, T.R.; Van Niel, T.G.; Jupp, D.; Pearlman, J.S. Preprocessing EO-1 Hyperion hyperspectral data to support the application of agricultural indexes. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1246–1259. [Google Scholar] [CrossRef]
- Kumar, S.; Gautam, G.; Saha, S.K. Hyperspectral remote sensing data derived spectral indices in characterizing salt-affected soils: A case study of Indo-Gangetic plains of India. Environ. Earth Sci. 2015, 73, 3299–3308. [Google Scholar] [CrossRef]
- El-Shikha, D.M.; Waller, P.; Hunsaker, D.; Clarke, T.; Barnes, E. Ground-based remote sensing for assessing water and nitrogen status of broccoli. Agric. Water Manag. 2007, 92, 183–193. [Google Scholar] [CrossRef]
- Hunt, E.R., Jr.; Daughtry, C.S.T.; Eitel, J.U.H.; Long, D.S. Remote Sensing Leaf Chlorophyll Content Using a Visible Band Index. Agron. J. 2011, 103, 1090–1099. [Google Scholar] [CrossRef]
- Tucker, C.J.; Elgin, J.H., Jr.; McMurtrey, J.E.I.; Fan, C.J. Monitoring corn and soybean crop development with hand-held radiometer spectral data. Remote Sens. Environ. 1979, 8, 237–248. [Google Scholar] [CrossRef]
- Underwood, E.; Ustin, S.; DiPietro, D. Mapping nonnative plants using hyperspectral imagery. Remote Sens. Environ. 2003, 86, 150–161. [Google Scholar] [CrossRef]
- Wang, F.; Huang, J.; Tang, Y.; Wang, X. New Vegetation Index and Its Application in Estimating Leaf Area Index of Rice. Rice Sci. 2007, 14, 195–203. [Google Scholar] [CrossRef]
- Ehammer, A.; Fritsch, S.; Conrad, C.; Lamers, J.; Dech, S. Statistical derivation of fPAR and LAI for irrigated cotton and rice in arid Uzbekistan by combining multi-temporal RapidEye data and ground measurements. In Remote Sensing for Agriculture, Ecosystems, and Hydrology XII, Toulouse, France; SPIE: Bellingham, WA, USA, 2010; Volume 7824, p. 9. [Google Scholar]
- Ren, H.; Zhou, G.; Zhang, F. Using negative soil adjustment factor in soil-adjusted vegetation index (SAVI) for aboveground living biomass estimation in arid grasslands. Remote Sens. Environ. 2018, 209, 439–445. [Google Scholar] [CrossRef]
- Cao, Q.; Miao, Y.; Feng, G.; Gao, X.; Li, F.; Liu, B.; Yue, S.; Cheng, S.; Ustin, S.L.; Khosla, R. Active canopy sensing of winter wheat nitrogen status: An evaluation of two sensor systems. Comput. Electron. Agric. 2015, 112, 54–67. [Google Scholar] [CrossRef]
- Hu, H.; Zheng, K.; Zhang, X.; Lu, Y.; Zhang, H. Nitrogen Status Determination of Rice by Leaf Chlorophyll Fluorescence and Reflectance Properties. Sens. Lett. 2011, 9, 1207–1211. [Google Scholar] [CrossRef]
- Zhou, J.; Yungbluth, D.; Vong, C.N.; Scaboo, A.; Zhou, J. Estimation of the Maturity Date of Soybean Breeding Lines Using UAV-Based Multispectral Imagery. Remote Sens. 2019, 11, 2075. [Google Scholar] [CrossRef]
- Cao, Q.; Miao, Y.; Wang, H.; Huang, S.; Cheng, S.; Khosla, R.; Jiang, R. Non-destructive estimation of rice plant nitrogen status with Crop Circle multispectral active canopy sensor. Field Crop Res. 2013, 154, 133–144. [Google Scholar] [CrossRef]
- Sripada, R.P.; Heiniger, R.W.; White, J.G.; Meijer, A.D. Aerial color infrared photography for determining early in-season nitrogen requirements in corn. Agron. J. 2006, 98, 968–977. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating par absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Ren, H.; Zhou, G. Determination of green aboveground biomass in desert steppe using litter-soil-adjusted vegetation index. Eur. J. Remote Sens. 2014, 47, 611–625. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Gong, P.; Pu, R.L.; Biging, G.S.; Larrieu, M.R. Estimation of forest leaf area index using vegetation indices derived from Hyperion hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1355–1362. [Google Scholar] [CrossRef]
- Karbasi, M.; Jamei, M.; Ahmadianfar, I.; Asadi, A. Toward the accurate estimation of elliptical side orifice discharge coefficient applying two rigorous kernel-based data-intelligence paradigms. Sci. Rep. 2021, 11, 19784. [Google Scholar] [CrossRef]
- Huang, N.; Wang, L.; Song, X.; Black, T.A.; Jassal, R.S.; Myneni, R.B.; Wu, C.; Wang, L.; Song, W.; Ji, D.; et al. Spatial and temporal variations in global soil respiration and their relationships with climate and land cover. Sci. Adv. 2020, 6, eabb8508. [Google Scholar] [CrossRef]
- Nakagome, S.; Trieu, P.L.; He, Y.; Ravindran, A.S.; Contreras-Vidal, J.L. An empirical comparison of neural networks and machine learning algorithms for EEG gait decoding. Sci. Rep. 2020, 10, 4372. [Google Scholar] [CrossRef]
- Galie, F.; Rospleszcz, S.; Keeser, D.; Beller, E.; Illigens, B.; Lorbeer, R.; Grosu, S.; Selder, S.; Auweter, S.; Schlett, C.L.; et al. Machine-learning based exploration of determinants of gray matter volume in the KORA-MRI study. Sci. Rep. 2020, 10, 8363. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Z.; Cheng, Q.; Duan, F.; Sui, R.; Huang, X.; Xu, H. UAV-Based Hyperspectral and Ensemble Machine Learning for Predicting Yield in Winter Wheat. Agronomy 2022, 12, 202. [Google Scholar] [CrossRef]
- Coburn, C.A.; Smith, A.M.; Logie, G.S.; Kennedy, P. Radiometric and spectral comparison of inexpensive camera systems used for remote sensing. Int. J. Remote Sens. 2018, 39, 4869–4890. [Google Scholar] [CrossRef]
- Furukawa, F.; Laneng, L.A.; Ando, H.; Yoshimura, N.; Kaneko, M.; Morimoto, J. Comparison of RGB and Multispectral Unmanned Aerial Vehicle for Monitoring Vegetation Coverage Changes on a Landslide Area. Drones 2021, 5, 97. [Google Scholar] [CrossRef]
- Wang, F.; Yi, Q.; Hu, J.; Xie, L.; Yao, X.; Xu, T.; Zheng, J. Combining spectral and textural information in UAV hyperspectral images to estimate rice grain yield. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102397. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, H.; Yue, J.; Li, Z.; Yang, G.; Song, X.; Yang, X.; Zhao, Y. Remote-sensing estimation of potato above-ground biomass based on spectral and spatial features extracted from high-definition digital camera images. Comput. Electron. Agric. 2022, 198, 107089. [Google Scholar] [CrossRef]
- AlSuwaidi, A.; Grieve, B.; Yin, H. Combining spectral and texture features in hyperspectral image analysis for plant monitoring. Meas. Sci. Technol. 2018, 29, 104001. [Google Scholar] [CrossRef]
- Xu, X.; Fan, L.; Li, Z.; Meng, Y.; Feng, H.; Yang, H.; Xu, B. Estimating Leaf Nitrogen Content in Corn Based on Information Fusion of Multiple-Sensor Imagery from UAV. Remote Sens. 2021, 13, 340. [Google Scholar] [CrossRef]
- Peterson, K.T.; Sagan, V.; Sidike, P.; Hasenmueller, E.A.; Sloan, J.J.; Knouft, J.H. Machine Learning-Based Ensemble Prediction of Water-Quality Variables Using Feature-Level and Decision-Level Fusion with Proximal Remote Sensing. Photogramm. Eng. Remote Sens. 2019, 85, 269–280. [Google Scholar] [CrossRef]
- Wang, J.; Shi, T.; Yu, D.; Teng, D.; Ge, X.; Zhang, Z.; Yang, X.; Wang, H.; Wu, G. Ensemble machine-learning-based framework for estimating total nitrogen concentration in water using drone-borne hyperspectral imagery of emergent plants: A case study in an arid oasis, NW China. Environ. Pollut. 2020, 266, 115412. [Google Scholar] [CrossRef]
- Rossel, R.A.V.; Webster, R.; Bui, E.N.; Baldock, J.A. Baseline map of organic carbon in Australian soil to support national carbon accounting and monitoring under climate change. Glob. Chang. Biol. 2014, 20, 2953–2970. [Google Scholar] [CrossRef] [PubMed]
- Frame, J.; Merrilees, D.W. The effect of tractor wheel passes on herbage production from diploid and tetraploid ryegrass swards. Grass Forage Sci. 1996, 51, 13–20. [Google Scholar] [CrossRef]
- Zhou, K.; Yang, Y.; Qiao, Y.; Xiang, T. Domain Adaptive Ensemble Learning. IEEE Trans. Image Process. 2021, 30, 8008–8018. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Sun, J.; Wang, Y.; Yang, W.; He, H.; Liu, L.; Huang, L.; Fan, D.; Gao, E. Evaluation of LAI Estimation of Mangrove Communities Using DLR and ELR Algorithms With UAV, Hyperspectral, and SAR Images. Front. Mar. Sci. 2022, 9, 944454. [Google Scholar] [CrossRef]
- Osco, L.P.; Marques Ramos, A.P.; Faita Pinheiro, M.M.; Saito Moriya, E.A.; Imai, N.N.; Estrabis, N.; Ianczyk, F.; de Araujo, F.F.; Liesenberg, V.; de Castro Jorge, L.A.; et al. A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements. Remote Sens. 2020, 12, 906. [Google Scholar] [CrossRef]
- Osco, L.P.; Marcato Junior, J.; Marques Ramos, A.P.; Garcia Furuya, D.E.; Santana, D.C.; Ribeiro Teodoro, L.P.; Goncalves, W.N.; Rojo Baio, F.H.; Pistori, H.; Da Silva Junior, C.A.; et al. Leaf Nitrogen Concentration and Plant Height Prediction for Maize Using UAV-Based Multispectral Imagery and Machine Learning Techniques. Remote Sens. 2020, 12, 3237. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, Z.; Sassoubre, L.; Yu, G.; Wang, Y. Improving the robustness of beach water quality modeling using an ensemble machine learning approach. Sci. Total Environ. 2020, 765, 142760. [Google Scholar] [CrossRef]
- Jyab, C.; Ma, C.; Eb, D.; Zhu, L.A. Bayesian machine learning ensemble approach to quantify model uncertainty in predicting groundwater storage change. Sci. Total Environ. 2021, 769, 144715. [Google Scholar]
- Singh, H.; Roy, A.; Setia, R.K.; Pateriya, B. Estimation of nitrogen content in wheat from proximal hyperspectral data using machine learning and explainable artificial intelligence (XAI) approach. Model. Earth Syst. Environ. 2022, 8, 2505–2511. [Google Scholar] [CrossRef]










| Treatments | Jointing Stage (kg·hm−2) | Heading Stage (kg·hm−2) | Fertiliser Types |
|---|---|---|---|
| N1 | 200 | 100 | Urea |
| N2 | 120 | 60 | Urea |
| N3 | 40 | 20 | Urea |
| Data Type | Feature | Formula | Source |
|---|---|---|---|
| RGB | r | / | |
| g | / | ||
| b | / | ||
| Visible atmospherically resistant index | [35] | ||
| Ground-level image index | [36] | ||
| Green, red vegetation index | [37] | ||
| Excess red index | [38] | ||
| Normalised difference index | [39] | ||
| g/r | [40] | ||
| r/b | [40] | ||
| Grey-level co-occurrence matrix | ME, HO, DI, EN, SE, VA, CO, COR | [41] |
| Data Type | Feature | Formula | Source |
|---|---|---|---|
| MS | Chlorophyll vegetation index | [42] | |
| Colouration index | [43] | ||
| Canopy chlorophyll content index | [44] | ||
| Chlorophyll index Red-edge | [45] | ||
| Green difference vegetation index | [46] | ||
| Normalised difference vegetation index | [47] | ||
| Green NDVI | [48] | ||
| Normalised difference red-edge | [49] | ||
| Green soil adjusted vegetation index | [50] | ||
| Green optimised soil adjusted vegetation index | [51] | ||
| Nitrogen reflectance index | [52] | ||
| Green ratio vegetation index | [53] | ||
| Normalised red-edge index | [54] | ||
| Normalised NIR index | [55] | ||
| Modified normalised difference index | [54] | ||
| Difference vegetation index | [37] | ||
| Renormalised difference vegetation index | [56] | ||
| Soil-adjusted vegetation index | [57] | ||
| Optimised SAVI | [58] | ||
| MERIS terrestrial chlorophyll index | [59] | ||
| ModifiedNon-linear index | [60] | ||
| Grey-level co-occurrence matrix | ME, HO, DI, EN, SE, VA, CO, COR | [41] |
| Category | Observations | Min | Max | Mean | SD | Q25 | Q50 | Q75 | CV |
|---|---|---|---|---|---|---|---|---|---|
| All datasets | 180 | 8.26 | 31.63 | 20.07 | 5.70 | 16.12 | 19.28 | 24.74 | 0.28 |
| N1 dataset | 60 | 15.33 | 31.63 | 23.66 | 4.38 | 20.44 | 24.50 | 26.97 | 0.19 |
| N2 dataset | 60 | 12.20 | 30.72 | 21.28 | 5.00 | 17.40 | 20.68 | 25.47 | 0.23 |
| N3 dataset | 60 | 8.26 | 26.34 | 15.28 | 4.07 | 11.66 | 15.56 | 18.01 | 0.27 |
| Sensor Type | Feature Type | Metrics | GPR | RFR | RR | ENR | Stacking (RR) |
|---|---|---|---|---|---|---|---|
| RGB | Spectral | R2 | 0.493 | 0.382 | 0.481 | 0.479 | 0.511 |
| RMSE (mg·g−1) | 4.273 | 4.591 | 4.303 | 4.401 | 4.216 | ||
| MSE (mg·g−1) | 18.259 | 21.077 | 18.516 | 19.369 | 17.775 | ||
| RPD | 1.386 | 1.279 | 1.374 | 1.342 | 1.384 | ||
| RPIQ | 2.083 | 1.962 | 2.069 | 2.026 | 2.125 | ||
| MS | Spectral | R2 | 0.541 | 0.465 | 0.515 | 0.505 | 0.551 |
| RMSE (mg·g−1) | 4.013 | 4.205 | 4.149 | 4.174 | 3.978 | ||
| MSE (mg·g−1) | 16.104 | 17.682 | 17.214 | 17.422 | 15.824 | ||
| RPD | 1.468 | 1.373 | 1.420 | 1.405 | 1.468 | ||
| RPIQ | 2.194 | 2.068 | 2.113 | 2.104 | 2.198 | ||
| RGB + RGB | Spectral + textural | R2 | 0.494 | 0.531 | 0.509 | 0.507 | 0.562 |
| RMSE (mg·g−1) | 4.179 | 3.955 | 4.138 | 4.158 | 3.947 | ||
| MSE (mg·g−1) | 17.464 | 15.642 | 17.123 | 17.289 | 15.579 | ||
| RPD | 1.401 | 1.466 | 1.413 | 1.395 | 1.469 | ||
| RPIQ | 2.156 | 2.262 | 2.178 | 2.165 | 2.280 | ||
| MS + MS | Spectral + textural | R2 | 0.625 | 0.650 | 0.630 | 0.625 | 0.672 |
| RMSE (mg·g−1) | 3.610 | 3.536 | 3.584 | 3.608 | 3.415 | ||
| MSE (mg·g−1) | 13.032 | 12.503 | 12.845 | 13.018 | 11.662 | ||
| RPD | 1.641 | 1.686 | 1.657 | 1.645 | 1.738 | ||
| RPIQ | 2.483 | 2.543 | 2.490 | 2.478 | 2.625 | ||
| RGB + MS + RGB | Spectral + spectral + textural | R2 | 0.570 | 0.554 | 0.554 | 0.546 | 0.597 |
| RMSE (mg·g−1) | 3.936 | 3.881 | 3.942 | 3.991 | 3.788 | ||
| MSE (mg·g−1) | 15.492 | 15.062 | 15.539 | 15.928 | 14.349 | ||
| RPD | 1.504 | 1.504 | 1.484 | 1.468 | 1.540 | ||
| RPIQ | 2.256 | 2.278 | 2.239 | 2.224 | 2.337 | ||
| RGB + RGB + MS | Spectral + textural + textural | R2 | 0.651 | 0.651 | 0.671 | 0.662 | 0.686 |
| RMSE (mg·g−1) | 3.599 | 3.553 | 3.468 | 3.495 | 3.386 | ||
| MSE (mg·g−1) | 12.953 | 12.624 | 12.027 | 12.215 | 11.465 | ||
| RPD | 1.689 | 1.680 | 1.742 | 1.719 | 1.765 | ||
| RPIQ | 2.508 | 2.494 | 2.576 | 2.544 | 2.628 | ||
| RGB + MS + MS | Spectral + spectral + textural | R2 | 0.659 | 0.675 | 0.668 | 0.664 | 0.699 |
| RMSE (mg·g−1) | 3.487 | 3.466 | 3.433 | 3.438 | 3.300 | ||
| MSE (mg·g−1) | 12.159 | 12.013 | 11.785 | 11.820 | 10.890 | ||
| RPD | 1.720 | 1.714 | 1.745 | 1.734 | 1.802 | ||
| RPIQ | 2.568 | 2.534 | 2.579 | 2.562 | 2.668 | ||
| MS + RGB + MS | Spectral + textural + textural | R2 | 0.666 | 0.675 | 0.671 | 0.675 | 0.710 |
| RMSE (mg·g−1) | 3.504 | 3.404 | 3.435 | 3.416 | 3.257 | ||
| MSE (mg·g−1) | 12.278 | 11.587 | 11.799 | 11.669 | 10.608 | ||
| RPD | 1.719 | 1.713 | 1.738 | 1.734 | 1.802 | ||
| RPIQ | 2.605 | 2.637 | 2.639 | 2.643 | 2.746 | ||
| RGB + MS + RGB + MS | Spectral + spectral + textural + textural | R2 | 0.670 | 0.697 | 0.700 | 0.692 | 0.726 |
| RMSE (mg·g−1) | 3.456 | 3.365 | 3.352 | 3.362 | 3.203 | ||
| MSE (mg·g−1) | 11.944 | 11.323 | 11.236 | 11.303 | 10.259 | ||
| RPD | 1.735 | 1.769 | 1.822 | 1.798 | 1.867 | ||
| RPIQ | 2.647 | 2.731 | 2.724 | 2.708 | 2.827 |
| Feature | t | p-Value |
|---|---|---|
| N1 vs. N2 | 3.847 | 0.000 |
| N1 vs. N3 | 9.416 | 0.000 |
| N2 vs. N3 | 5.654 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Zhou, X.; Cheng, Q.; Fei, S.; Chen, Z. A Machine-Learning Model Based on the Fusion of Spectral and Textural Features from UAV Multi-Sensors to Analyse the Total Nitrogen Content in Winter Wheat. Remote Sens. 2023, 15, 2152. https://doi.org/10.3390/rs15082152
Li Z, Zhou X, Cheng Q, Fei S, Chen Z. A Machine-Learning Model Based on the Fusion of Spectral and Textural Features from UAV Multi-Sensors to Analyse the Total Nitrogen Content in Winter Wheat. Remote Sensing. 2023; 15(8):2152. https://doi.org/10.3390/rs15082152
Chicago/Turabian StyleLi, Zongpeng, Xinguo Zhou, Qian Cheng, Shuaipeng Fei, and Zhen Chen. 2023. "A Machine-Learning Model Based on the Fusion of Spectral and Textural Features from UAV Multi-Sensors to Analyse the Total Nitrogen Content in Winter Wheat" Remote Sensing 15, no. 8: 2152. https://doi.org/10.3390/rs15082152
APA StyleLi, Z., Zhou, X., Cheng, Q., Fei, S., & Chen, Z. (2023). A Machine-Learning Model Based on the Fusion of Spectral and Textural Features from UAV Multi-Sensors to Analyse the Total Nitrogen Content in Winter Wheat. Remote Sensing, 15(8), 2152. https://doi.org/10.3390/rs15082152






