UAV-Based Estimation of Grain Yield for Plant Breeding: Applied Strategies for Optimizing the Use of Sensors, Vegetation Indices, Growth Stages, and Machine Learning Algorithms
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Trials
2.2. UAV Imagery Acquisition and Preprocessing
2.3. Postprocessing of Spectral Data
| Spectral Parameter | Equation | Description | Reference |
|---|---|---|---|
| Multispectral (MS) Camera | |||
| M_G | Green band, 530 nm 1 | ||
| M_NDRE1 | Normalized difference red edge index 1 | [45] | |
| M_NDRE2 | Normalized difference red edge index 2 | [45] | |
| M_NDVI | Normalized difference vegetation index | [46] | |
| M_NIR1 | NIR band 1; 780 nm 1 | ||
| M_NIR2 | NIR band 2; 900 nm 1 | ||
| M_PSRI | Plant senescence reflectance index | [47] | |
| M_R | Red band, 670 nm 1 | ||
| M_RE1 | Red edge band 1; 700 nm 1 | ||
| M_RE2 | Red edge band 2; 730 nm 1 | ||
| RGB Camera | |||
| R_BN | Normalized blue band | ||
| R_EVI2_green | Enhanced Vegetation Index 2-Green | [39] | |
| R_GLI | Green leaf index | [42] | |
| R_GN | Normalized green band | ||
| R_PH | Plant height derived from the digital surface model (DEM) at Day i (Di), corrected by the DEM at areference day (Dr) | ||
| R_RN | Normalized red band | ||
| R_TGI | Triangular greenness index | [41] | |
| R_VARI | Visible Atmospherically Resistant Index | [42] | |
2.4. Grain Yield Modeling
3. Results
3.1. Descriptive Statistics
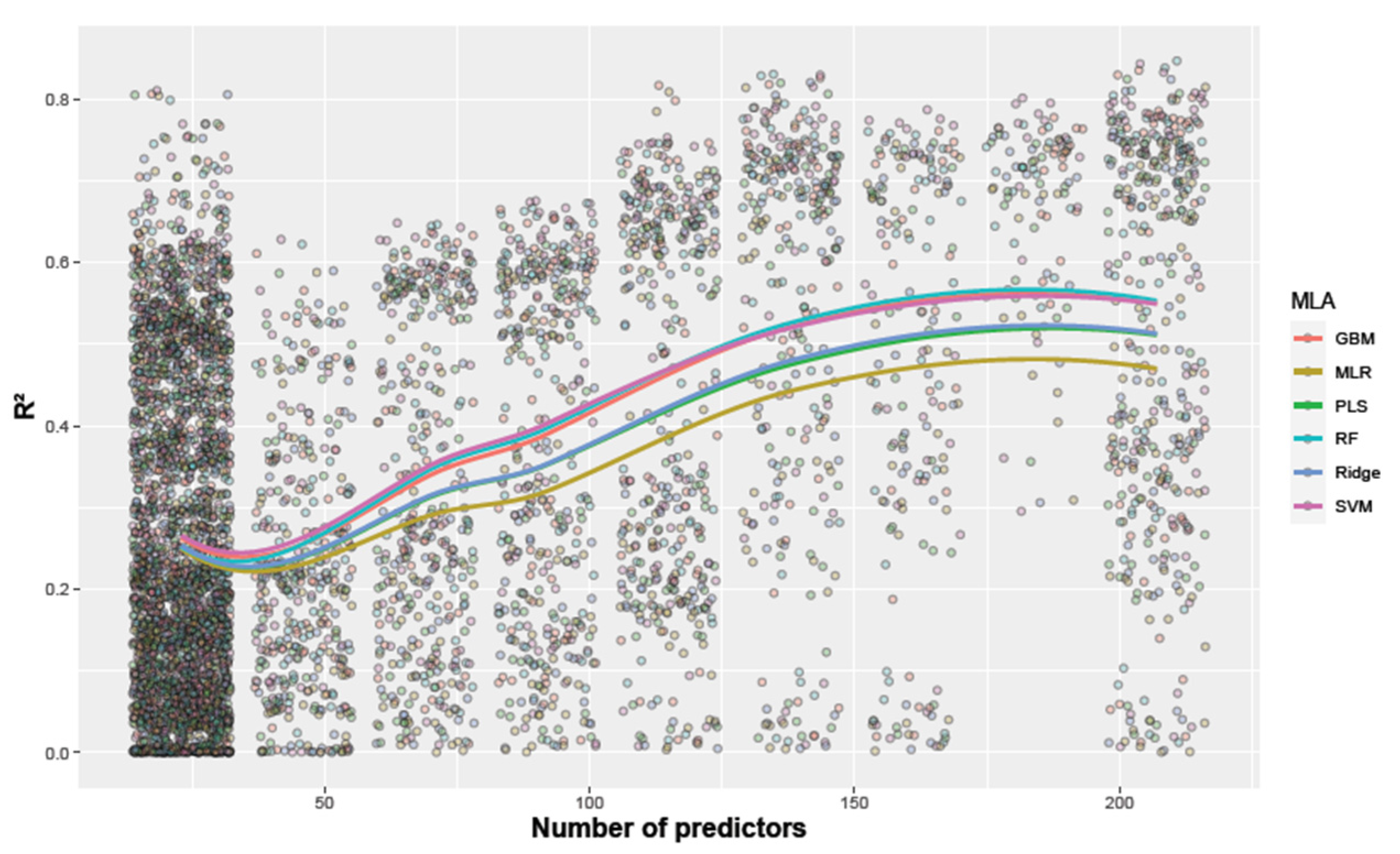
3.2. The Selection of Machine Learning Algorithms
3.3. The Selection of Individual Measurement Dates
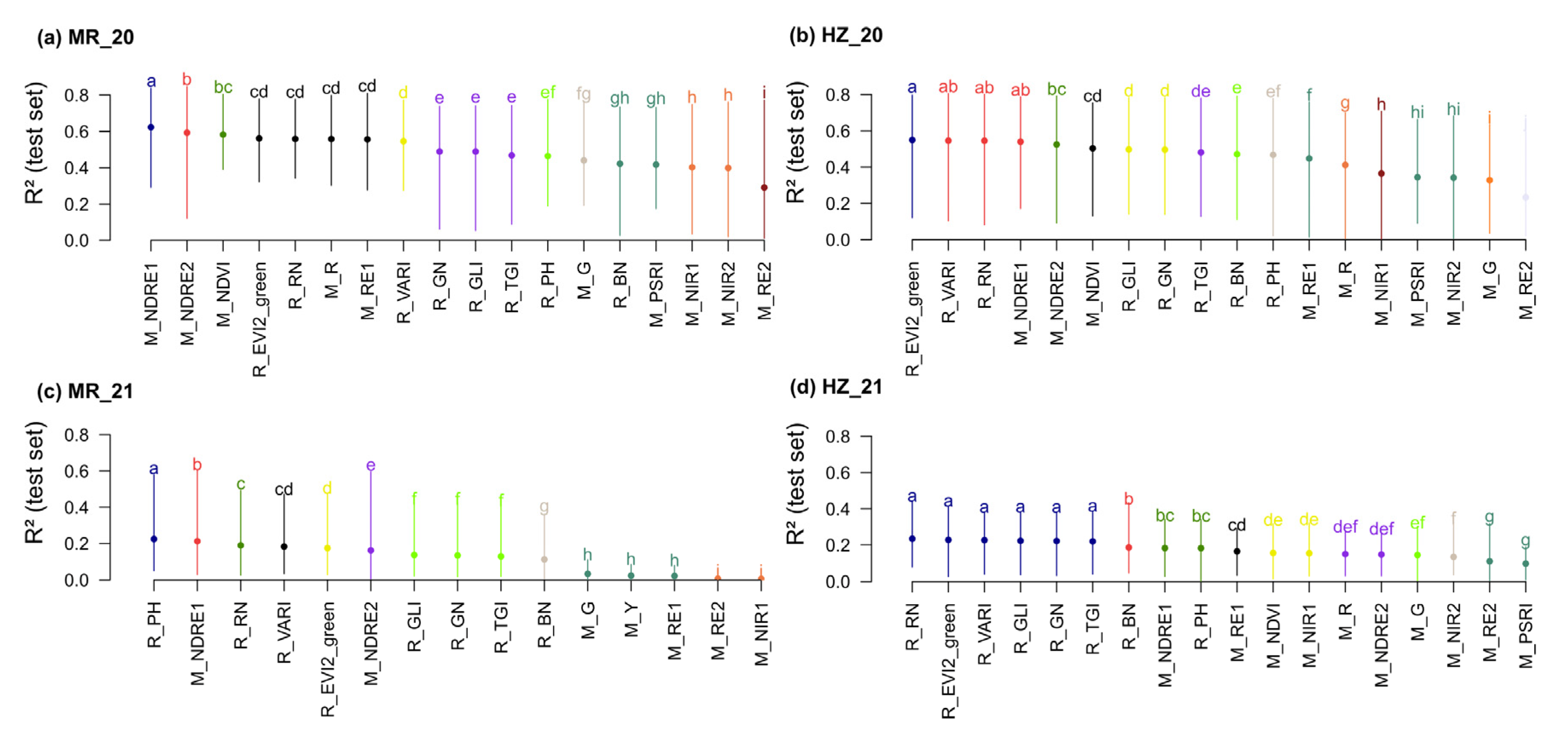
3.4. The Selection of Spectral Parameters
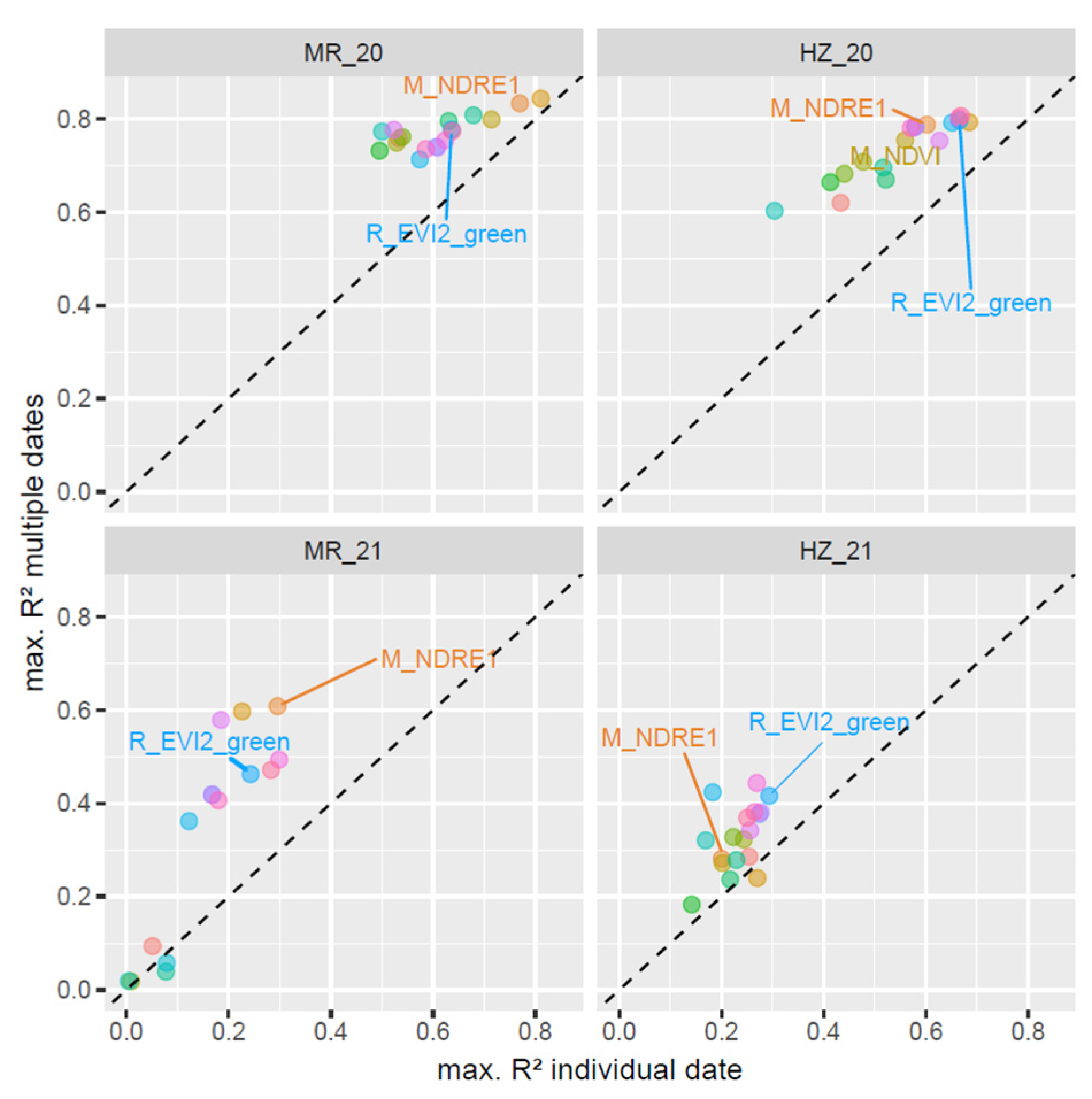
3.5. The Use of Incremental Date Combinations
4. Discussion
4.1. The Influence of Growth Stage and Trial Conditions
4.2. The Advantage of Multi-Temporal Data
4.3. The Comparison of Machine Learning Algorithms
4.4. The Selection of Sensors and Spectral Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate Trends and Global Crop Production Since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global Food Demand and the Sustainable Intensification of Agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.I.; Hu, H.; Gill, M.; Batley, J.; Edwards, D. Crop Breeding for a Changing Climate: Integrating Phenomics and Genomics with Bioinformatics Crop Breeding for a Changing Climate: Integrating Phenomics and Genomics with Bioinformatics. Theor. Appl. Genet. 2021, 134, 1677–1690. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Cairns, J.E. Field High-Throughput Phenotyping: The New Crop Breeding Frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Houle, D.; Govindaraju, D.R.; Omholt, S. Phenomics: The next Challenge. Nat. Rev. Genet. 2010, 11, 855–866. [Google Scholar] [CrossRef]
- Basu, P.S.; Srivastava, M.; Singh, P.; Porwal, P.; Kant, R.; Singh, J. High-Precision Phenotyping under Controlled versus Natural Environments. In Phenomics in Crop Plants: Trends, Options and Limitations; Kumar, J., Pratap, A., Kumar, S., Eds.; Springer: New Delhi, India, 2015; pp. 27–40. [Google Scholar]
- Roitsch, T.; Cabrera-Bosquet, L.; Fournier, A.; Ghamkhar, K.; Jiménez-Berni, J.; Pinto, F.; Ober, E.S. Review: New Sensors and Data-Driven Approaches—A Path to next Generation Phenomics. Plant Sci. 2019, 282, 2–10. [Google Scholar] [CrossRef]
- Prey, L.; Schmidhalter, U. Simulation of Satellite Reflectance Data Using High-Frequency Ground Based Hyperspectral Canopy Measurements for in-Season Estimation of Grain Yield and Grain Nitrogen Status in Winter Wheat. ISPRS J. Photogramm. Remote Sens. 2019, 149, 176–187. [Google Scholar] [CrossRef]
- Herzig, P.; Borrmann, P.; Knauer, U.; Klück, H.; Kilias, D.; Seiffert, U. Evaluation of RGB and Multispectral Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping and Yield Prediction in Barley Breeding. Remote Sens. 2021, 13, 2670. [Google Scholar] [CrossRef]
- Xie, C.; Yang, C. A Review on Plant High-Throughput Phenotyping Traits Using UAV-Based Sensors. Comput. Electron. Agric. 2020, 178, 105731. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating Biomass of Barley Using Crop Surface Models (CSMs) Derived from UAV-Based RGB Imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef]
- Rasmussen, J.; Ntakos, G.; Nielsen, J.; Svensgaard, J.; Poulsen, R.N.; Christensen, S. Are Vegetation Indices Derived from Consumer-Grade Cameras Mounted on UAVs Sufficiently Reliable for Assessing Experimental Plots? Eur. J. Agron. 2016, 74, 75–92. [Google Scholar] [CrossRef]
- Deery, D.; Jimenez-Berni, J.; Jones, H.; Sirault, X.; Furbank, R. Proximal Remote Sensing Buggies and Potential Applications for Field-Based Phenotyping. Agronomy 2014, 4, 349–379. [Google Scholar] [CrossRef]
- Oehlschläger, J.; Schmidhalter, U.; Noack, P.O. UAV-Based Hyperspectral Sensing for Yield Prediction in Winter Barley. In Proceedings of the 2018 9th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 23–26 September 2018; pp. 1–4. [Google Scholar]
- de Souza, R.; Buchhart, C.; Heil, K.; Plass, J.; Padilla, F.M.; Schmidhalter, U. Effect of Time of Day and Sky Conditions on Different Vegetation Indices Calculated from Active and Passive Sensors and Images Taken from Uav. Remote Sens. 2021, 13, 1691. [Google Scholar] [CrossRef]
- Hu, Y.; Knapp, S.; Schmidhalter, U. Advancing High-Throughput Phenotyping of Wheat in Early Selection Cycles. Remote Sens. 2020, 12, 574. [Google Scholar] [CrossRef]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A Review on UAV-Based Applications for Precision Agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef]
- Kefauver, S.C.; Vicente, R.; Vergara-Díaz, O.; Fernandez-Gallego, J.A.; Kerfal, S.; Lopez, A.; Melichar, J.P.E.; Serret Molins, M.D.; Araus, J.L. Comparative UAV and Field Phenotyping to Assess Yield and Nitrogen Use Efficiency in Hybrid and Conventional Barley. Front. Plant Sci. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Krajewski, P.; Chen, D.; Ćwiek, H.; van Dijk, A.D.J.; Fiorani, F.; Kersey, P.; Klukas, C.; Lange, M.; Markiewicz, A.; Nap, J.P.; et al. Towards Recommendations for Metadata and Data Handling in Plant Phenotyping. J. Exp. Bot. 2015, 66, 5417–5427. [Google Scholar] [CrossRef]
- Reynolds, D.; Reynolds, D.; Baret, F.; Welcker, C.; Bostrom, A.; Ball, J.; Cellini, F.; Lorence, A.; Chawade, A.; Kha, M.; et al. What Is Cost-Efficient Phenotyping ? Optimizing Costs for Different Scenarios. Plant Sci. 2019, 282, 14–22. [Google Scholar] [CrossRef]
- Garriga, M.; Romero-Bravo, S.; Estrada, F.; Escobar, A.; Matus, I.A.; del Pozo, A.; Astudillo, C.A.; Lobos, G.A. Assessing Wheat Traits by Spectral Reflectance: Do We Really Need to Focus on Predicted Trait-Values or Directly Identify the Elite Genotypes Group ? Front. Plant Sci. 2017, 8, 280. [Google Scholar] [CrossRef]
- Tucker, C.J.; Holben, B.N.; Elgin Jr, J.H.; McMurtrey III, J.E. Relationship of Spectral Data to Grain Yield Variation. Photogramm. Eng. Remote Sens. 1980, 46, 657–666. [Google Scholar]
- Babar, M.A.; Reynolds, M.P.; Van Ginkel, M.; Klatt, A.R.; Raun, W.R.; Stone, M.L. Spectral Reflectance Indices as a Potential Indirect Selection Criteria for Wheat Yield under Irrigation. Crop Sci. 2006, 46, 578–588. [Google Scholar] [CrossRef]
- Freeman, K.W.; Raun, W.R.; Johnson, G.V.; Mullen, R.W.; Stone, M.L.; Solie, J.B. Late-Season Prediction of Wheat Grain Yield and Grain Protein. Commun. Soil Sci. Plant Anal. 2003, 34, 1837–1852. [Google Scholar] [CrossRef]
- Prey, L.; Hu, Y.; Schmidhalter, U. High-Throughput Field Phenotyping Traits of Grain Yield Formation and Nitrogen Use Efficiency: Optimizing the Selection of Vegetation Indices and Growth Stages. Front. Plant Sci. 2020, 10, 1672. [Google Scholar] [CrossRef] [PubMed]
- Uav, C.M.; Wang, F.; Yang, M.; Ma, L.; Zhang, T.; Qin, W.; Li, W.; Zhang, Y.; Sun, Z.; Wang, Z.; et al. Estimation of Above-Ground Biomass of Winter Wheat Based. Remote Sens. 2022, 14, 1251. [Google Scholar] [CrossRef]
- Song, Y.; Wang, J.; Shan, B. Estimation of Winter Wheat Yield from Uav-Based Multi-Temporal Imagery Using Crop Allometric Relationship and Safy Model. Drones 2021, 5, 78. [Google Scholar] [CrossRef]
- Lu, N.; Zhou, J.; Han, Z.; Li, D.; Cao, Q.; Yao, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cheng, T. Improved Estimation of Aboveground Biomass in Wheat from RGB Imagery and Point Cloud Data Acquired with a Low-Cost Unmanned Aerial Vehicle System. Plant Methods 2019, 15, 1–16. [Google Scholar] [CrossRef]
- Amorim, J.G.A.; Schreiber, L.V.; de Souza, M.R.Q.; Negreiros, M.; Susin, A.; Bredemeier, C.; Trentin, C.; Vian, A.L.; de Oliveira Andrades-Filho, C.; Doering, D.; et al. Biomass Estimation of Spring Wheat with Machine Learning Methods Using UAV-Based Multispectral Imaging. Int. J. Remote Sens. 2022, 43, 4758–4773. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Zhu, X.; Dong, Z.; Guo, W. Estimation of Biomass in Wheat Using Random Forest Regression Algorithm and Remote Sensing Data. Crop J. 2016, 4, 212–219. [Google Scholar] [CrossRef]
- Teodoro, P.E.; Teodoro, L.P.R.; Baio, F.H.R.; da Silva Junior, C.A.; Dos Santos, R.G.; Ramos, A.P.M.; Pinheiro, M.M.F.; Osco, L.P.; Gonçalves, W.N.; Carneiro, A.M.; et al. Predicting Days to Maturity, Plant Height, and Grain Yield in Soybean: A Machine and Deep Learning Approach Using Multispectral Data. Remote Sens. 2021, 13, 4632. [Google Scholar] [CrossRef]
- Marszalek, M.; Körner, M.; Schmidhalter, U. Prediction of Multi-Year Winter Wheat Yields at the Field Level with Satellite and Climatological Data. Comput. Electron. Agric. 2022, 194, 106777. [Google Scholar] [CrossRef]
- Shafiee, S.; Lied, L.M.; Burud, I.; Dieseth, J.A.; Alsheikh, M.; Lillemo, M. Sequential Forward Selection and Support Vector Regression in Comparison to LASSO Regression for Spring Wheat Yield Prediction Based on UAV Imagery. Comput. Electron. Agric. 2021, 183, 106036. [Google Scholar] [CrossRef]
- Landesbetrieb Geoinformation und Vermessung MetaVer. Available online: https://metaver.de/ingrid-webmap-client/frontend/prd/?lang=de&topic=themen&bgLayer=sgx_geodatenzentrum_de_web_light_grau_EU_EPSG_25832_TOPPLUS&E=676481.34&N=5700778.57&zoom=8 (accessed on 7 October 2022).
- Bayerisches Landesamt für Umwelt Umwelt Atlas Bayern. Available online: https://www.umweltatlas.bayern.de/mapapps/resources/apps/umweltatlas/index.html?lang=de (accessed on 7 October 2022).
- Anderegg, J.; Yu, K.; Aasen, H.; Walter, A.; Liebisch, F.; Hund, A. Spectral Vegetation Indices to Track Senescence Dynamics in Diverse Wheat Germplasm. Front. Plant Sci. 2020, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Peng, G.; Meng, R.; Man, J.; Li, W.; Xu, B.; Lv, Z.; Sun, R. Wheat Yield Prediction Based on Unmanned Aerial Vehicles-Collected Red–Green–Blue Imagery. Remote Sens. 2021, 13, 2937. [Google Scholar] [CrossRef]
- Taruna, B.; Putra, W.; Soni, P. Enhanced Broadband Greenness in Assessing Chlorophyll a and b, Carotenoid, and Nitrogen in Robusta Coffee. Precis. Agric. 2017, 19, 238–256. [Google Scholar] [CrossRef]
- Louhaichi, M.; Borman, M.M.; Johnson, D.E. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar] [CrossRef]
- Hunt, E.R.; Doraiswamy, P.C.; McMurtrey, J.E.; Daughtry, C.S.T.; Perry, E.M.; Akhmedov, B. A Visible Band Index for Remote Sensing Leaf Chlorophyll Content at the Canopy Scale. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 103–112. [Google Scholar] [CrossRef]
- Lussem, U.; Bolten, A.; Gnyp, M.L.; Jasper, J.; Bareth, G. Evaluation of rgb-based vegetation indices from uav imagery to estimate forage yield in grassland. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, XLII–3, 1215–1219. [Google Scholar] [CrossRef]
- van Rossum, G.; de Boer, J. Interactively Testing Remote Servers Using the Python Programming Language. CWI Q. 1991, 4, 283–303. [Google Scholar]
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model Dev. Discuss. 2015, 8, 2271–2312. [Google Scholar] [CrossRef]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident Detection of Crop Water Stress, Nitrogen Status and Canopy Density Using Ground Based Multispectral Data. In Proceedings of the Fifth International Conference on Precision Agriculture, Bloomington, MN, USA, 16–19 July 2000; Volume 1619, pp. 16–19. [Google Scholar]
- Rouse, J.W.; Hass, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. Third Earth Resour. Technol. Satell. (ERTS) Symp. 1973, 1, 309–317. [Google Scholar]
- Sims, D.A.; Gamon, J.A. Relationships between Leaf Pigment Content and Spectral Reflectance across a Wide Range of Species, Leaf Structures and Developmental Stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- R Core Team: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012.
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Joseph, V.R. Optimal Ratio for Data Splitting. Stat. Anal. Data Min. 2022, 15, 531–538. [Google Scholar] [CrossRef]
- Breunig, M.M.; Kriegel, H.-P.; Ng, R.T.; Sander, J.; Abasi, Z.; Abedian, Z. LOF: Identifying Density-Based Local Outliers. In Proceedings of the 2000 ACM SIGMOD International Conference on Management of Data, Dallas, TX, USA, 15–18 May 2000; Volume 107, pp. 93–104. [Google Scholar]
- Privé, F. Utility Functions for Large-Scale Data. R Package Version 0.3.4 2021. Available online: https://CRAN.R-project.org/package=bigutilsr (accessed on 13 December 2022).
- Alghushairy, O.; Alsini, R.; Soule, T.; Ma, X. A Review of Local Outlier Factor Algorithms for Outlier Detection in Big Data Streams. Big Data Cogn. Comput. 2021, 5, 1–24. [Google Scholar] [CrossRef]
- Kuhn, M. A Short Introduction to the Caret Package. R Found Stat Comput. 2015, 10, 1–10. Available online: https://cran.microsoft.com/snapshot/2015-08-17/web/packages/caret/vignettes/caret.pdf (accessed on 21 May 2021).
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. 2020. Available online: https://CRAN.R-project.org/package=agricolae (accessed on 29 December 2020).
- Prey, L.; Hu, Y.; Schmidhalter, U. Temporal Dynamics and the Contribution of Plant Organs in a Phenotypically Diverse Population of High-Yielding Winter Wheat: Evaluating Concepts for Disentangling Yield Formation and Nitrogen Use Efficiency. Front. Plant Sci. 2019, 10, 1295. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Slafer, G.A.; Davies, W.J.; Berry, P.M.; Sylvester-Bradley, R.; Martre, P.; Calderini, D.F.; Griffiths, S.; Reynolds, M.P. Raising Yield Potential of Wheat. III. Optimizing Partitioning to Grain While Maintaining Lodging Resistance. J. Exp. Bot. 2011, 62, 469–486. [Google Scholar] [CrossRef]
- Prey, L.; von Bloh, M.; Schmidhalter, U. Evaluating RGB Imaging and Multispectral Active and Hyperspectral Passive Sensing for Assessing Early Plant Vigor in Winter Wheat. Sensors 2018, 18, 2931. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Gitelson, A.A.; Schepers, J.S.; Walthall, C.L. Application of Spectral Remote Sensing for Agronomic Decisions. Agron. J. 2008, 100, 117–131. [Google Scholar] [CrossRef]
- Babar, M.A.; Van Ginkel, M.; Reynolds, M.P.; Prasad, B.; Klatt, A.R. Heritability, Correlated Response, and Indirect Selection Involving Spectral Reflectance Indices and Grain Yield in Wheat. Aust. J. Agric. Res. 2007, 58, 432–442. [Google Scholar] [CrossRef]
- Gutierrez, M.; Reynolds, M.P.; Klatt, A.R. Association of Water Spectral Indices with Plant and Soil Water Relations in Contrasting Wheat Genotypes. J. Exp. Bot. 2010, 61, 3291–3303. [Google Scholar] [CrossRef] [PubMed]
- Christopher, J.T.; Veyradier, M.; Borrell, A.K.; Harvey, G.; Fletcher, S.; Chenu, K. Phenotyping Novel Stay-Green Traits to Capture Genetic Variation in Senescence Dynamics. Funct. Plant Biol. 2014, 41, 1035. [Google Scholar] [CrossRef] [PubMed]
- Spano, G.; Di Fonzo, N.; Perrotta, C.; Platani, C.; Ronga, G.; Lawlor, D.W.; Napier, J.A.; Shewry, P.R. Physiological Characterization of “stay Green” Mutants in Durum Wheat. J. Exp. Bot. 2003, 54, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Berdugo, C.A.; Mahlein, A.K.; Steiner, U.; Dehne, H.W.; Oerke, E.C. Sensors and Imaging Techniques for the Assessment of the Delay of Wheat Senescence Induced by Fungicides. Funct. Plant Biol. 2013, 40, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, N.; Villegas, D.; Casadesus, J.; Araus, J.L.; Royo, C. Spectral Vegetation Indices as Nondestructive Tools for Determining Durum Wheat Yield. Agron. J. 2000, 92, 83–91. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting Grain Yield in Rice Using Multi-Temporal Vegetation Indices from UAV-Based Multispectral and Digital Imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Cavalaris, C.; Megoudi, S.; Maxouri, M.; Anatolitis, K.; Sifakis, M.; Levizou, E.; Kyparissis, A. Modeling of Durum Wheat Yield Based on Sentinel-2 Imagery. Agronomy 2021, 11, 1486. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of Machine-Learning Classification in Remote Sensing: An Applied Review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef]
- Ramezan, C.A.; Warner, T.A.; Maxwell, A.E.; Price, B.S. Effects of Training Set Size on Supervised Machine-Learning Land-Cover Classification of Large-Area High-Resolution Remotely Sensed Data. Remote Sens. 2021, 13, 368. [Google Scholar] [CrossRef]
- Nguy-Robertson, A.; Gitelson, A.; Peng, Y.; Viña, A.; Arkebauer, T.; Rundquist, D. Green Leaf Area Index Estimation in Maize and Soybean: Combining Vegetation Indices to Achieve Maximal Sensitivity. Agron. J. 2012, 104, 1336–1347. [Google Scholar] [CrossRef]
- Kefauver, S.C.; El-Haddad, G.; Vergara-Diaz, O.; Araus, J.L. RGB Picture Vegetation Indexes for High-Throughput Phenotyping Platforms (HTPPs). In Proceedings of the Remote Sensing for Agriculture, Ecosystems, and Hydrology XVII, Toulouse, France, 14 October 2015. [Google Scholar] [CrossRef]
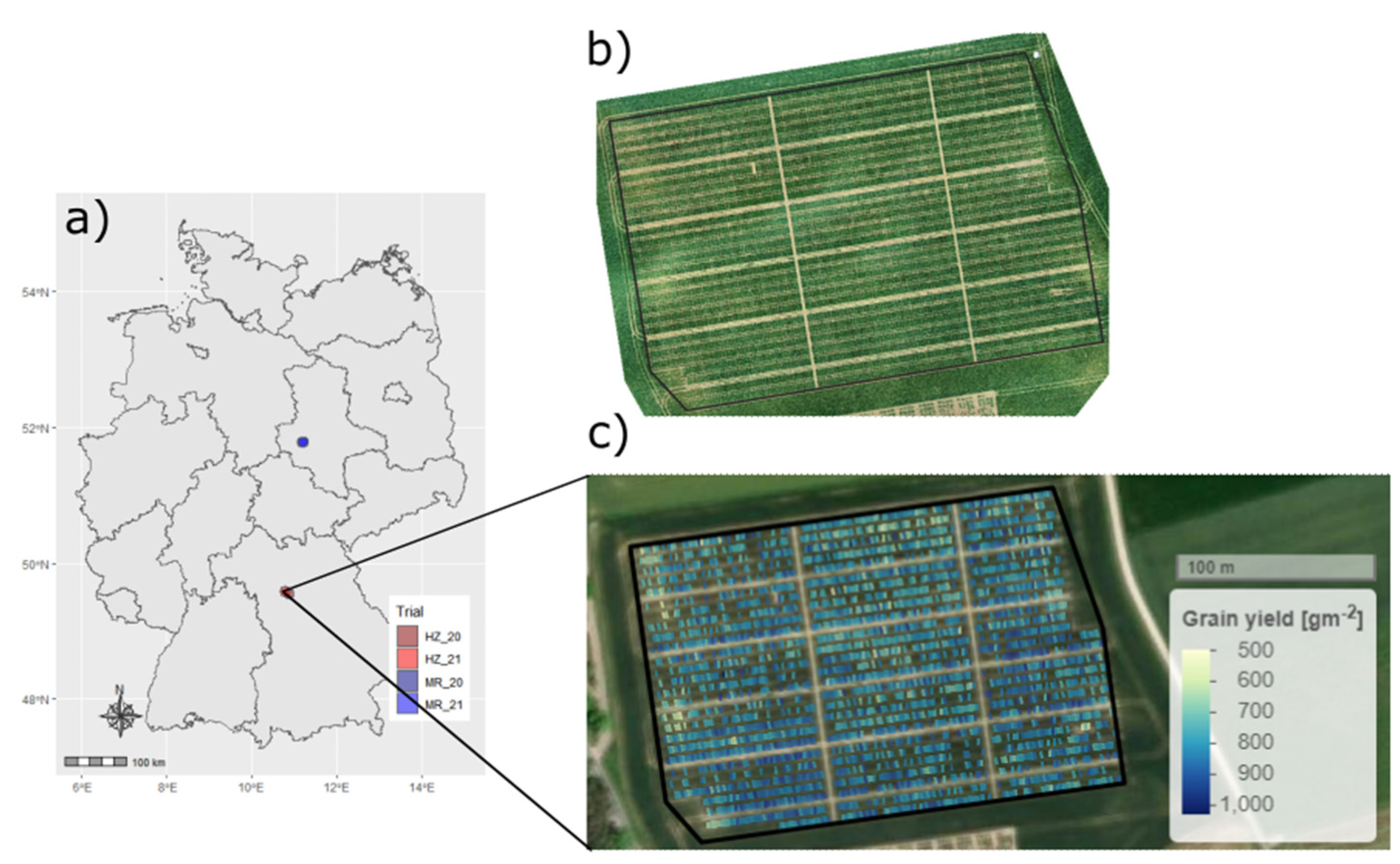


| Trial | Date | Growth Stage | GDD | Notes |
|---|---|---|---|---|
| MR_20 | 03 March 2020 | 21 | 256 | 1 |
| 01 April 2020 | 25 | 338 | ||
| 23 April 2020 | 30 | 472 | ||
| 08 May 2020 | 33 | 574 | ||
| 19 May 2020 | 43 | 647 | ||
| 08 June 2020 | 72 | 839 | ||
| 24 June 2020 | 75 | 1061 | ||
| 09 July 2020 | 83 | 1275 | ||
| HZ_20 | 20 February 2020 | 21 | 137 | 1 |
| 26 March 2020 | 25 | 233 | ||
| 09 April 2020 | 30 | 309 | ||
| 06 May 2020 | 33 | 516 | ||
| 20 May 2020 | 43 | 635 | ||
| 29 May 2020 | 65 | 717 | ||
| 08 June 2020 | 75 | 812 | ||
| 19 June 2020 | 81 | 943 | ||
| 26 June 2020 | 84 | 1040 | ||
| 08 July 2020 | 85 | 1205 | ||
| MR_21 | 02 March 2021 | 21 | 213 | 2 |
| 24 March 2021 | 24 | 249 | ||
| 20 April 2021 | 30 | 356 | ||
| 28 April 2021 | 31 | 389 | ||
| 03 June 2021 | 63 | 670 | ||
| 17 June 2021 | 72 | 871 | ||
| 06 July 2021 | 79 | 1149 | ||
| 20 July 2021 | 85 | 1349 | ||
| HZ_21 | 25 March 2021 | 22 | 190 | 2 |
| 15 April 2021 | 27 | 274 | ||
| 30 April 2021 | 33 | 343 | ||
| 14 May 2021 | 37 | 422 | ||
| 08 June 2021 | 57 | 638 | ||
| 13 July 2021 | 85 | 1132 |
| Trial | Mean | Minimum | Maximum | SD | CV | n |
|---|---|---|---|---|---|---|
| MR_20 | 792 | 223 | 1131 | 126 | 16% | 4423 |
| HZ_20 | 715 | 290 | 1031 | 108 | 15% | 4349 |
| MR_21 | 849 | 300 | 1116 | 91 | 11% | 2787 |
| HZ_21 | 818 | 500 | 1027 | 72 | 9% | 2711 |
| Trial | MLA | Mean R2 | Maximum R2 | Group | Mean RMSE [kg plot−1] | Minimum RMSE [kg plot−1] |
|---|---|---|---|---|---|---|
| MR_20 | GBM | 0.51 | 0.84 | ab | 0.52 | 0.32 |
| MLR | 0.47 | 0.81 | c | 0.54 | 0.33 | |
| PLS | 0.48 | 0.82 | bc | 0.54 | 0.32 | |
| RF | 0.50 | 0.85 | abc | 0.53 | 0.31 | |
| Ridge | 0.48 | 0.82 | bc | 0.53 | 0.32 | |
| SVM | 0.52 | 0.84 | a | 0.51 | 0.31 | |
| HZ_20 | GBM | 0.47 | 0.78 | a | 0.45 | 0.29 |
| MLR | 0.42 | 0.75 | c | 0.48 | 0.31 | |
| PLS | 0.44 | 0.76 | abc | 0.46 | 0.30 | |
| RF | 0.46 | 0.78 | ab | 0.45 | 0.29 | |
| Ridge | 0.44 | 0.76 | bc | 0.47 | 0.30 | |
| SVM | 0.46 | 0.81 | ab | 0.45 | 0.27 | |
| MR_21 | GBM | 0.12 | 0.57 | a | 0.47 | 0.33 |
| MLR | 0.10 | 0.46 | a | 0.48 | 0.37 | |
| PLS | 0.11 | 0.49 | a | 0.48 | 0.36 | |
| RF | 0.12 | 0.57 | a | 0.47 | 0.33 | |
| Ridge | 0.11 | 0.50 | a | 0.47 | 0.36 | |
| SVM | 0.13 | 0.61 | a | 0.47 | 0.32 | |
| HZ_21 | GBM | 0.19 | 0.41 | ab | 0.34 | 0.29 |
| MLR | 0.15 | 0.35 | c | 0.37 | 0.31 | |
| PLS | 0.17 | 0.42 | b | 0.35 | 0.29 | |
| RF | 0.19 | 0.41 | ab | 0.34 | 0.29 | |
| Ridge | 0.17 | 0.44 | b | 0.34 | 0.29 | |
| SVM | 0.19 | 0.44 | a | 0.34 | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prey, L.; Hanemann, A.; Ramgraber, L.; Seidl-Schulz, J.; Noack, P.O. UAV-Based Estimation of Grain Yield for Plant Breeding: Applied Strategies for Optimizing the Use of Sensors, Vegetation Indices, Growth Stages, and Machine Learning Algorithms. Remote Sens. 2022, 14, 6345. https://doi.org/10.3390/rs14246345
Prey L, Hanemann A, Ramgraber L, Seidl-Schulz J, Noack PO. UAV-Based Estimation of Grain Yield for Plant Breeding: Applied Strategies for Optimizing the Use of Sensors, Vegetation Indices, Growth Stages, and Machine Learning Algorithms. Remote Sensing. 2022; 14(24):6345. https://doi.org/10.3390/rs14246345
Chicago/Turabian StylePrey, Lukas, Anja Hanemann, Ludwig Ramgraber, Johannes Seidl-Schulz, and Patrick Ole Noack. 2022. "UAV-Based Estimation of Grain Yield for Plant Breeding: Applied Strategies for Optimizing the Use of Sensors, Vegetation Indices, Growth Stages, and Machine Learning Algorithms" Remote Sensing 14, no. 24: 6345. https://doi.org/10.3390/rs14246345
APA StylePrey, L., Hanemann, A., Ramgraber, L., Seidl-Schulz, J., & Noack, P. O. (2022). UAV-Based Estimation of Grain Yield for Plant Breeding: Applied Strategies for Optimizing the Use of Sensors, Vegetation Indices, Growth Stages, and Machine Learning Algorithms. Remote Sensing, 14(24), 6345. https://doi.org/10.3390/rs14246345







