New Hyperspectral Procedure to Discriminate Intertidal Macroalgae
Abstract
:1. Introduction
2. Material and Methods
2.1. Biological Material and Data Acquisitions
2.1.1. Biological Material
2.1.2. Radiometric Acquisition and Hyperspectral Treatment
2.1.3. Pigment Extraction
2.2. Similarity Indices and Hierarchical Cluster Analyses
2.2.1. Pigment Classification
2.2.2. Spectral Data Classification
2.2.3. Classification of Reflectance Data Using SAM
2.2.4. Robustness Analysis and Monthly Monitoring
3. Results
3.1. Pigment Analysis
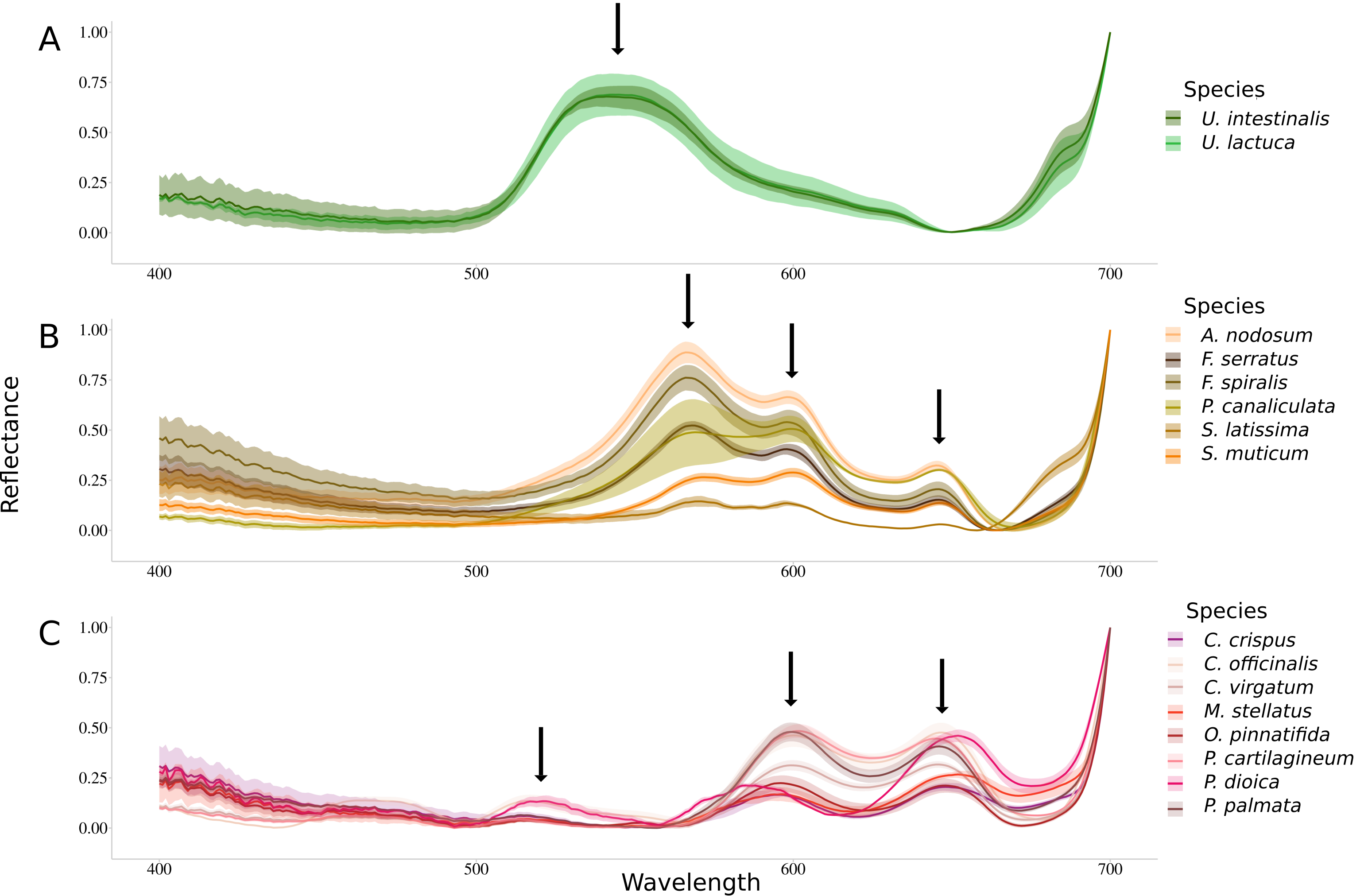
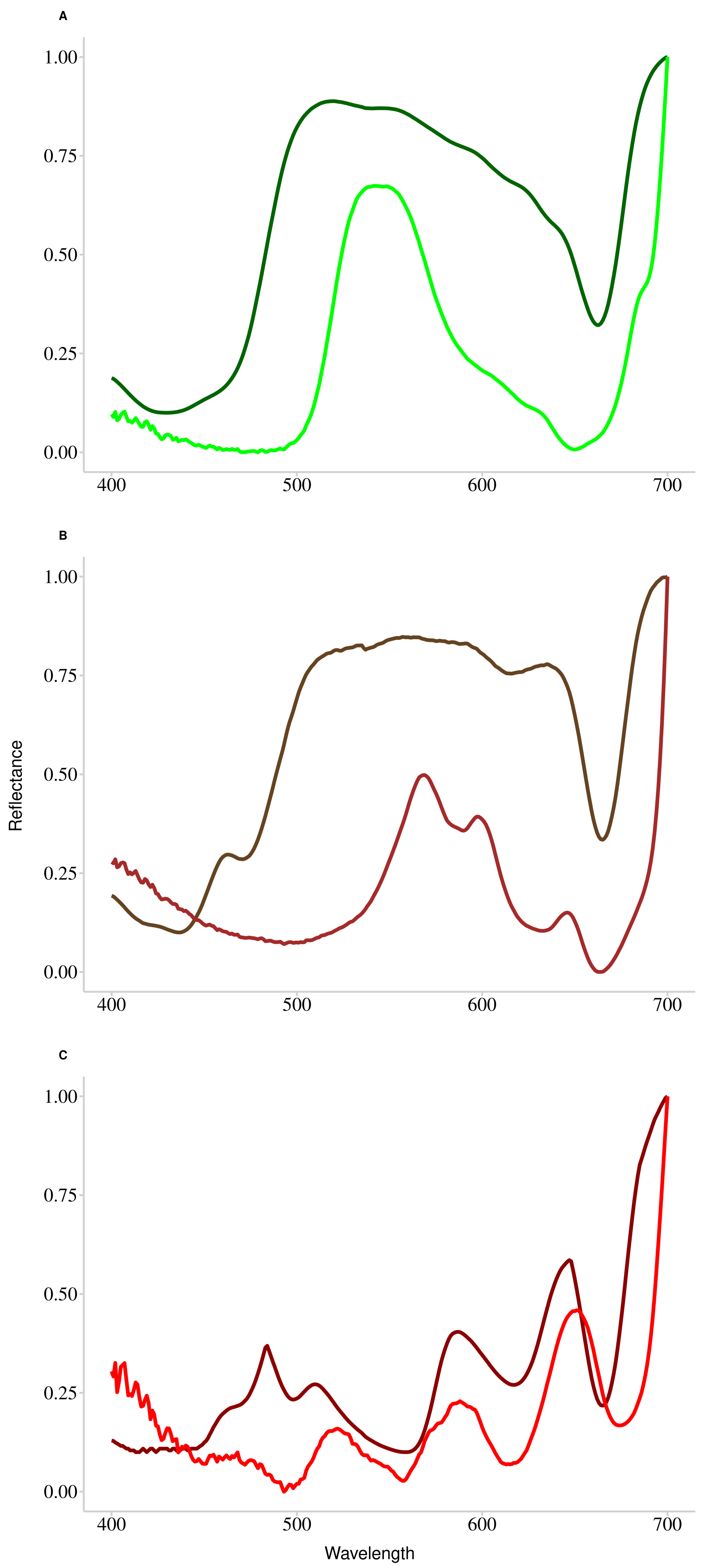
3.2. Raw Reflectance Spectra Analysis
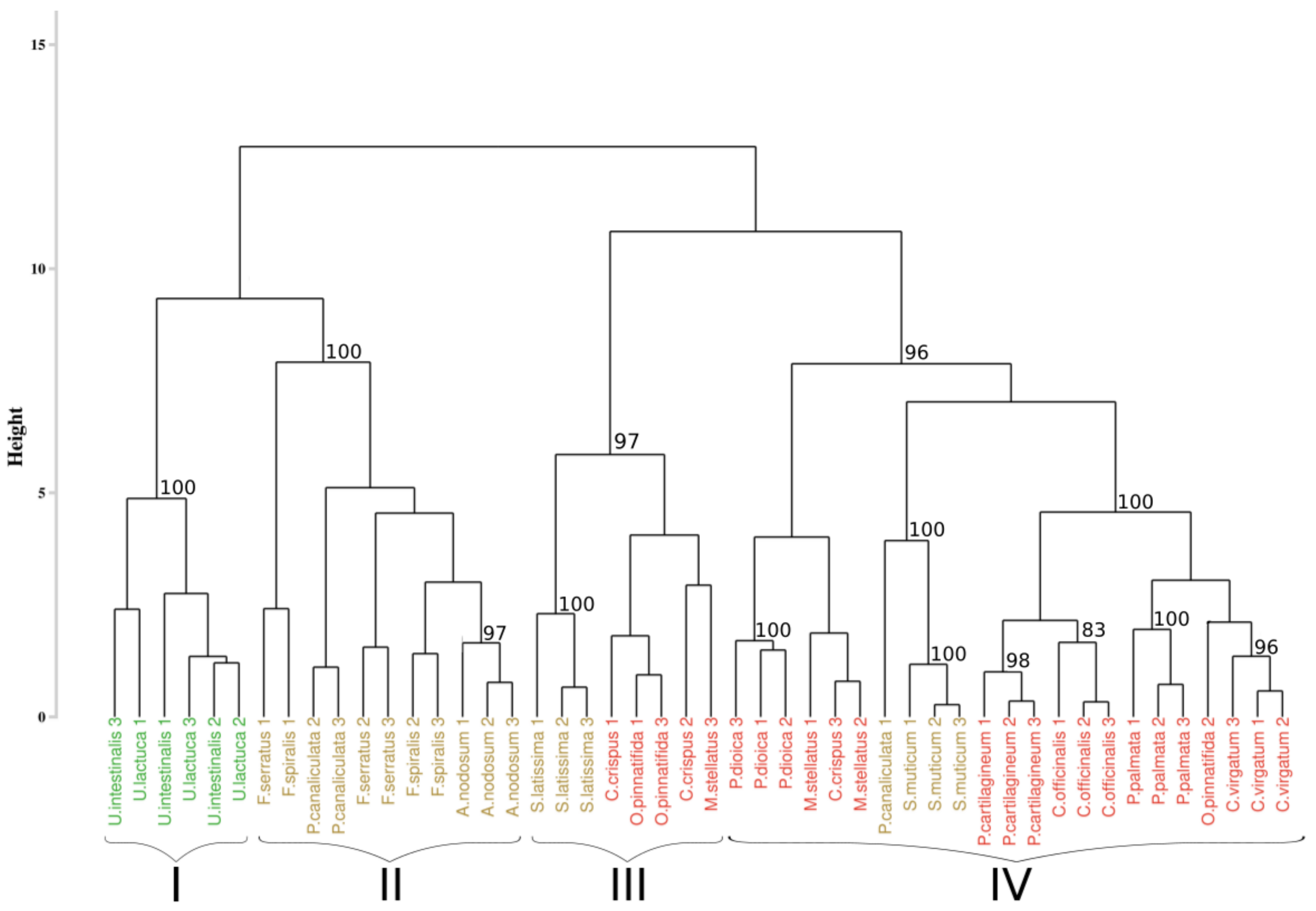
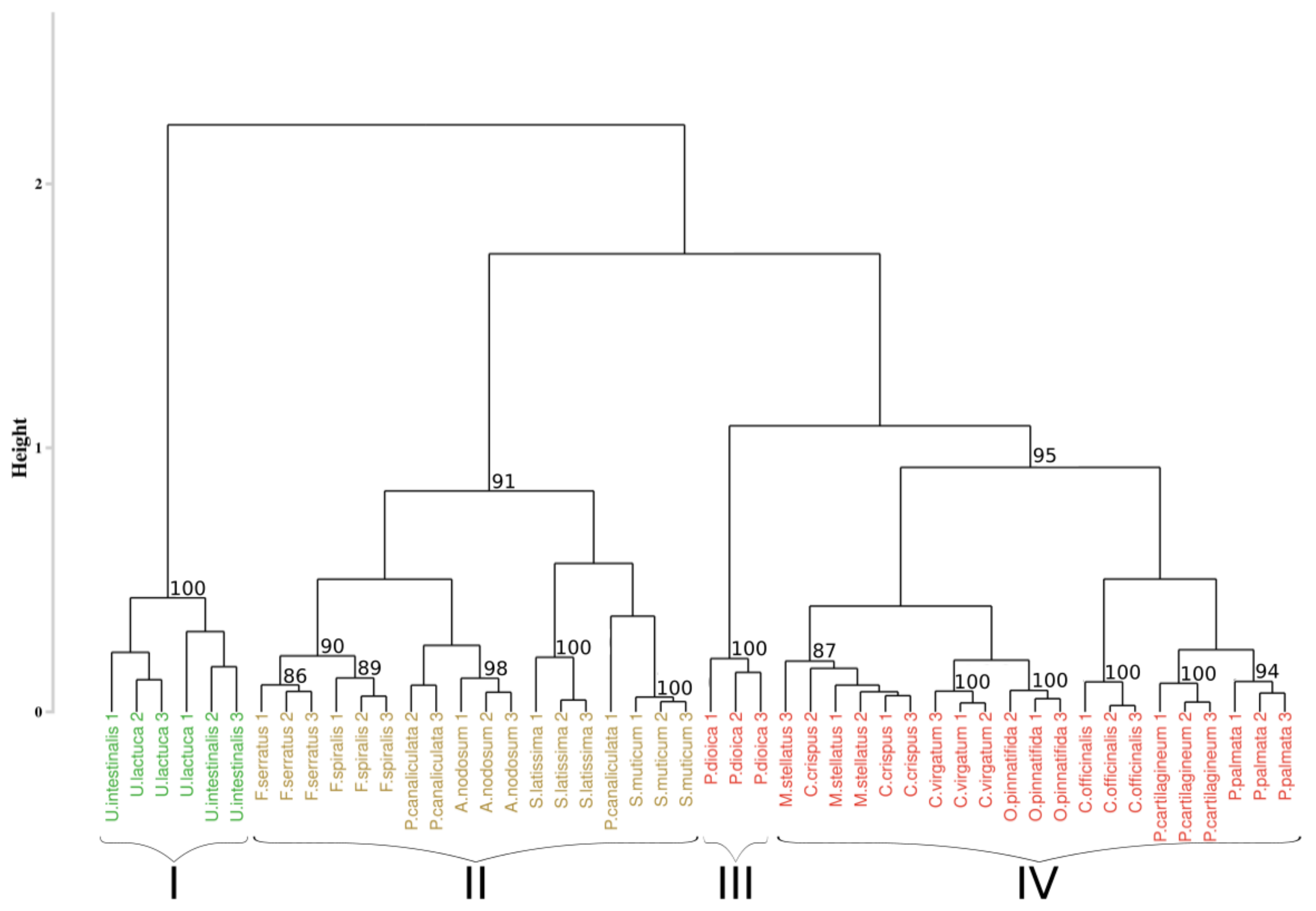
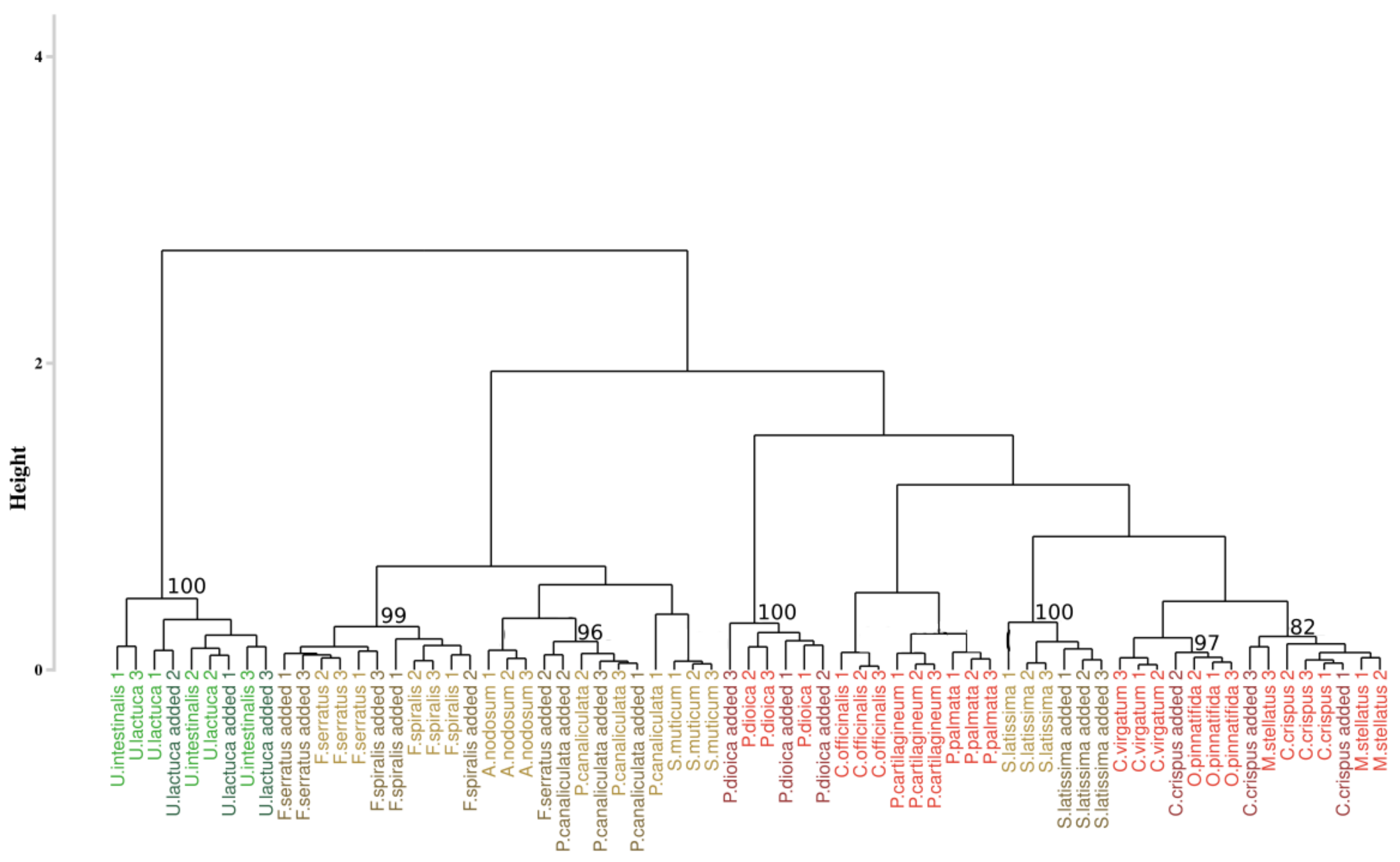
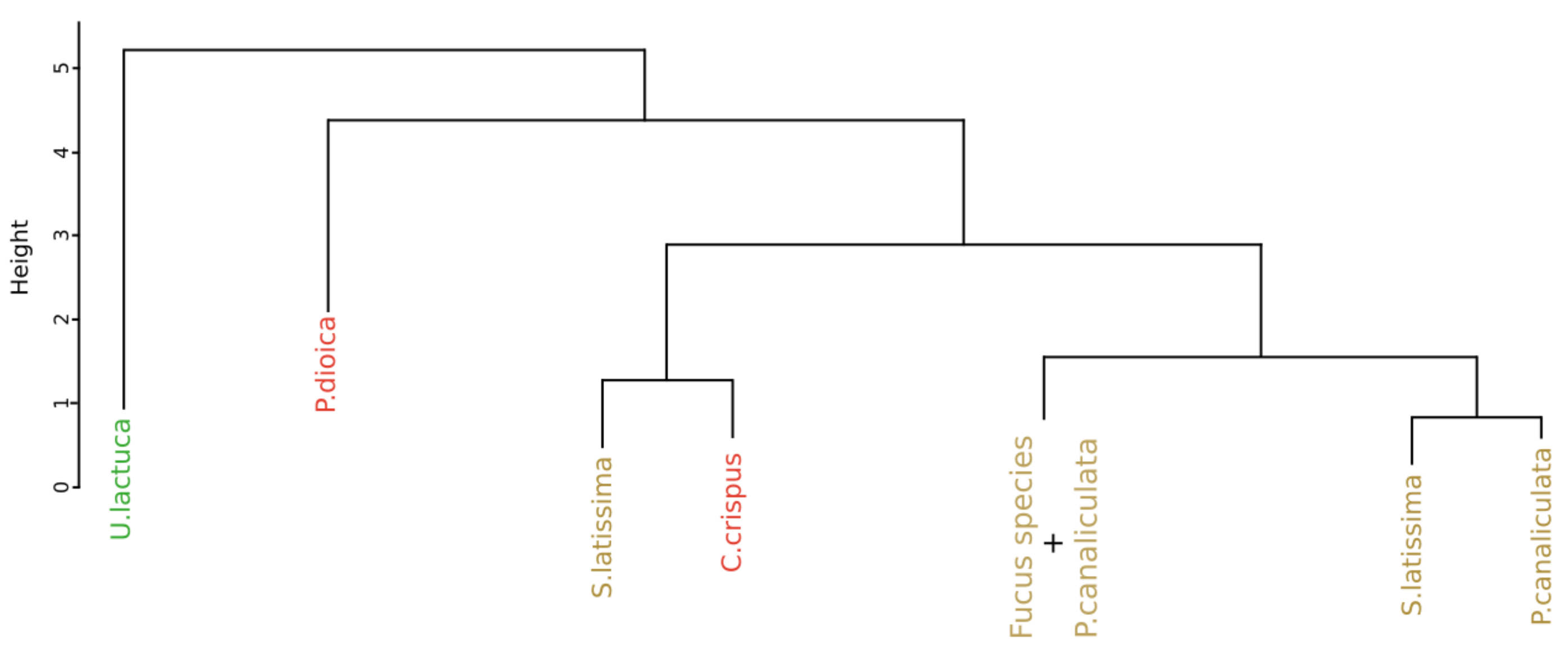
3.3. Reflectance Data Classification
3.3.1. Application of the Euclidean Technique
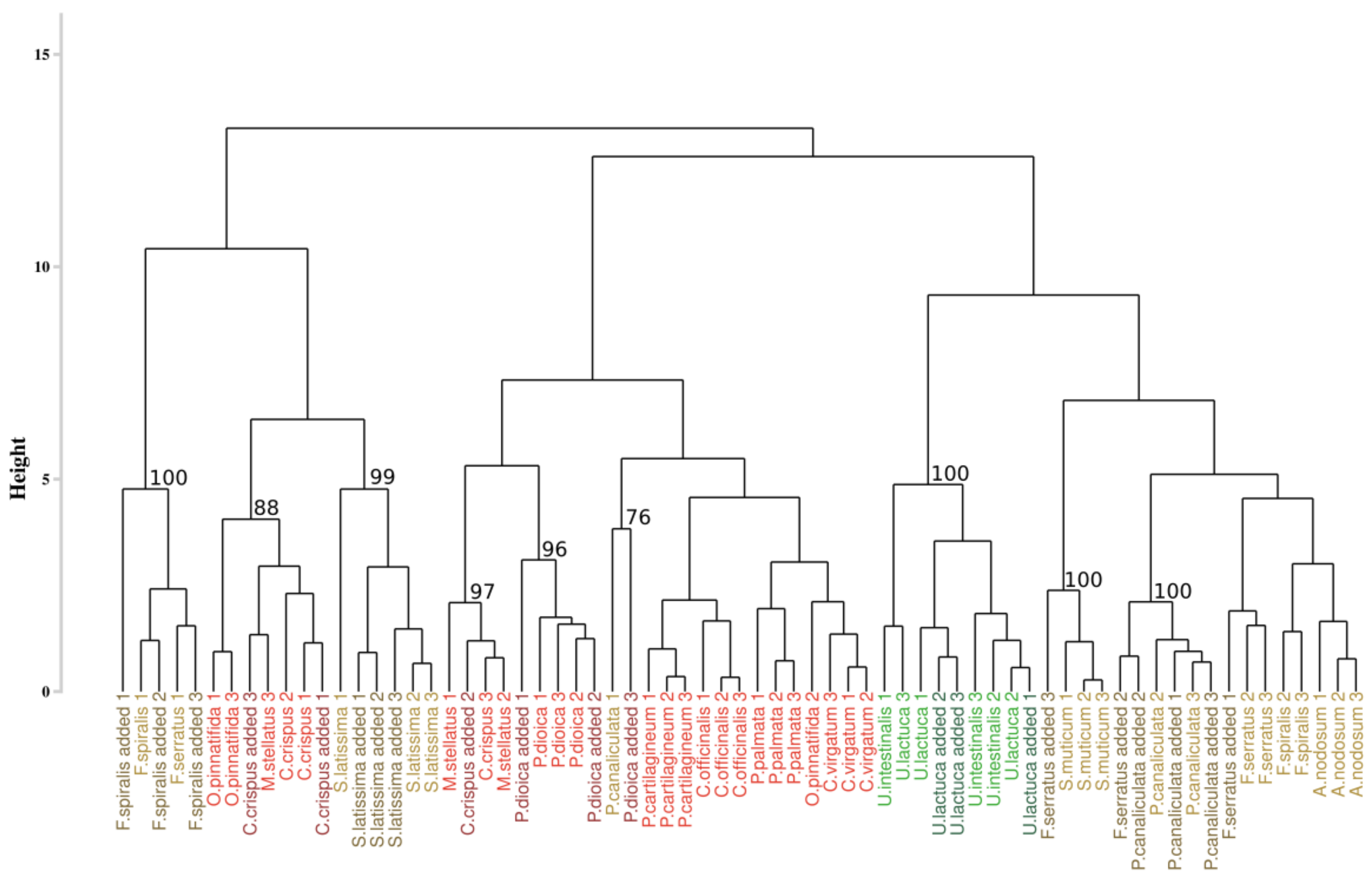
3.3.2. Application of the Spectral Angle Mapper Approach
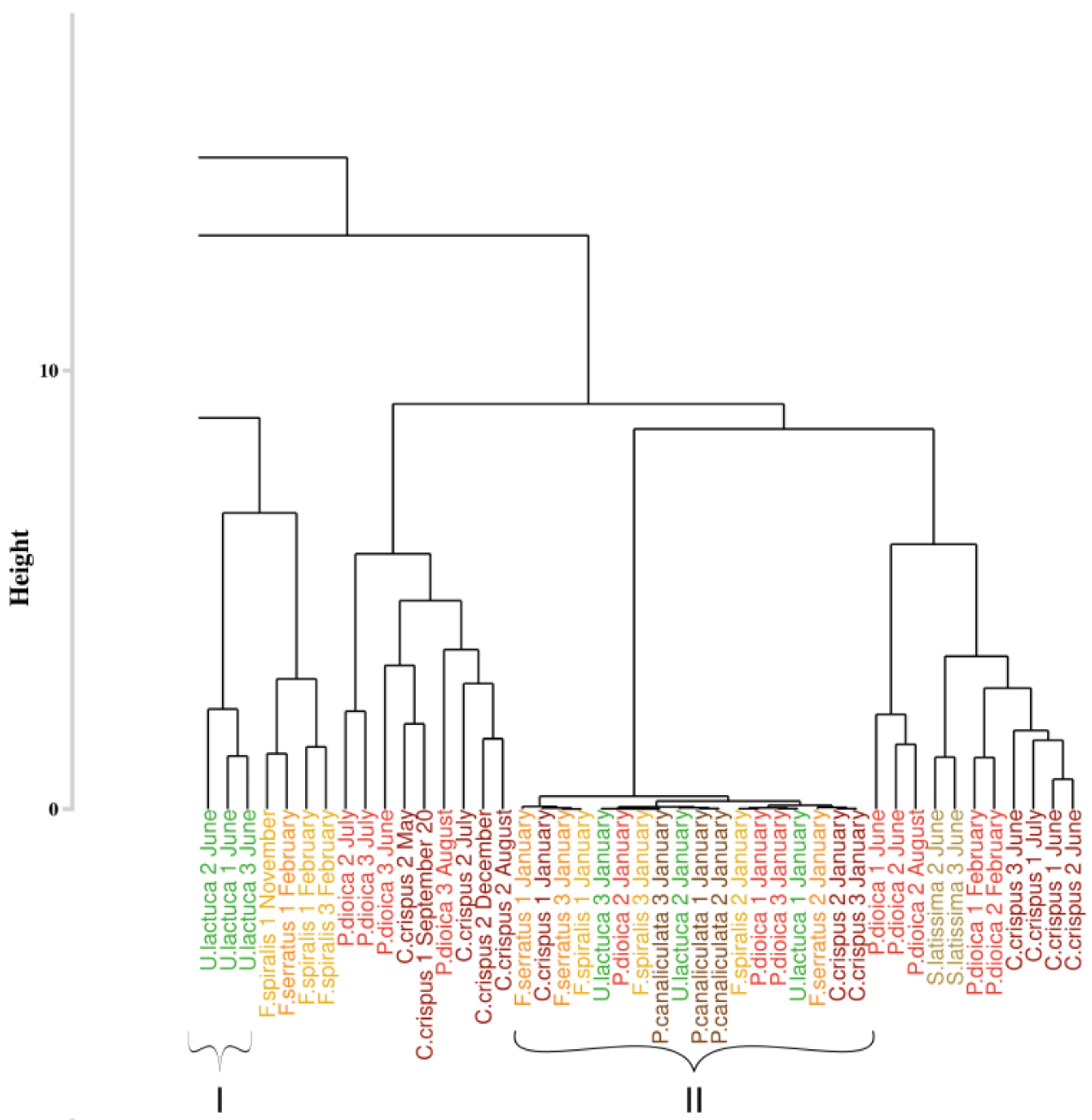
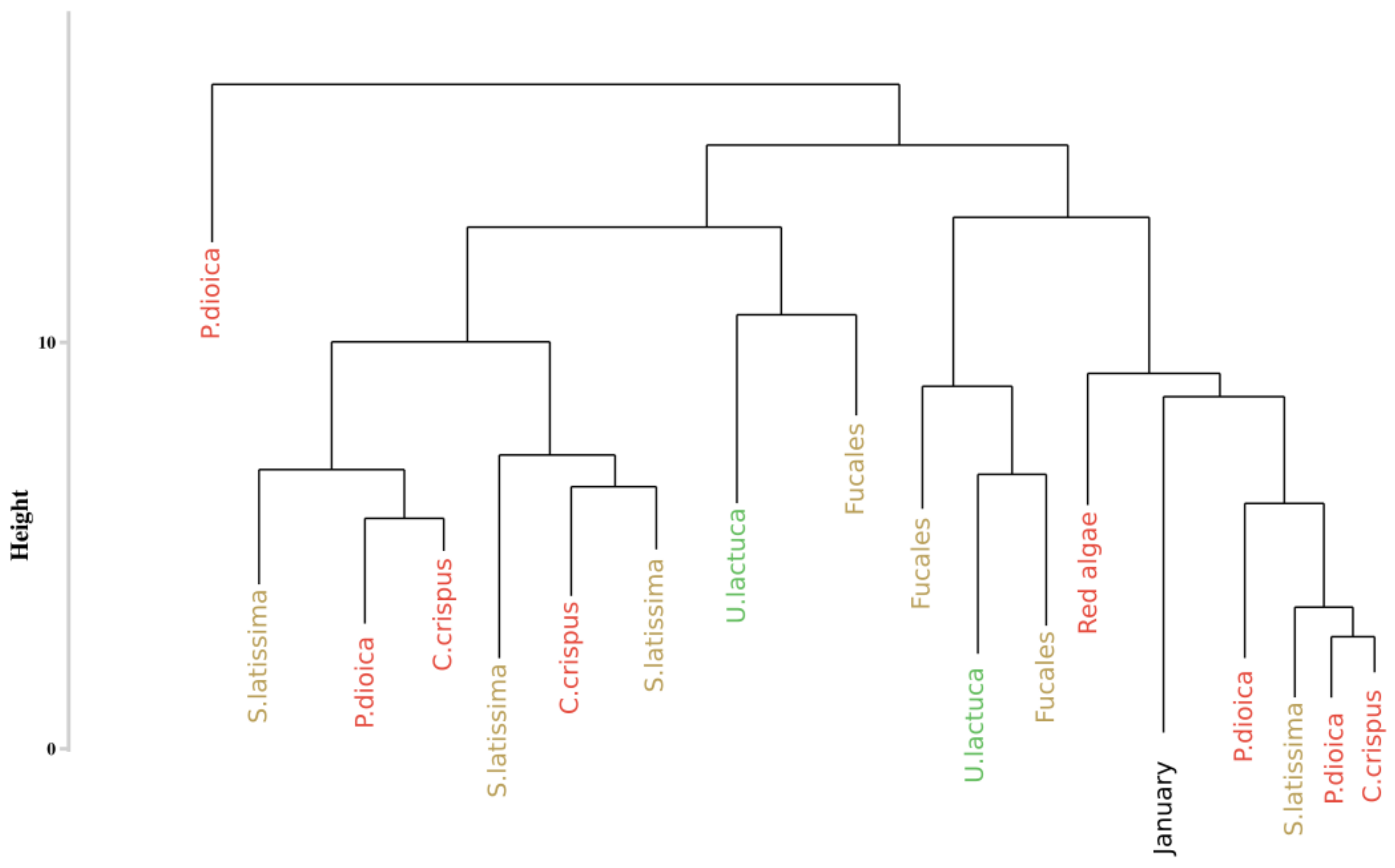
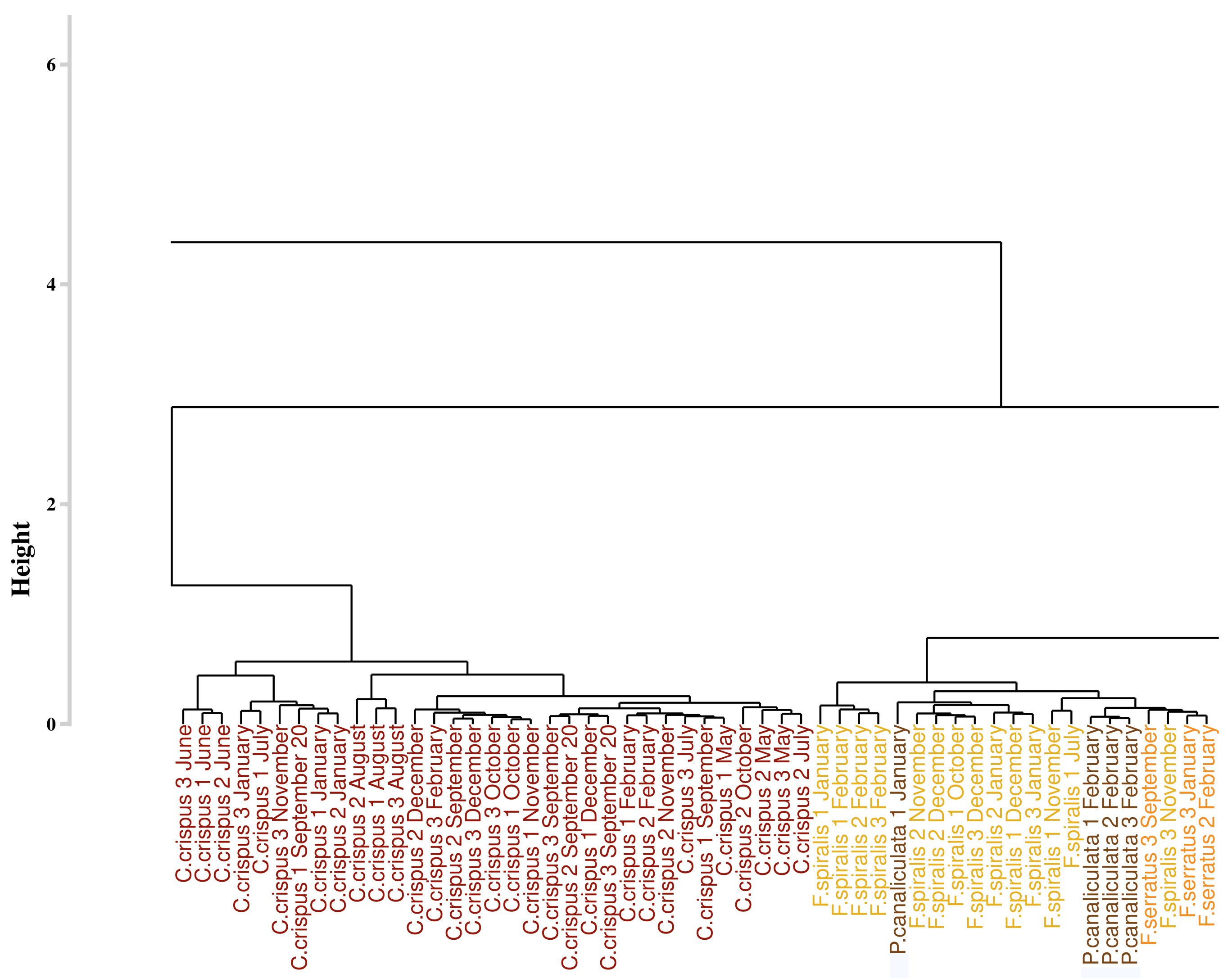
3.4. Assessment of the Approaches Reproducibility over Time
3.4.1. Application of the Euclidean Technique
3.4.2. Application of the Spectral Angle Mapper Approach
4. Discussion
4.1. Macroalgae Discrimination
4.1.1. Pigment
4.1.2. Spectral Signatures Classification
4.2. Robustness
4.3. Monthly Monitoring
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A


References
- Borg, Å.; Pihl, L.; Wennhage, H. Habitat Choice by Juvenile Cod (Gadus Morhua L.) on Sandy Soit Bottoms with Different Vegetation Types. Helgol. Meeresunters. 1997, 51, 197–212. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, A. Preferential Use of Nearshore Kelp Habitats by Juvenile Salmon and Forage Fish. In Proceedings of the Georgia Basin/Puget Sound Research Conference, Vancouver, BC, Canada, 31 March–3 April 2003. [Google Scholar]
- Lorentsen, S.H.; Grémillet, D.; Nymoen, G.H. Annual Variation in Diet of Breeding Great Cormorants: Does It Reflect Varying Recruitment of Gadoids? Waterbirds 2004, 27, 161–169. [Google Scholar] [CrossRef]
- Buschmann, A. Intertidal Macroalgae as Refuge and Food for Amphipoda in Central Chile. Aquat. Bot. 1990, 36, 237–245. [Google Scholar] [CrossRef]
- Rassweiler, A.; Arkema, K.K.; Reed, D.C.; Zimmerman, R.C.; Brzezinski, M.A. Net Primary Production, Growth, and Standing Crop of Macrocystis Pyrifera Southern California. Ecology 2008, 89, 2068. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.I.; Fredriksen, S. Production, Respiration and Exudation of Dissolved Organic Matter by the Kelp LaminariaHyperborea Along West Coast Norway. J. Mar. Biol. Assoc. UK 2004, 84, 887–894. [Google Scholar] [CrossRef]
- Ryther, J.H. Geographic Variations in Productivity. In The Sea: Ideas and Observations on Progress in the Study of the Seas; Hill, M.N., Ed.; Wiley-Interscience: New York, NY, USA, 1963; pp. 347–380. [Google Scholar]
- Tomanek, L.; Helmuth, B. Physiological Ecology of Rocky Intertidal Organisms: A Synergy of Concepts. Integr. Comp. Biol. 2002, 42, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Chappuis, E.; Terradas, M.; Cefalì, M.E.; Mariani, S.; Ballesteros, E. Vertical Zonation Is the Main Distribution Pattern of Littoral Assemblages on Rocky Shores at a Regional Scale. Estuar. Coast. Shelf Sci. 2014, 147, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Raffaelli, D.; Hawkins, S. Intertidal Ecology; Springer: Dordrecht, The Netherlands, 1999. [Google Scholar] [CrossRef]
- Vadas Sr, R.L.; Wright, W.A.; Beal, B.F. Biomass and Productivity of Intertidal Rockweeds (Ascophyllum Nodosum LeJolis) in Cobscook Bay. Northeast. Nat. 2004, 11, 123–142. [Google Scholar] [CrossRef]
- Mann, K.H. Seaweeds: Their Productivity and Strategy for Growth. Science 1973, 182, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Pereira, T.R.; Engelen, A.H.; Pearson, G.A.; Valero, M.; Serrão, E.A. Response of Kelps from Different Latitudes to Consecutive Heat Shock. J. Exp. Mar. Biol. Ecol. 2015, 463, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Wernberg, T.; Russell, B.D.; Thomsen, M.S.; Gurgel, C.F.D.; Bradshaw, C.J.; Poloczanska, E.S.; Connell, S.D. Seaweed Communities in Retreat from Ocean Warming. Curr. Biol. 2011, 21, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Raybaud, V.; Beaugrand, G.; Goberville, E.; Delebecq, G.; Destombe, C.; Valero, M.; Davoult, D.; Morin, P.; Gevaert, F. Decline in Kelp in West Europe and Climate. PLoS ONE 2013, 8, e66044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díez, I.; Muguerza, N.; Santolaria, A.; Ganzedo, U.; Gorostiaga, J. Seaweed Assemblage Changes in the Eastern Cantabrian Sea and Their Potential Relationship to Climate Change. Estuar. Coast. Shelf Sci. 2012, 99, 108–120. [Google Scholar] [CrossRef]
- de la Hoz, C.F.; Ramos, E.; Puente, A.; Juanes, J.A. Climate Change Induced Range Shifts in Seaweeds Distributions in Europe. Mar. Environ. Res. 2019, 148, 1–11. [Google Scholar] [CrossRef]
- Straub, S.C.; Wernberg, T.; Thomsen, M.S.; Moore, P.J.; Burrows, M.T.; Harvey, B.P.; Smale, D.A. Resistance, Extinction, and Everything in between—The Diverse Responses of Seaweeds to Marine Heatwaves. Front. Mar. Sci. 2019, 6, 763. [Google Scholar] [CrossRef]
- Araújo, R.M.; Assis, J.; Aguillar, R.; Airoldi, L.; Bárbara, I.; Bartsch, I.; Bekkby, T.; Christie, H.; Davoult, D.; Derrien-Courtel, S.; et al. Status, Trends and Drivers of Kelp Forests in Europe: An Expert Assessment. Biodivers. Conserv. 2016, 25, 1319–1348. [Google Scholar] [CrossRef] [Green Version]
- Christie, H.; Andersen, G.S.; Bekkby, T.; Fagerli, C.W.; Gitmark, J.K.; Gundersen, H.; Rinde, E. Shifts Between Sugar Kelp and Turf Algae in Norway: Regime Shifts or Fluctuations Between Different Opportunistic Seaweed Species? Front. Mar. Sci. 2019, 6, 72. [Google Scholar] [CrossRef] [Green Version]
- Connell, S.; Foster, M.; Airoldi, L. What Are Algal Turfs? Towards a Better Description of Turfs. Mar. Ecol. Prog. Ser. 2014, 495, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Filbee-Dexter, K.; Wernberg, T. Rise of Turfs: A New Battlefront for Globally Declining Kelp Forests. BioScience 2018, 68, 64–76. [Google Scholar] [CrossRef] [Green Version]
- Coelho, S.M.; Rijstenbil, J.W.; Brown, M.T. Impacts of Anthropogenic Stresses on the Early Development Stages of Seaweeds. J. Aquat. Ecosyst. Stress Recovery 2000, 7, 317–333. [Google Scholar] [CrossRef]
- Hiscock, K.; Southward, A.; Tittley, I.; Hawkins, S. Effects of Changing Temperature on Benthic Marine Life in Britain and Ireland. Aquat. Conserv. Mar. Freshw. Ecosyst. 2004, 14, 333–362. [Google Scholar] [CrossRef]
- Bajjouk, T.; Guillaumont, B.; Populus, J. Application of Airborne Imaging Spectrometry System Data to Intertidal Seaweed Classification and Mapping. Hydrobiologia 1996, 326/327, 463–471. [Google Scholar] [CrossRef]
- Nijland, W. Satellite Remote Sensing of Canopy-Forming Kelp on a Complex Coastline: A Novel Procedure Using the Landsat Image Archive. Remote Sens. Environ. 2019, 220, 41–50. [Google Scholar] [CrossRef]
- Olmedo-Masat, O.M.; Raffo, M.P.; Rodríguez-Pérez, D.; Arijón, M.; Sánchez-Carnero, N. How Far Can We Classify Macroalgae Remotely? An Example Using a New Spectral Library of Species from the South West Atlantic (Argentine Patagonia). Remote Sens. 2020, 12, 3870. [Google Scholar] [CrossRef]
- Nelson, S.A.; Cheruvelil, K.S.; Soranno, P.A. Satellite Remote Sensing of Freshwater Macrophytes and the Influence of Water Clarity. Aquat. Bot. 2006, 85, 289–298. [Google Scholar] [CrossRef]
- Malthus, T.J.; Georgeb, D.G. Airborne Remote Sensing of Macrophytes in Cefni Reservoir, Anglesey, UK. Aquat. Bot. 1997, 58, 317–332. [Google Scholar] [CrossRef]
- Jensen, J.R.; Hodgson, M.E.; Christensen, E. Remote Sensing Inland Wetlands: A Multispectral Approach. Photogramm. Eng. Remote Sens. 1986, 52, 87–100. [Google Scholar]
- Hochberg, E.J.; Atkinson, M.J. Capabilities of Remote Sensors to Classify Coral, Algae, and Sand as Pure and Mixed Spectra. Remote Sens. Environ. 2003, 85, 174–189. [Google Scholar] [CrossRef]
- Karpouzli, E.; Malthus, T.J.; Place, C.J. Hyperspectral Discrimination of Coral Reef Benthic Communities in the Western Caribbean. Coral Reefs 2004, 23, 141–151. [Google Scholar] [CrossRef]
- Kutser, T.; Dekker, A.G.; Skirving, W. Modeling Spectral Discrimination of Great Barrier Reef Benthic Communities by Remote Sensing Instruments. Limnol. Oceanogr. 2003, 48, 497–510. [Google Scholar] [CrossRef] [Green Version]
- Meinesz, A. Methods for Identifying and Tracking Seaweed Invasions. Bot. Mar. 2007, 50, 373–384. [Google Scholar] [CrossRef]
- Uhl, F.; Bartsch, I.; Oppelt, N. Submerged Kelp Detection with Hyperspectral Data. Remote Sens. 2016, 8, 487. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.; Rand, A.; Share, A.; Bolton, J. Mapping and Quantifying the South African Kelp Resource. Afr. J. Mar. Sci. 2007, 29, 369–378. [Google Scholar] [CrossRef]
- Stekoll, M.S.; Deysher, L.E.; Hess, M. A Remote Sensing Approach to Estimating Harvestable Kelp Biomass. J. Appl. Phycol. 2006, 18, 323–334. [Google Scholar] [CrossRef]
- Schroeder, S.B.; Boyer, L.; Juanes, F.; Costa, M. Spatial and Temporal Persistence of Nearshore Kelp Beds on the West Coast of British Columbia, Canada Using Satellite Remote Sensing. Remote Sens. Ecol. Conserv. 2020, 6, 327–343. [Google Scholar] [CrossRef] [Green Version]
- Manfreda, S.; McCabe, M.; Miller, P.; Lucas, R.; Pajuelo Madrigal, V.; Mallinis, G.; Ben Dor, E.; Helman, D.; Estes, L.; Ciraolo, G.; et al. On the Use of Unmanned Aerial Systems for Environmental Monitoring. Remote Sens. 2018, 10, 641. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.; Gaston, K.J. Lightweight Unmanned Aerial Vehicles Will Revolutionize Spatial Ecology. Front. Ecol. Environ. 2013, 11, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Rossiter, T.; Furey, T.; McCarthy, T.; Stengel, D.B. UAV-Mounted Hyperspectral Mapping of Intertidal Macroalgae. Estuar. Coast. Shelf Sci. 2020, 242, 106789. [Google Scholar] [CrossRef]
- Kislik, C.; Genzoli, L.; Lyons, A.; Kelly, M. Application of UAV Imagery to Detect and Quantify Submerged Filamentous Algae and Rooted Macrophytes in a Non-Wadeable River. Remote Sens. 2020, 12, 3332. [Google Scholar] [CrossRef]
- Kotta, J.; Remm, K.; Vahtmäe, E.; Kutser, T.; Orav-Kotta, H. In-Air Spectral Signatures of the Baltic Sea Macrophytes and Their Statistical Separability. J. Appl. Remote Sens. 2014, 8, 083634. [Google Scholar] [CrossRef] [Green Version]
- Kutser, T.; Vahtmäe, E.; Metsamaa, L. Spectral Library of Macroalgae and Benthic Substrates in Estonian Coastal Waters. Proc. Est. Acad. Sci. Biol. Ecol. 2006, 55, 329–340. [Google Scholar] [CrossRef]
- Kutser, T.; Vahtmäe, E.; Martin, G. Assessing Suitability of Multispectral Satellites for Mapping Benthic Macroalgal Cover in Turbid Coastal Waters by Means of Model Simulations. Estuar. Coast. Shelf Sci. 2006, 67, 521–529. [Google Scholar] [CrossRef]
- Vahtmäe, E.; Kutser, T.; Martin, G.; Kotta, J. Feasibility of Hyperspectral Remote Sensing for Mapping Benthic Macroalgal Cover in Turbid Coastal Waters—A Baltic Sea Case Study. Remote Sens. Environ. 2006, 101, 342–351. [Google Scholar] [CrossRef]
- Chao Rodríguez, Y.; Domínguez Gómez, J.; Sánchez-Carnero, N.; Rodríguez-Pérez, D. A Comparison of Spectral Macroalgae Taxa Separability Methods Using an Extensive Spectral Library. Algal Res. 2017, 26, 463–473. [Google Scholar] [CrossRef]
- Lubin, D. Spectral Signatures of Coral Reefs Features from Space. Remote Sens. Environ. 2001, 75, 127–137. [Google Scholar] [CrossRef]
- Fyfe, S.K. Spatial and Temporal Variation in Spectral Reflectance: Are Seagrass Species Spectrally Distinct? Limnol. Oceanogr. 2003, 48, 464–479. [Google Scholar] [CrossRef]
- O’Neill, J.D.; Costa, M.; Sharma, T. Remote Sensing of Shallow Coastal Benthic Substrates: In Situ Spectra and Mapping of Eelgrass (Zostera Marina) in the Gulf Islands National Park Reserve of Canada. Remote Sens. 2011, 3, 975–1005. [Google Scholar] [CrossRef] [Green Version]
- Kisevic, M.; Smailbegovic, A.; Gray, K.T.; Andricevic, R.; Craft, J.D.; Petrov, V.; Brajcic, D.; Dragicevic, I. Spectral Reflectance Profile of Caulerpa Racemosa Var. Cylindracea and Caulerpa Taxifolia in the Adriatic Sea. In Proceedings of the 2011 3rd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Lisbon, Portugal, 6–9 June 2011; pp. 1–4. [Google Scholar] [CrossRef] [Green Version]
- Arsalane, W.; Rousseau, B.; Duval, J.C. Influence of the Pool Size of the Xanthophyll Cycle on the Effects of the Light Stress in a Diatom: Competition Betwe’en Photoprotection and Photoinhibition. Photochem. Photobiol. 1994, 60, 237–243. [Google Scholar] [CrossRef]
- Beer, S.; Eshel, A. Determining Phycoerythrin and Phycocyanin Concentrations in Aqueous Crude Extracts of Red Algae. Mar. Freshw. Res. 1985, 36, 785. [Google Scholar] [CrossRef]
- Lv, W.; Wang, X. Overview of Hyperspectral Image Classification. J. Sens. 2020, 2020, 4817234. [Google Scholar] [CrossRef]
- R Development Core Team. A Language and Environment for Statistical Computing: Reference Index; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Suzuki, R.; Shimodaira, H. Pvclust: An R Package for Assessing the Uncertainty in Hiearchical Clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.; Yang, Z.; Ren, J.; Jiang, M.; Ling, W.K. Does Normalization Methods Play a Role for Hyperspectral Image Classification? arXiv 2017, arXiv:1710.02939. [Google Scholar]
- Demetriades-Shah, T.H.; Steven, M.D.; Clark, J.A. High Resolution Derivative Spectra in Remote Sensing. Remote Sens. Environ. 1990, 33, 55–64. [Google Scholar] [CrossRef]
- Ruffin, C.; King, R. The Analysis of Hyperspectral Data Using Savitzky-Golay Filtering-Theoretical Basis. 1. In Proceedings of the IEEE 1999 International Geoscience and Remote Sensing Symposium. IGARSS’99 (Cat. No.99CH36293), Hamburg, Germany, 28 June–2 July 1999; Volume 2, pp. 756–758. [Google Scholar] [CrossRef]
- Talsky, G. Higher-Order Derivative Spectrophotometry in Environmental Analytical Chemistry. Int. J. Environ. Anal. Chem. 1983, 14, 81–91. [Google Scholar] [CrossRef]
- Boardman, J. SIPS User’s Guide Spectral Image Processing System, Version 1.2; Center for the Study of Earth from Space: Boulder, CO, USA, 1992. [Google Scholar]
- Kruse, F.A.; Heidebrecht, K.B.; Shapiro, A.T.; Barloon, P.J.; Goetz, A.F.H. The Spectral Image Processing System (SIPS) Interactive Visualization and Analysis of Imaging Spectrometer Data. Remote Sens. Environ. 1993, 44, 145–163. [Google Scholar] [CrossRef]
- Ramirez-Lopez, L.; Stevens, A.; Viscarra Rossel, R.; Lobsez, C.; Wadoux, A.; Breure, T. Resemble: Regression and Similarity Evaluation for Memory-Based Learning in Spectral Chemometrics; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Rowan, K.S. Photosynthetic Pigments of Algae; Cambridge University Press: Cambridge, UK; New York, NY, USA, 1989. [Google Scholar]
- Casal, G.; Sánchez-Carnero, N.; Domínguez-Gómez, J.A.; Kutser, T.; Freire, J. Assessment of AHS (Airborne Hyperspectral Scanner) Sensor to Map Macroalgal Communities on the Ría de Vigo and Ría de Aldán Coast (NW Spain). Mar. Biol. 2012, 159, 1997–2013. [Google Scholar] [CrossRef]
- Ramus, J. A Form-Function Analysis of Photon Capture for Seaweeds. Hydrobiologia 1990, 204/205, 64–71. [Google Scholar] [CrossRef]
- Ramus, J. Seaweed Anatomy and Photosynthetic Performance: The Ecological Significance of Light Guides, Heterogeneous Absorption and Multiple Scatter. J. Phycol. 1978, 14, 352–362. [Google Scholar] [CrossRef]
- Enríquez, S.; Agustí, S.; Duarte, C.M. Light Absorption by Marine Macrophytes. Oecologia 1994, 98, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Méléder, V.; Laviale, M.; Jesus, B.; Mouget, J.L.; Lavaud, J.; Kazemipour, F.; Launeau, P.; Barillé, L. In Vivo Estimation of Pigment Composition and Optical Absorption Cross-Section by Spectroradiometry in Four Aquatic Photosynthetic Micro-Organisms. J. Photochem. Photobiol. B Biol. 2013, 129, 115–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beach, K.S.; Borgeas, H.B.; Nishimura, N.J.; Smith, C.M. In Vivo Absorbance Spectra and the Ecophysiology of Reef Macroalgae. Coral Reefs 1997, 16, 21–28. [Google Scholar] [CrossRef]
- Huang, J.; Wei, C.; Zhang, Y.; Blackburn, G.A.; Wang, X.; Wei, C.; Wang, J. Meta-Analysis of the Detection of Plant Pigment Concentrations Using Hyperspectral Remotely Sensed Data. PLoS ONE 2015, 10, e0137029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, S.; Packer, L. Light-Induced Changes in the Conformation and Configuration of the Thylakoid Membrane of Ulva Porphyra Chloroplasts Vivo. Plant Physiol. 1970, 45, 289–299. [Google Scholar] [CrossRef] [Green Version]
- Kirk, J.T.O. Light and Photosynthesis in Aquatic Ecosystems, 3rd ed.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2011. [Google Scholar]
- Korbee, N.; Figueroa, F.L.; Aguilera, J. Effect of Light Quality on the Accumulation of Photosynthetic Pigments, Proteins and Mycosporine-like Amino Acids in the Red Alga PorphyraLeucosticta (Bangiales, Rhodophyta). J. Photochem. Photobiol. B Biol. 2005, 80, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Cruces, E.; Rautenberger, R.; Cubillos, V.M.; Ramírez-Kushel, E.; Rojas-Lillo, Y.; Lara, C.; Montory, J.A.; Gómez, I. Interaction of Photoprotective and Acclimation Mechanisms in Ulva Rigida (Chlorophyta) in Response to Diurnal Changes in Solar Radiation in Southern Chile. J. Phycol. 2019, 55, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Gerasimenko, N.I.; Busarova, N.G.; Moiseenko, O.P. Seasonal Changes in the Content of Lipids, Fatty Acids, and Pigments in Brown Alga Costaria Costata. Russ. J. Plant Physiol. 2010, 57, 205–211. [Google Scholar] [CrossRef]
- García-Sánchez, M.; Korbee, N.; Pérez-Ruzafa, I.M.; Marcos, C.; Figueroa, F.L.; Pérez-Ruzafa, Á. Living in a Coastal Lagoon Environment: Photosynthetic and Biochemical Mechanisms of Key Marine Macroalgae. Mar. Environ. Res. 2014, 101, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Gevaert, F.; Creach, A.; Davoult, D.; Holl, A.C.; Seuront, L.; Lemoine, Y. Photo-Inhibition and Seasonal Photosynthetic Performance of the Seaweed LaminariaSaccharina A Simulated Tidal Cycle: Chlorophyll Fluoresc. Meas. Pigment Anal. Plant, Cell Environ. 2002, 25, 859–872. [Google Scholar] [CrossRef] [Green Version]
- Flores-Moya, A.; Fernandez, J.A.; Niell, F.X. Seasonal Variations of Photosynthetic Pigments, Total C, N, and P Content, and Photosynthesis in PhyllariopsisPurpurascens (Phaeophyta) Strait Gibraltar. J. Phycol. 1995, 31, 867–874. [Google Scholar] [CrossRef]
- Somers, B.; Asner, G. Tree Species Mapping in Tropical Forests Using Mult-Temporal Imaging Spectroscopy: Wavelength Adaptative Spectral Mixture Analysis. Int. J. Appl. Earth Obs. Geoinf. 2014, 31, 57–66. [Google Scholar] [CrossRef]
- Selvaraj, S.; Case, B.S.; White, W.L. Effects of Location and Season on Seaweed Spectral Signatures. Front. Ecol. Evol. 2021, 9, 581852. [Google Scholar] [CrossRef]
- Uhl, F.; Oppelt, N.; Bartsch, I. Spectral Mixture of Intertidal Marine Macroalgae around the Island of Helgoland (Germany, North Sea). Aquat. Bot. 2013, 111, 112–124. [Google Scholar] [CrossRef]

| Phylum | Species | Abbreviations |
|---|---|---|
| Chlorophyta | Ulva lactuca * | U. lactuca |
| Ulva intestinalis | U. intestinalis | |
| Ochrophyta | Ascophyllum nodosum | A. nodosum |
| Fucus serratus * | F. serratus | |
| Fucus spiralis * | F. spiralis | |
| Pelvetia canaliculata * | P. canaliculata | |
| Saccharina latissima * | S. latissima | |
| Sargassum muticum | S. muticum | |
| Rhodophyta | Ceramium virgatum | C. virgatum |
| Chondrus crispus * | C. crispus | |
| Corallina officinalis | C. officinalis | |
| Mastocarpus stellatus | M. stellatus | |
| Osmundea pinnatifida | O. pinnatifida | |
| Palmaria palmata | P. palmata | |
| Plocamium cartilagineum | P. cartilagineum | |
| Porphyra dioica * | P. dioica |
| Species | Chlb | Chlc | Vio | Ant | Fuc | Zea | Car | Lut | Neo | PE | PC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| U. intestinalis | 39.4 | 0 | 6.63 | 0.22 | 0 | 0 | 7.68 | 17.86 | 4.89 | 0 | 0 |
| (1.62) | (0.84) | (0.38) | (2.25) | (0.58) | (0.5) | ||||||
| U. lactuca | 49.96 | 0 | 1.43 | 0.35 | 0 | 0 | 2.42 | 11.67 | 0.7 | 0 | 0 |
| (1.86) | (0.43) | (0.21) | (0.77) | (1.34) | (0.39) | ||||||
| A. nodosum | 0 | 5.33 | 12.48 | 1.85 | 29.4 | 1.37 | 3.21 | 0 | 0 | 0 | 0 |
| (0.64) | (1.22) | (0.29) | (2.70) | (0.62) | (2.05) | ||||||
| F. serratus | 0 | 7.69 | 11.69 | 1.22 | 32.88 | 0.58 | 4.57 | 0 | 0 | 0 | 0 |
| (0.29) | (0.87) | (0.02) | (1.29) | (0.14) | (0.74) | ||||||
| F. spiralis | 0 | 6.12 | 9.52 | 1.99 | 26.1 | 2.6 | 4.99 | 0 | 0 | 0 | 0 |
| (0.63) | (0.53) | (0.11) | (1.87) | (0.62) | (0.36) | ||||||
| P. canaliculata | 0 | 8.18 | 14.22 | 1.17 | 33.21 | 1.88 | 4.39 | 0 | 0 | 0 | 0 |
| (1.62) | (3.64) | (0.38) | (6.93) | (1.34) | (1.81) | ||||||
| S. latissima | 0 | 10.17 | 2.6 | 0.15 | 39.64 | 0.01 | 1.10 | 0 | 0 | 0 | 0 |
| (3.07) | (0.31) | (0.03) | (5.89) | (0.01) | (0.83) | ||||||
| S. muticum | 0 | 12.49 | 6.88 | 0 | 43.59 | 0.16 | 5.78 | 0 | 0 | 0 | 0 |
| (0.24) | (0.66) | (2.64) | (0.02) | (0.39) | |||||||
| C. virgatum | 0 | 0 | 0.66 | 6.32 | 0 | 0 | 9.87 | 9.23 | 0 | 8.11 | 0.96 |
| (0.19) | (4.31) | (2.8) | (2.86) | (1.44) | (0.6) | ||||||
| C. crispus | 0 | 0 | 0 | 0 | 0 | 0 | 7.23 | 20.05 | 0 | 0.79 | 0.11 |
| (1.39) | (3.48) | (.14) | (0.01) | ||||||||
| C. officinalis | 0 | 0 | 0.52 | 9.32 | 0 | 2.31 | 15.53 | 1.87 | 0 | 15.07 | 1.69 |
| (0.14) | (1.35) | (1.23) | (4.05) | (1.11) | (0.72) | (0.72) | |||||
| M. stellatus | 0 | 0 | 0 | 0 | 0 | 0 | 5.62 | 26.94 | 0 | 12.5 | 5.7 |
| (3.99) | (0.81) | (3.63) | (1.06) | ||||||||
| O. pinnatifida | 0 | 0 | 0 | 0.37 | 0 | 5.80 | 6.73 | 1.04 | 0 | 43.19 | 6.23 |
| (0.18) | (2.24) | (1.93) | (0.36) | (1.51) | (1.07) | ||||||
| P. palmata | 0 | 0 | 0 | 0 | 0 | 0 | 12.8 | 23.6 | 0 | 7.8 | 1.12 |
| (0.76) | (3.16) | (1.06) | (0.25) | ||||||||
| P. cartilagineum | 0 | 0 | 0 | 0 | 0 | 0 | 9.36 | 11.66 | 0 | 84.58 | 11.96 |
| (1.34) | (2.68) | (17.57) | (1.24) | ||||||||
| P. dioica | 0 | 0 | 0 | 0 | 0 | 0 | 9.77 | 24.09 | 0 | 4.85 | 2.04 |
| (2.11) | (2.04) | (0.75) | (0.43) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Douay, F.; Verpoorter, C.; Duong, G.; Spilmont, N.; Gevaert, F. New Hyperspectral Procedure to Discriminate Intertidal Macroalgae. Remote Sens. 2022, 14, 346. https://doi.org/10.3390/rs14020346
Douay F, Verpoorter C, Duong G, Spilmont N, Gevaert F. New Hyperspectral Procedure to Discriminate Intertidal Macroalgae. Remote Sensing. 2022; 14(2):346. https://doi.org/10.3390/rs14020346
Chicago/Turabian StyleDouay, Florian, Charles Verpoorter, Gwendoline Duong, Nicolas Spilmont, and François Gevaert. 2022. "New Hyperspectral Procedure to Discriminate Intertidal Macroalgae" Remote Sensing 14, no. 2: 346. https://doi.org/10.3390/rs14020346
APA StyleDouay, F., Verpoorter, C., Duong, G., Spilmont, N., & Gevaert, F. (2022). New Hyperspectral Procedure to Discriminate Intertidal Macroalgae. Remote Sensing, 14(2), 346. https://doi.org/10.3390/rs14020346






