1. Introduction
The palm oil industry is now the fourth largest contributor to the Malaysian economy [
1] and plays an important role in other countries, such as Indonesia [
2], Thailand [
3], and Africa [
4]. It is well documented that oil palm trees need a comprehensive and ongoing understanding of their current state, since oil palms are subjected to major pathogens that can threaten the production of palm oil. Among the crucial diseases that requires attention, Basal Stem Rot (BSR) is a root disease that has been estimated to cost as much as USD 500 million a year to oil palm producers in some Southeast Asian countries [
5]. Hence, in order to improve planning and management decisions, information on BSR infected oil palms is needed. The identification of the disease, however, is very difficult due to the fact that there is no symptom at the early stages of infection. It is only when the disease has reached a critical stage that symptoms of infection usually manifest, which include the following: rot of the base of the stem from where basidiocarps of Ganoderma Boninense rise, as well as the rot of the roots and chlorosis. At the advanced level, wilting and skirting of the older fronds could be observed [
6], along with leveling of the crown and unopened spears and cracking of the stem [
7].
Currently, the early identification of infected palms can be performed by some techniques, such as (I) the colorimetric strategy utilizing Ethylenediamine Tetra-acetic Acid (EDTA) [
8], (II) Ganoderma-semi-selective media on agar plates [
9], (III) Ganoderma-semi-selective media [
10], (IV) Polymerase Chain Response (PCR) [
11], (V) Ganoderma Selective Media (GSM) testing for any tainted tissues [
12], and Volatile Organic Compounds (VOCs) [
10]. Nevertheless, the majority of these techniques are tedious, costly, and not feasible for applications in plantation areas. A perfect strategy for the detection of diseases requires minimal sample preparation, preciseness, and the quality of being non-destructive [
13].
In contrast to other remote sensing applications, a UAV-based method can be applied as a practical substitute due to lower cost funds, widespread utilization, better resolution (from many meters to a few centimeters), and, more noteworthily, adaptability in selecting reasonable payloads and fitting time and/or space resolutions [
14]. Rendana et al. [
15] recommended the use of a UAV system in small estates for the best results, given its reasonably precise coverage with low-altitude aircraft for large area coverage. By means of UAV imagery, it is possible to provide the best policies and strategies for agriculture towards desired scenarios with timely and consistent information to support stakeholder’s decisions because of their flexibility in flight control, accuracy in data and signal processing, off-board sensors, and lower cost than other existing tools [
16,
17,
18,
19,
20,
21].
Approaches utilizing UAV imagery for tree health monitoring has gained popularity in recent years. There were several investigations in agricultural areas and test fields that were performed on forestry inventory [
19,
22,
23] relative to fruit orchards [
24,
25,
26,
27,
28,
29]. Most of the imagery research utilized color and multispectral images, although the latter showed higher calibration ability and more accurate measurements for disease detection [
30]. The use of UAVs has avoided many limitations related to satellite data, such as low spatial resolution, cloud spots, and long waiting times in relatively small geographical areas; nevertheless, its uses for large-scale application can be limited because it is time consuming and costly [
31].
In disease-related studies employing unmanned or manned aerial systems, Calderon et al. [
25] investigated the capability of thermal and hyperspectral imagery obtained by using a manned aircraft to detect early stage verticillium wilt infection in olive caused by Verticillium dahliae Kleb. The acquired images were analysed using Linear Discriminant Analysis (LDA) and Support Vector Machine (SVM) classifiers. The severity classes were divided into five categories: asymptomatic, initial, low, moderate, and severe symptoms. Their results depicted that SVM obtained higher overall accuracy (79.2%) compared to LDA (59.0%); however, the latter was better in discriminating trees at their initial stage of disease. López-López et al. [
26] demonstrated the possibility of early detecting red leaf blotch (Prunus amigdalus), which is a fungal foliar disease of almond using high-resolution hyperspectral and thermal imagery, also retrieved by an aircraft. In order to discriminate between different levels of Prunus Amygdalus Dulcis severity classes (asymptomatic, initial, moderate, and high-severity), linear and nonlinear classification methods based on the forward stepwise discriminant were integrated with vegetation indices. Based on their results, linear and nonlinear models were efficient in separating healthy and severely infected trees and could be used to discriminate healthy plants from those at early disease stages. The model correctly classified 59.6% of the total sampled trees, whereas the individual classification accuracy was 64.3%, 20.0%, 70.6%, and 72.7% for asymptomatic, initial, moderate, and high-severity categories, respectively.
The spatial and spectral requirements for rapid and accurate detection of the lethal Laurel wilt disease that infected Avocado (Persea americana) were investigated by De Castro et al. [
27]. They assessed a Multiple Camera Array (MCA)-6 Tetracam camera with applied filters (580, 650, 740, 750, 760, and 850 nm) from a helicopter at three different altitudes (180, 250, and 300 m) in an avocado field with two classes (healthy and infected) and four classes (healthy, early, intermediate, and late severity) systems. They also tested more than 20 vegetation indices that were potentially used to differentiate between these infection classes. For the aforementioned research purpose, they reported that the ideal flight altitude was 250 m, which resulted in 15.3 cm pixel sizes, and the optimum vegetation index was the Transformed Chlorophyll Absorption Reflectance Index 760–650 (TCARI 760–650). A research study by Smigaj et al. [
28] showed the merit of using an affordable fixed-wing UAV with a thermal sensor for monitoring canopy temperature induced by needle blight infection in a diseased Scot pine. Their results indicated that the UAV-borne camera could detect the disease through temperature differences with acceptable accuracy (
). These studies have demonstrated that canopy physiological indices, such as Normalized Difference Vegetation Index (NDVI) [
32], chlorophyll fluorescence, and temperature, were connected with physiological stress brought about by diseases.
García-Ruiz et al. [
29] compared the use of a hyperspectral sensor AISA EAGLE VNIR Sensor (Specim Ltd., Oulu, Finland) onboard a manned aircraft, with a multiband imaging UAV-based sensor, miniMCA6 (Tetracam, Inc., Chatsworth, CA, USA). The former has 397 to 998 nm spectral range and 128 spectral bands, while the latter was a six narrow-band multispectral camera with image resolution of
pixels. Their study on citrus Huanglongbing diseases indicated the applicability of visible-near infrared spectroscopy at 710 nm and Near-Infrared (NIR)-R index values to discriminate between healthy or infected citrus plants, along with SVM with kernels. The accuracy of classification from the UAV based image was in the range of 67% to 85%, while for aircraft-based data, the corresponding values were slightly lower, which were from 61% to 74%.
In BSR disease-related research, the results derived from hyperspectral reflectance (non-imagery) indicated that disease infection caused by Ganoderma boninese resulted in significant reflectance changes, especially within the near infrared spectra [
30,
33,
34,
35,
36], where the reflectance changes associated with this disease might as well be detected with a digital camera equipped with a NIR filter. To date, there is no report yet on the early detection of Ganoderma-infected palms from UAV platforms.
Moreover, Artificial Neural Network (ANN) has a remarkable capability to estimate complex nonlinear functions with an unknown model precisely, owing to its high effectiveness to characterize parallel distributed processing, non-linear mapping, and adaptive learning. In agricultural applications, ANN has been used to predict crop yield [
37,
38,
39] among others to model fertilizer requirement [
40]. In plant diseases recognition and prediction, ANNs have exhibited powerful discriminant capabilities given the sets of the best trainer [
41,
42,
43,
44,
45]. Moshou et al. [
45], for example, classified spectrograph-acquired spectral images to discriminate between healthy and yellow rusted wheat plants using quadratic discriminant analysis and ANN based algorithms. The classification accuracies of the latter were found to be better than the former, despite the fact that the overall accuracies of both classifiers were more than 90%. Wang et al. [
43] compared ANN and Partial Least-Squares (PLS) models for classifying visible-IR reflectance data to identify healthy and multiple fungal-damaged soybean seeds such as downy mildew and soybean mosaic virus. The authors reported that while the PLS model could differentiate only between healthy and damaged seeds with 99% accuracy, ANN could discriminate between various fungal damages with accuracies between 84% to 100%. Sannakki et al. [
46] used ANN models to classify and distinguish between downy mildew and powdery mildew on grape leaves. Through a series of image-processing techniques such as image segmentation, feature extraction, and back propagation ANN classification performed on Red–Green–Blue (RGB) images, they achieved 100% training accuracy. Zhang et al. [
47] compared spectral based models developed using statistical and ANN approaches for predicting rice neck blasts severity levels. They reported that the latter provided better accuracies of disease index in differentiating disease severity levels. Laurindo et al. [
48] investigated the efficiency of several ANN algorithms for the early detection of tomato blight disease by utilizing an area under the disease progress curve as the disease indicator. Satisfactory results were produced by using the ANN given that most of the tested networks resulted in correlation values more than 0.90 between actual and predicted values.
Many scientists conducted research to detect BSR [
30,
31,
32,
33,
34,
35]; however, the lack of an algorithm and principles for conducting the detection of this disease within plantation areas remains a major setback. To overcome the aforementioned limitations, this study seeks to evaluate operational methods to facilitate extraction of Ganoderma-infected palms at their early stage by using a combination of UAV imagery and ANN techniques. This work is an extension of the previously published article by Ahmadi et al. [
49]. The previous article applied the ANN analysis technique (Multilayer and Back-Propagation) for discriminating fungal infections at leaf scale and frond scale using spectroradiometer reflectance in oil palm trees at an early stage. However, the current work explores the potential of canopy level spectral measurements acquired from UAV imagery with the help of ANN (Levenberg–Marquardt). Moreover, the current study reveals the automatic extraction of tree crowns to calculate the mean reflectance of each individual palm. The rest of this article is organized as follows. In
Section 2, the study region, datasets, and methodology are described. The results are presented in
Section 3. In
Section 4, a discussion is made in light of other studies; finally,
Section 5 concludes this article.
3. Results
Since our main objective is to detect Ganoderma-infected palms early or to technically discriminate between T1 and T2 palms, we emphasize our discussion on the classification accuracies of the T1 and T2 classes only. According to the classification accuracy for 287 oil palm samples that were classified into three disease levels (T1, T2, and T3), the best classification result was generated by the green and NIR bands with a circle radius of 35 pixels, 1/8 threshold limit, and ANN network of 219 hidden neurons, with a classification error of 14.29% (highlighted in
Table 4).
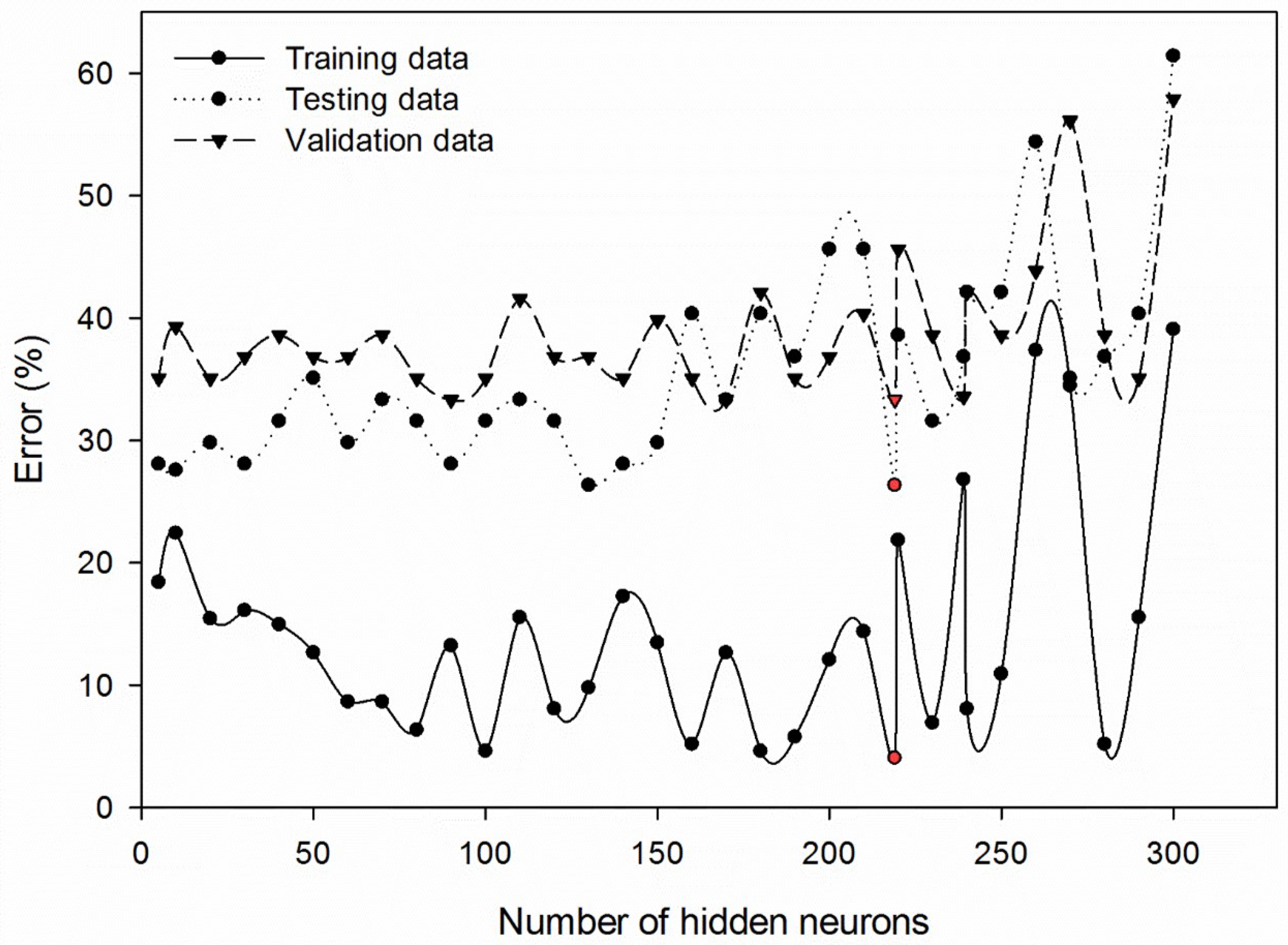
Additionally, the number of ANN hidden neurons that would result in the minimum training, testing, and validation errors was simultaneously tested with the combination of the best image configurations, as previously discussed. For the sake of simplicity, we illustrated training, testing, and validation errors for a number of hidden neurons at 5, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210, 219, 220, 230, 239, 240, 250, 260, 270, 280, 290, and 300 for the combination of 1/8 threshold limit, 35 pixels circle radius, and green and NIR bands (
Figure 2). The neuron numbers of 219 depicted the lowest errors of training, validation, and testing were 4.05%, 33.33%, and 26.32%, respectively (see the red-colored markers).
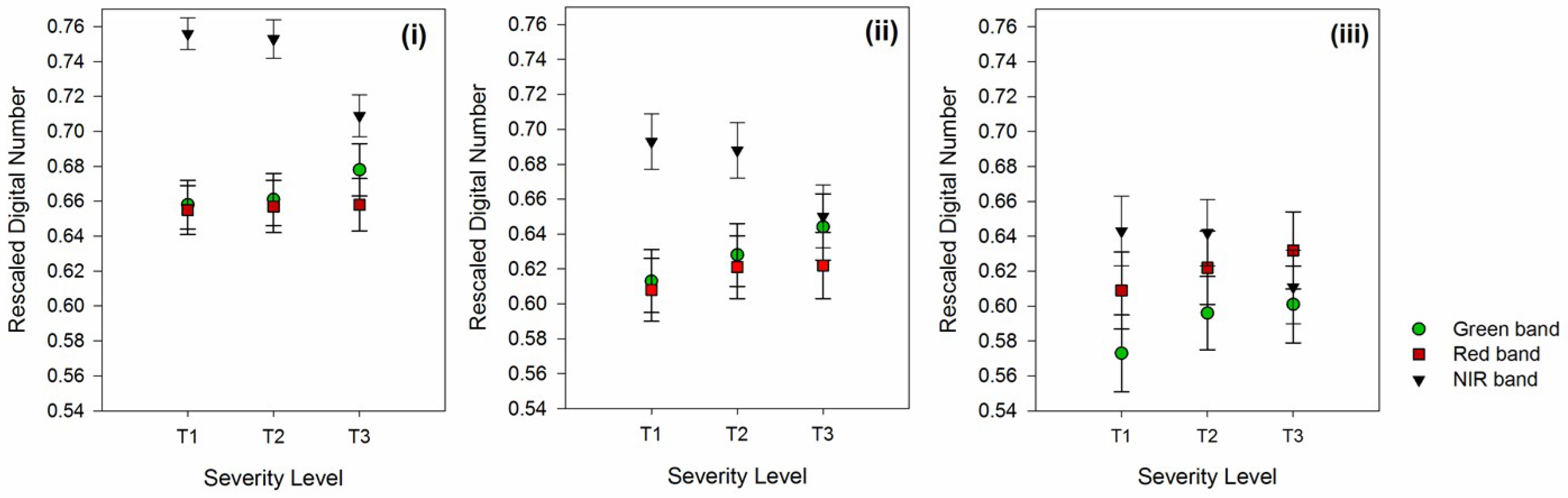
Further investigations on the effects of different circle radii on the mean and standard deviation of pixel values at those selected bands were performed, as shown in
Figure 3. As mentioned above, all DNs were normalized and rescaled to double precision. In general, the image processing toolbox functions in the software utilized prefer DNs in the range of 0 to 1, hence explaining the magnitude of the mean values that were less than 1. There are two prominent observations that could be inferred from the table: (i) mean DN values in all bands decreased with increasing circle radius, while the standard deviations among various severity levels are approximately the same, and (ii) the maximum separation values among severity classes were noticeable at a larger circle radius. For the circle radius of 25 pixels, the mean DN value for T1 and T2 palms in all bands illustrated minimal differences, which were 0.658 and 0.661 in the green band, 0.655 and 0.657 in the red band, and 0.756 and 0.753 in the NIR band, for instance (
Figure 3i). A similar observation was made for the circle radius that equaled 45 pixels, especially in the NIR band where the mean DN for T1 and T2 was 0.643 and 0.642, respectively. However, in green and red bands, the differences were slightly larger, which were 0.573 and 0.596 and 0.609 and 0.622, respectively (
Figure 3iii). These values provided an inadequate separability between severity classes, especially for T1 and T2, which allows for an early detection strategy. Nevertheless, the largest separability between the mean value of T1 and T2 was observed in the circle radius of 35 pixels, for instance, 0.613 and 0.628 in the green band, 0.608 and 0.621 in the red band, and 0.693 and 0.688 in the NIR band (
Figure 3ii). It was also noticeable that standard deviation increased by increasing pixel size, despite the values between T1 and T2 being almost similar, such as 0.014 and 0.015 in the green band, for the circle radius of 25 pixels and 0.018 and 0.018 in the green band, for the circle radius of 35 pixels.
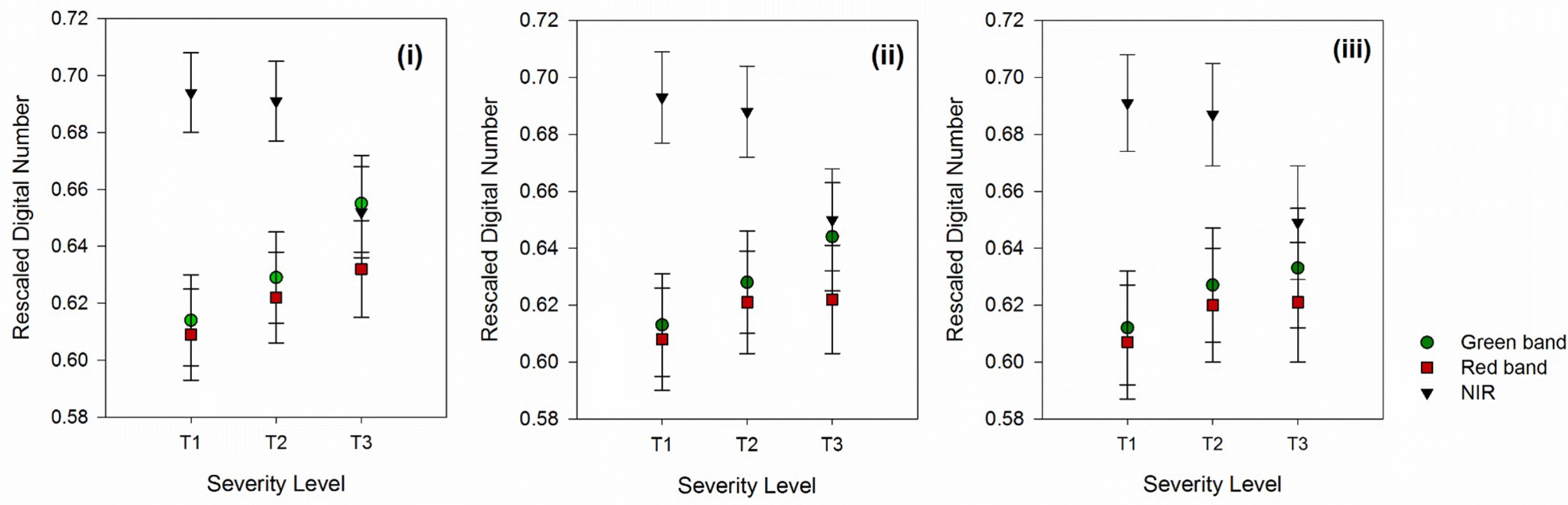
Investigation on threshold limit effects on the mean and standard deviation of pixel values at selected bands (
Figure 4) illustrated that although increasing the threshold limit resulted in larger mean and smaller standard deviation values, such differences were negligible among the threshold limits. For instance, for the green band, the mean DN values for T1 for the threshold limit of 1/7, 1/8, and 1/9 were 0.614, 0.613, and 0.612, respectively (
Figure 4i–iii). Likewise, for T2, the mean DN was 0.629, 0.628, and 0.627 for the threshold limit of 1/7, 1/8, and 1/9, respectively. It is also shown in
Table 4 that the adjustment of the threshold limit did not result in better classification results.
For training and testing samples, total classification accuracies of 97.52% and 72.73% were obtained, respectively. The training model with the explained properties could detect healthy or T1 palms with 99.27% accuracy and 88.50% for T2 palms (
Table 5). Nevertheless, the testing result was lower for the corresponding classes, which were 75.00% for T1 and 57.14% for T2 (
Table 6).
4. Discussion
The findings depicted that, for image configurations, the best combination of spectral bands to identify early infected palms was green and NIR. These results are in accordance with studies indicating that diseased or stressed vegetation showed a reflectance increment in the visible band and a decrease in NIR reflectance [
64,
65]. We hypothesize that the green band was more beneficial than the red one in detecting early Ganoderma-infected palms because the latter is confined to moderate to high chlorophyll content [
65] and becomes easily saturated at intermediate values of leaf area index [
66]. Considering that oil palm is a perennial crop and, therefore, can have a wide range of chlorophyll contents and leaf areas and the fact that BSR disease will lead to chlorosis [
67] through reductions in the palms’ nutrient and water uptake, the use of a green band resulted in better discrimination accuracies because it is not as easily saturated as the red band [
6].
The observed patterns between the mean and standard deviations values with the circle radius and threshold limit (
Figure 3 and
Figure 4) suggested that the determination of the threshold limit might not be as essential as for optimizing the circle’s radius. The best classification results indicated that the circle radius has to be 140 times GSD of the original UAV image or 35 times the pixel size of the resampled UAV image, which is approximately 3.6 m from the center of the canopy, where oil palm is commonly cultivated in 9 m planting distance. Physiologically, these pixels signified that the more developed fronds could be the best approach to the early detection of BSR disease. In an oil palm tree, a newly developed frond will emerge from the most top center of the canopy. The fronds will grow in a spiral form, either in a clockwise or counter-clockwise direction. Over time, more developed fronds will ascend to the lower part of the canopy. The younger fronds usually have a vertical angle, while more developed fronds lie horizontally. With the distance of 3.6 m from the center of canopy, signals related to Ganoderma infection are more representative from the more developed fronds that form the first spiral layer of the canopy than the young ones.
The intergroup classification accuracy of healthy palms (T1) in comparison to early infected ones (T2) was considerably better since healthier palms have more distinctive reflectance responses affected by chlorophyll quantity and structure of leaf cells that were depicted through low visible and high NIR reflectance. Therefore, with an increasing severity level of BSR disease, the accuracy of the model and the possibility of prediction by the UAV image became lower. At first glance, the validation accuracy for T2 suggested a mediocre classification capability. However, half of the T2 palms were misclassified as T1 possibly because, at an early stage of infection, a disease has not had much effect on physiological properties of plants such as on chlorophyll content [
67]. Nonetheless, even with limited palm properties such as affected chlorophyll content and the absence of any visual symptoms, the ANN model and the modified NIR camera could still detect more than half of infected palms correctly.
We also compared our results to those obtained by Kresnawaty et al. [
33], Liaghat et al. [
35], and Lelong et al. [
30], who reported 100%, 89%, and 92% accuracies for early detection of Ganoderma-infected oil palms. The latter obtained leaf spectra from a non-imaging spectroradiometer and the former utilized canopy reflectance that was also acquired from a non-imaging spectroradiometer. Better accuracies derived from these studies resulted from ground, destructive, individual leaf sampling that was time consuming and laborious compared to our rapid, non-destructive approach, yet it gained reasonable accuracies. While the use of UAV image in this study cannot justify the detection of BSR as good as ground hyperspectral equipment, this method is appropriate for field applications involving mass screening of oil palm plantations that are commonly cultivated in thousands of hectares in seeking potentially infected individual palms. Furthermore, based on another study conducted by the authors, the Support vector machine (SVM) algorithm, which is the best alternative method for non-parametric approaches and large data sets, has not achieved high accuracy for UAV image classification.
As stated earlier, previous studies were able to present high discrimination accuracy between healthy and moderately infected samples compared to the results by Shafri et al. [
34] and Liaghat et al. [
35]. However, the focus of this research is in mild or early stage infection where there is no visible symptom of infection that has yet appeared. To our best knowledge, no extensive study has been made to test the spectral indices on airborne hyperspectral image for the early detection of Ganoderma.
These results account for the results of previous studies that did not provide good performance in the classification of early-stage infection. Satellite multispectral remote sensing [
68] had the adequate spatial resolution (2.4 m) for the crown’s extraction However, its spectral resolution (60–140 nm) was inadequate for the detailed analysis of the red-edge region to allow for early detection. The aircraft-based hyperspectral remote sensing [
69,
70] had an adequately high spectral resolution (<10 nm) for the investigation of spectral features in the red-edge region. However, the spatial resolution (1 m) was too high to conduct pixel-based classification. If object-based classification was performed using the central portion of tree crowns, the aircraft-based hyperspectral remote sensing could have a better result for early detection. Lastly, the previous research method by Ahmadi et al. [
49] was time consuming and destructive compared to the current study, which is rapid, non-destructive, and on the canopy level.