Clustering Arid Rangelands Based on NDVI Annual Patterns and Their Persistence
Abstract
1. Introduction
2. Materials and Methods
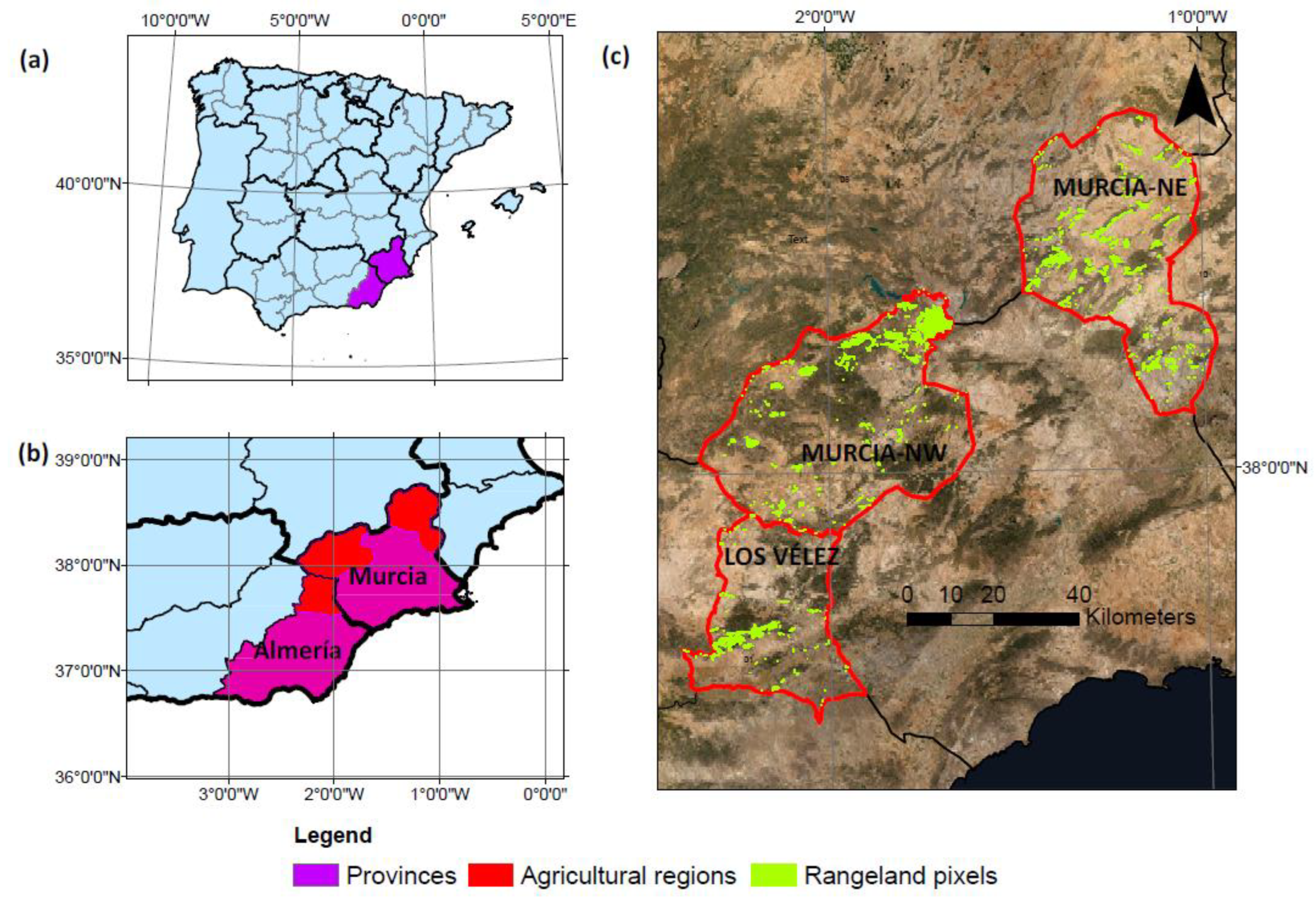
2.1. Area of Study
2.2. Data Collection
2.2.1. NDVI Data
2.2.2. GIS Data
2.3. Fractal Analysis
2.3.1. Rescale Ranged Hurst Exponent
2.3.2. Multifractal Detrended Fluctuation Analysis
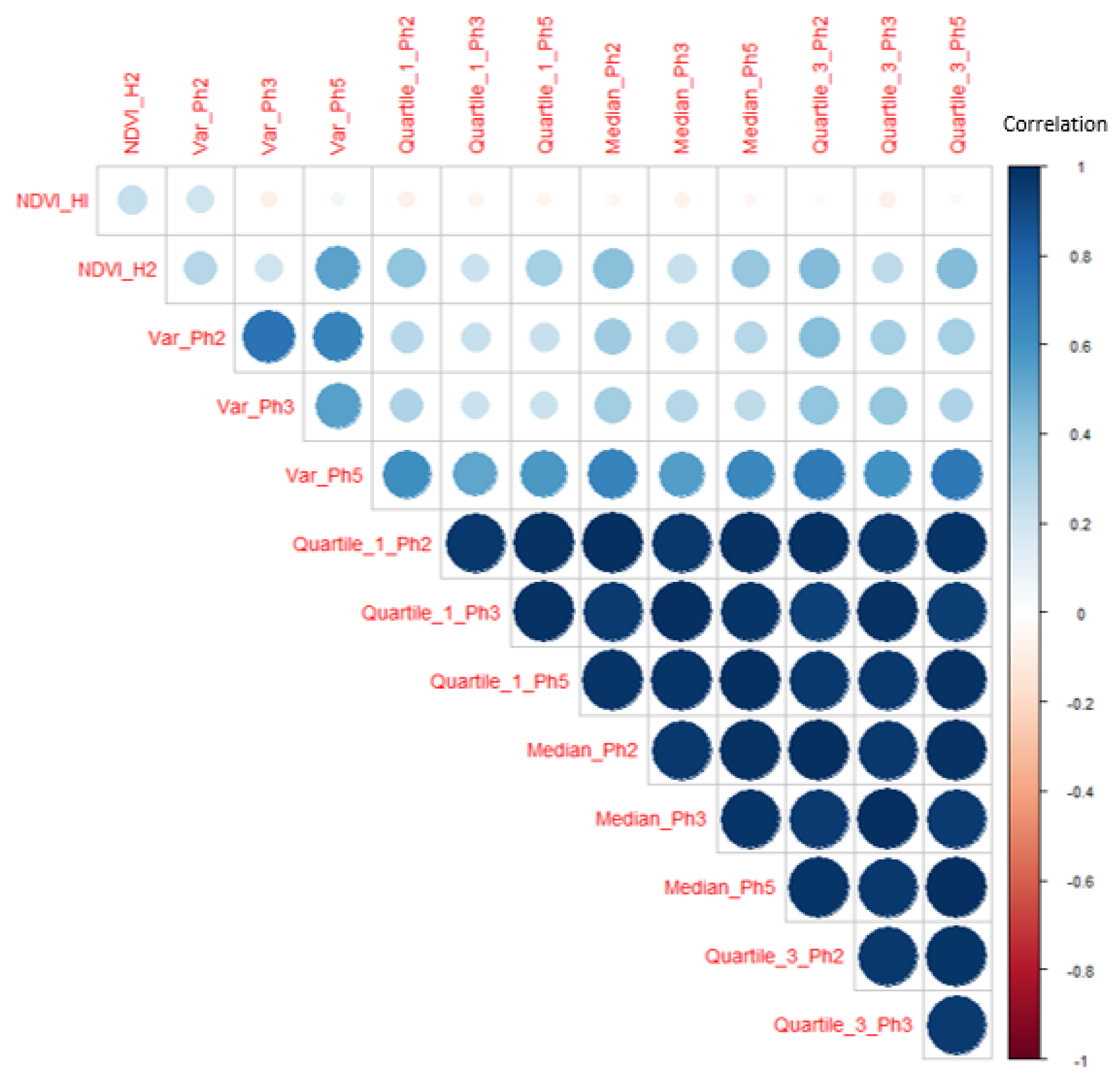
2.4. Variable Selection for Clustering
2.5. Clustering
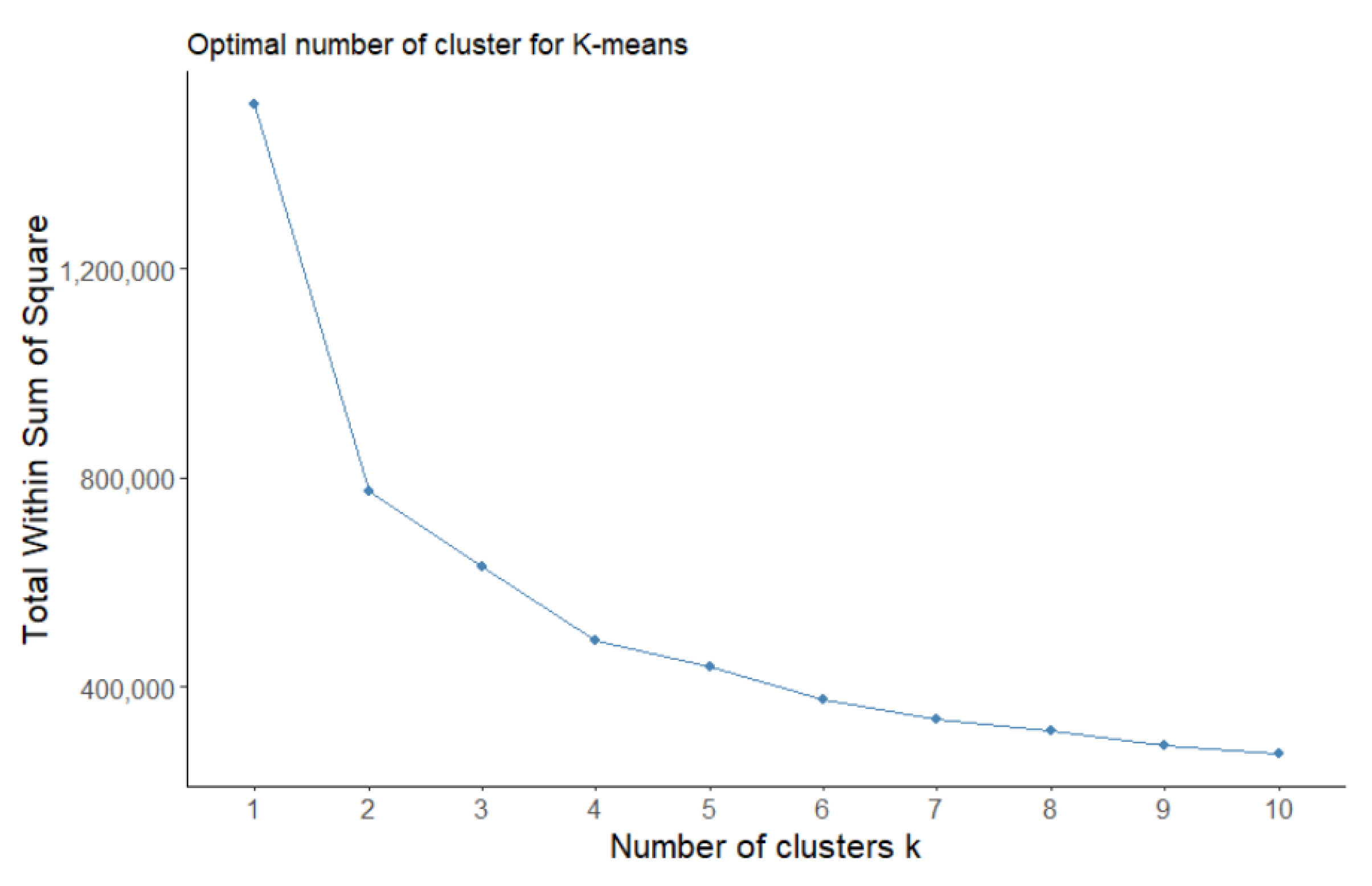
2.5.1. K-Means
2.5.2. Unsupervised Random Forest
3. Results
3.1. Variable Selection Approach
3.2. Clustering Analysis
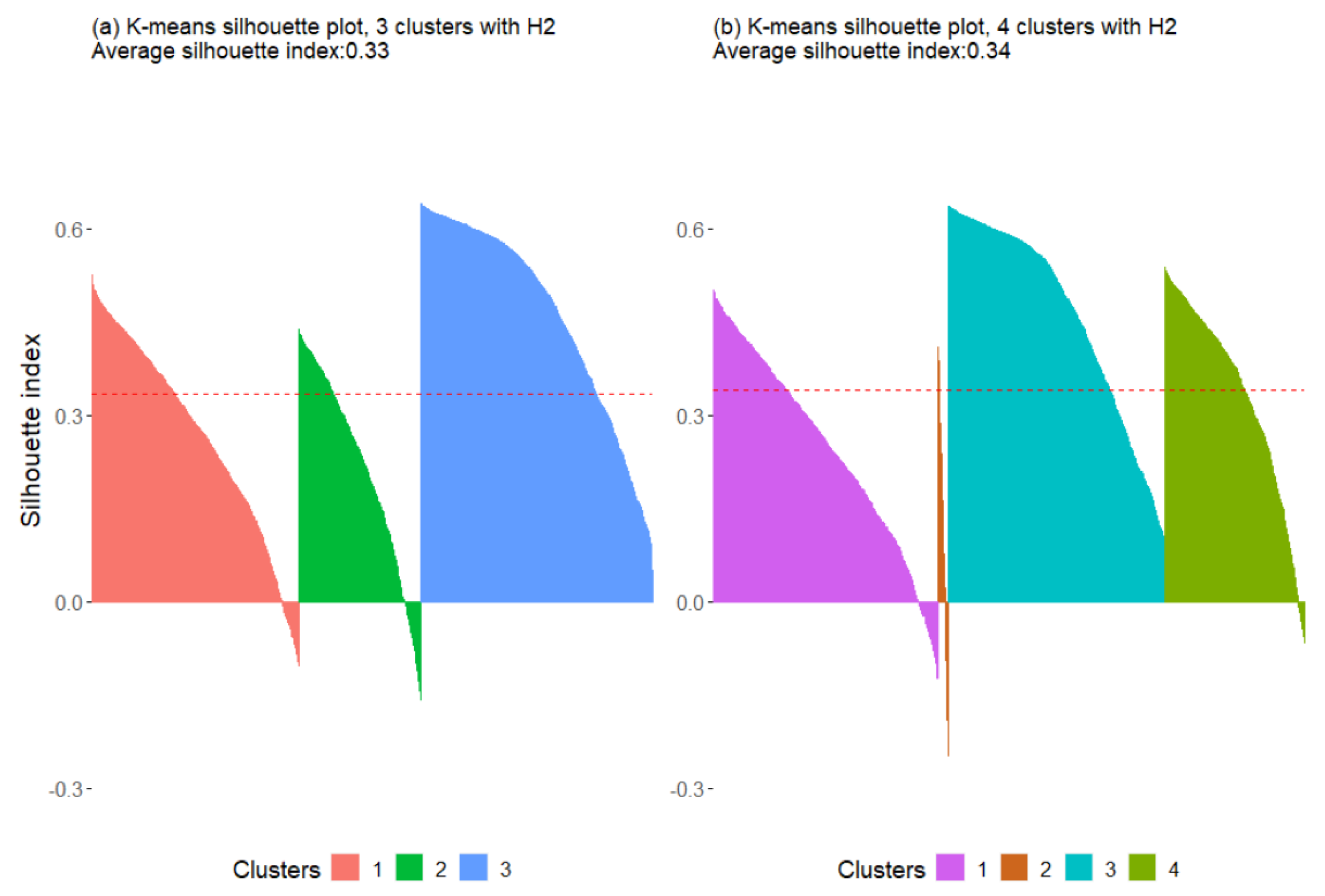
3.2.1. K-Means
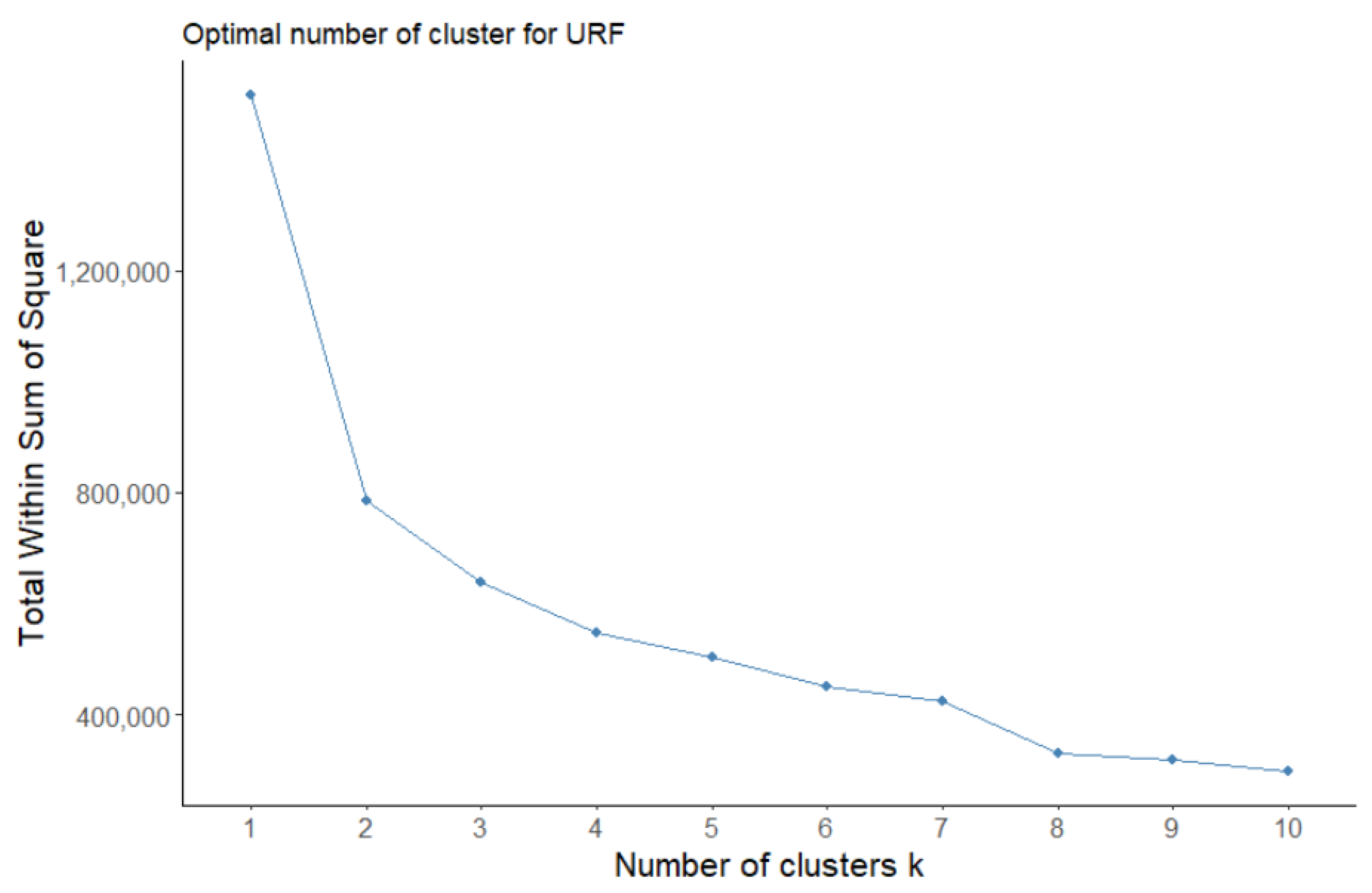
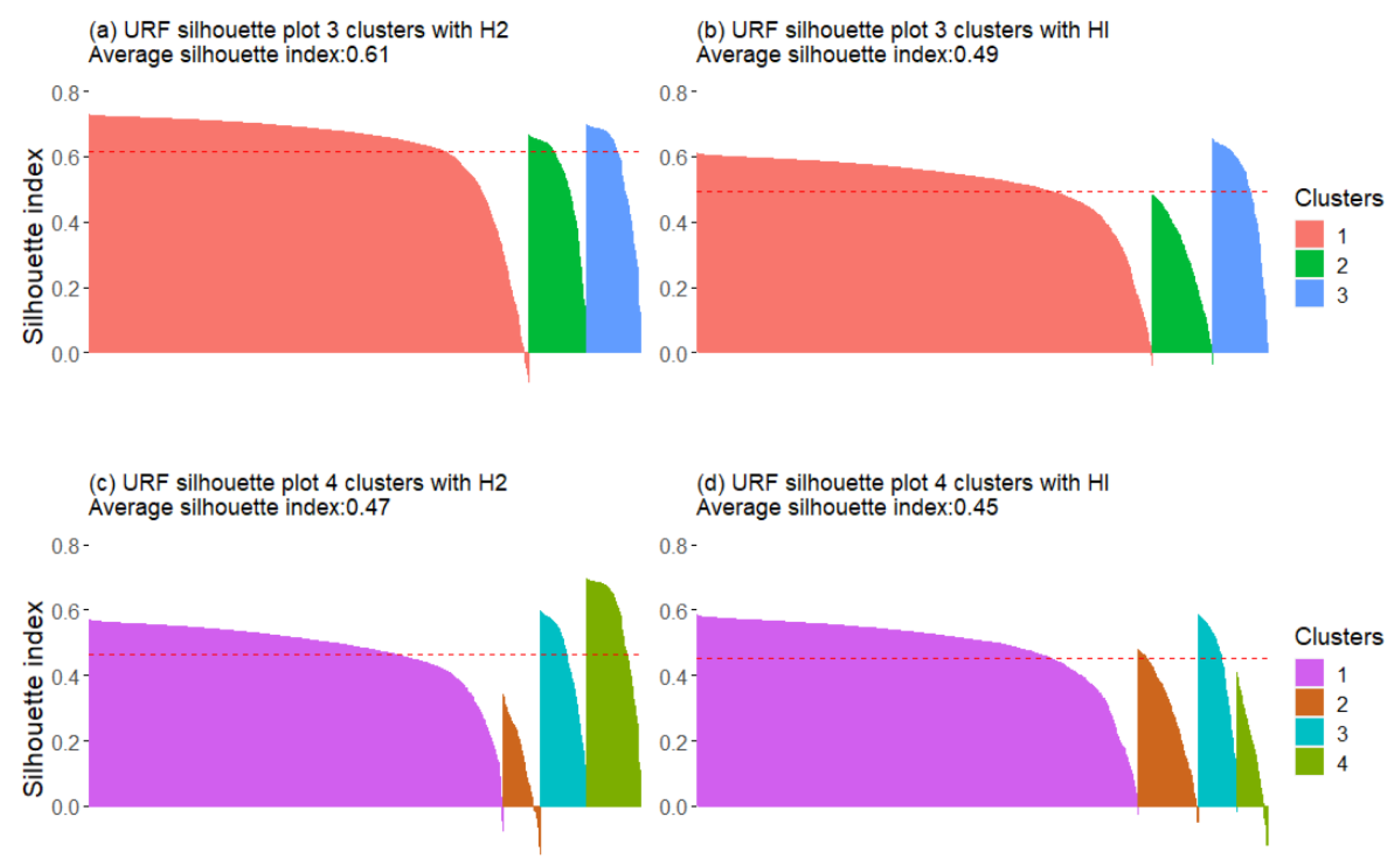
3.2.2. Unsupervised Random Forest
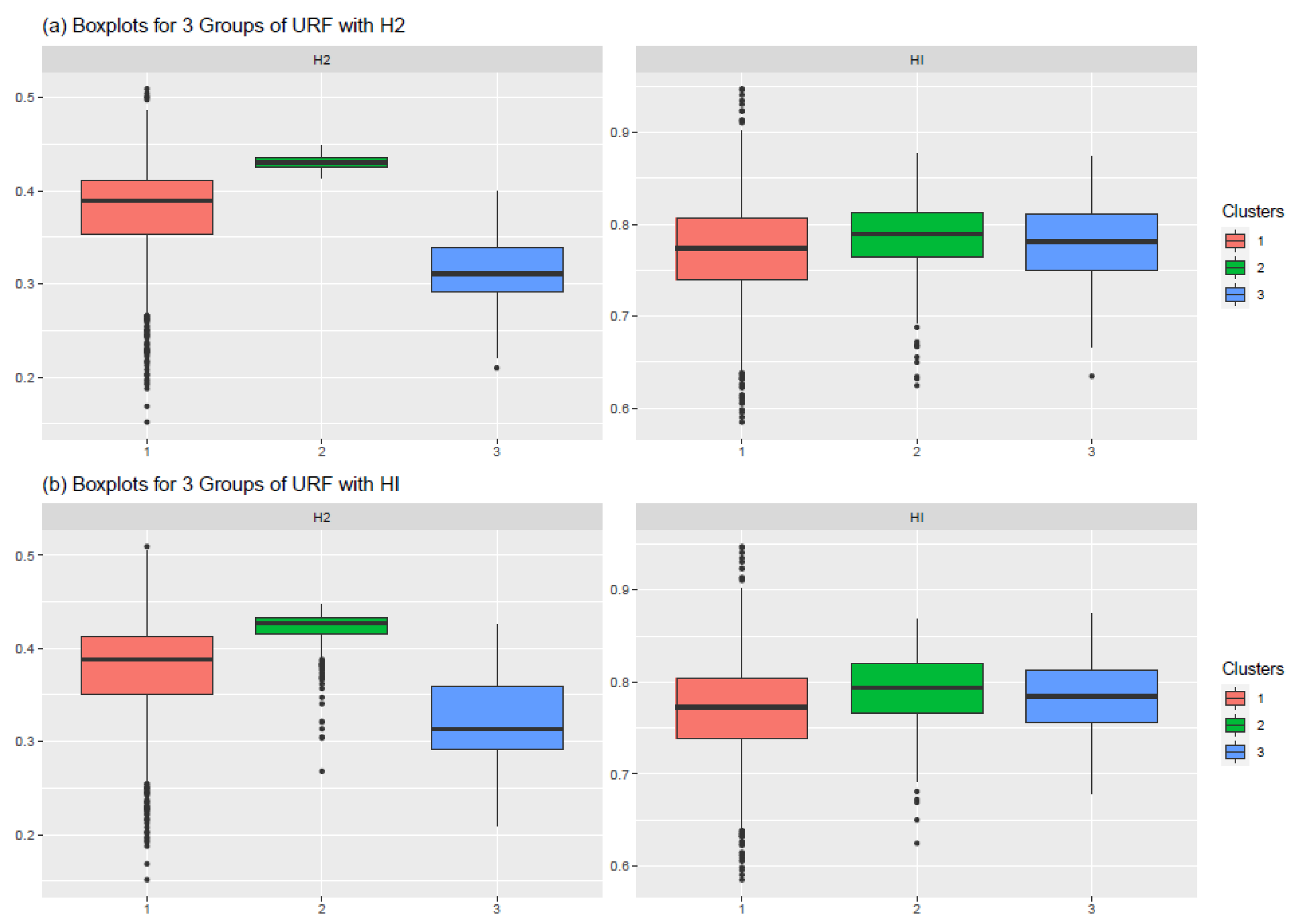
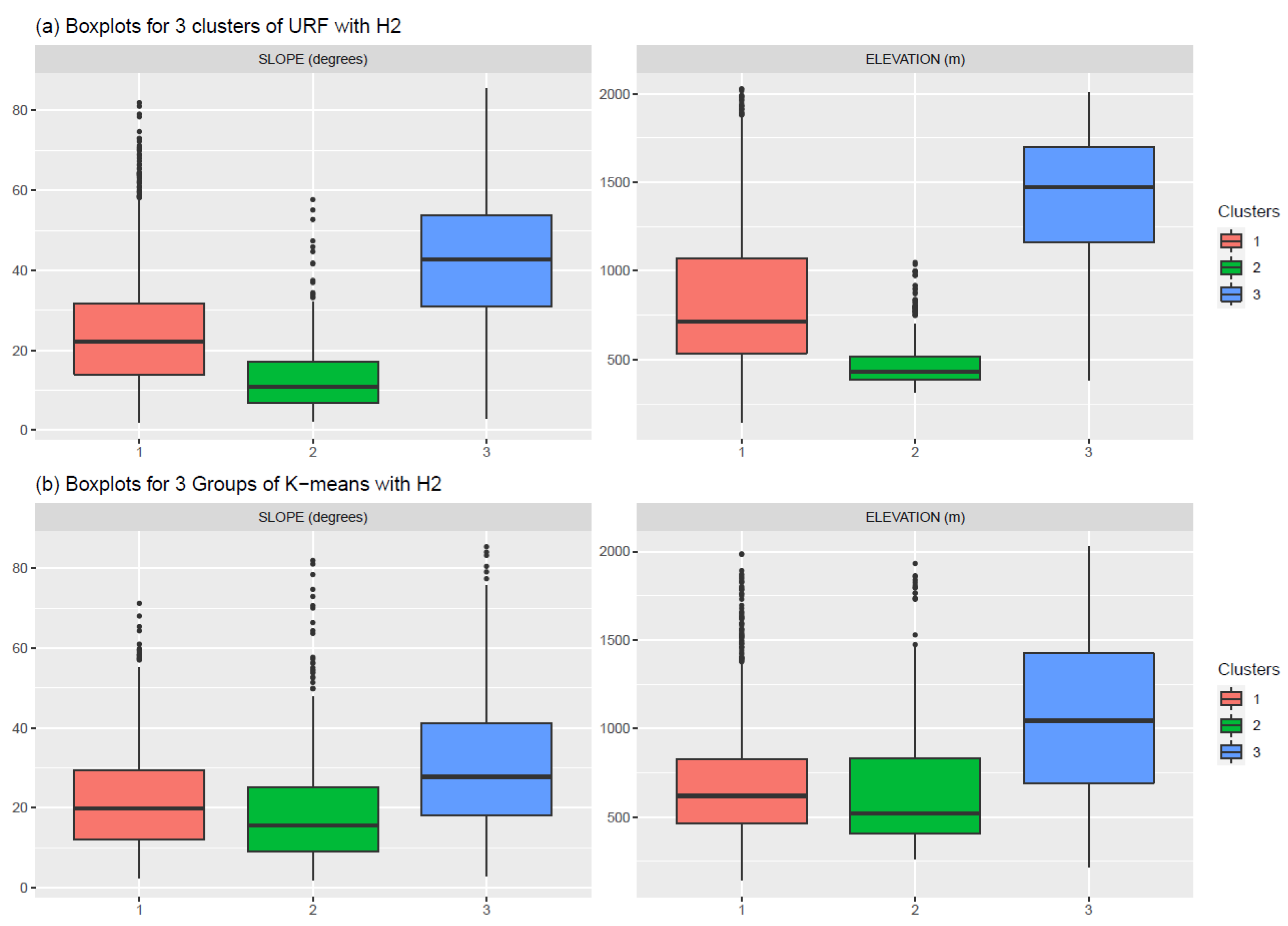
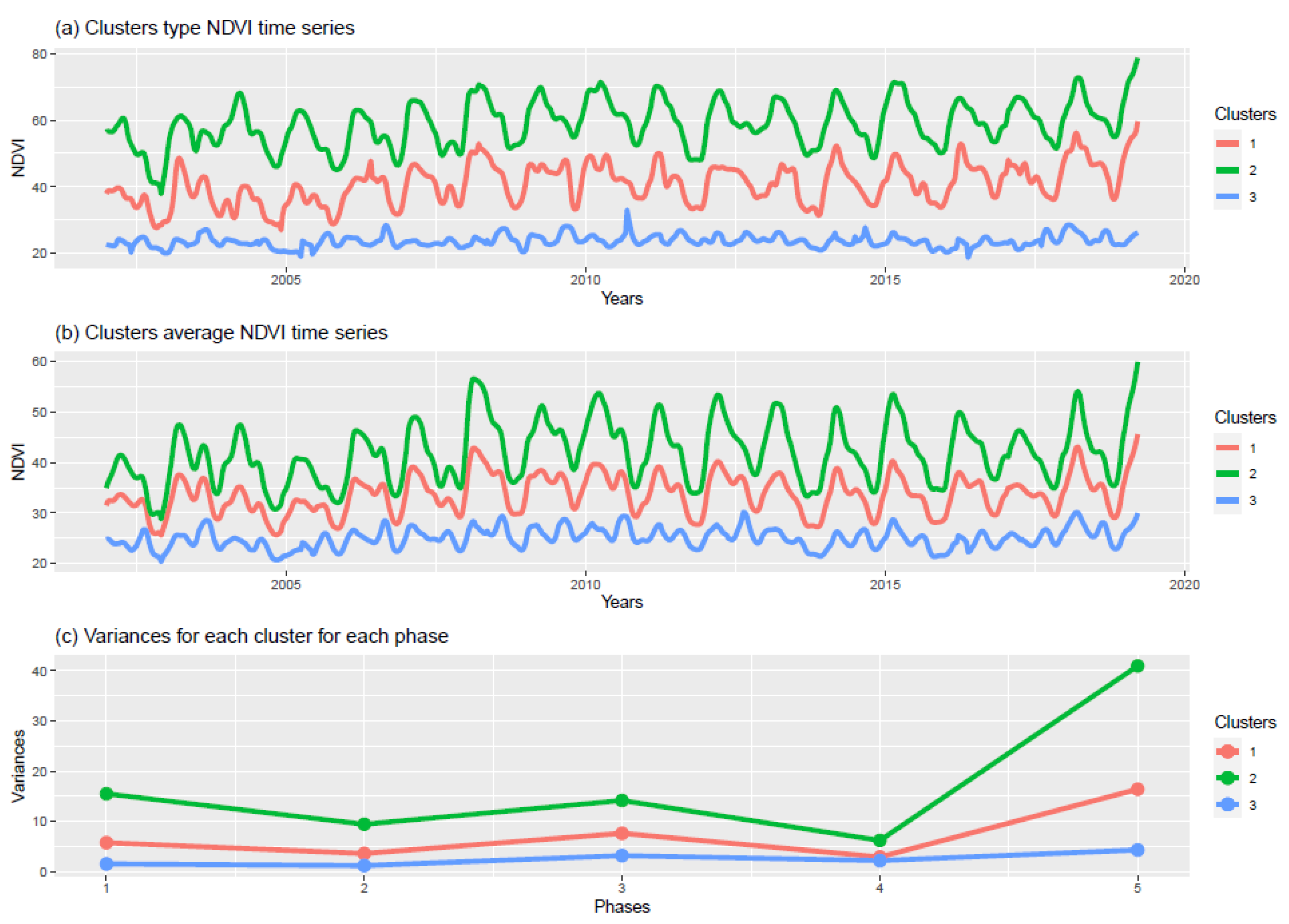
3.2.3. Cluster Characterisation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A. Principal Component Analyses Made to Select the Most Explanatory Variables
| Variables | PC1 | PC2 | PC3 |
|---|---|---|---|
| NDVI_H2 | 0.14 | −0.27 | 0.84 |
| Var_Ph2 | 0.13 | −0.59 | −0.22 |
| Var_Ph3 | 0.13 | −0.56 | −0.37 |
| Var_Ph5 | 0.23 | −0.38 | 0.17 |
| Quartile_1_Ph2 | 0.32 | 0.01 | 0.03 |
| Quartile_1_Ph3 | 0.31 | 0.19 | −0.13 |
| Quartile_1_Ph5 | 0.31 | 0.16 | 001 |
| Median_Ph2 | 0.32 | 0.05 | 0.2 |
| Median_Ph3 | 0.31 | 0.14 | −0.14 |
| Median_Ph5 | 0.31 | 0.11 | 0.04 |
| Quartile_3_Ph2 | 0.32 | −0.01 | 0.01 |
| Quartile_3_Ph3 | 0.32 | 0.08 | −0.17 |
| Quartile_3_Ph5 | 0.33 | 0.05 | 0.07 |
| Standard deviation | 3.10 | 1.36 | 0.95 |
| Proportion of Variance | 0.74 | 0.14 | 0.07 |
| Cumulative Proportion | 0.74 | 0.88 | 0.94 |
| Variables | PC1 | PC2 | PC3 |
|---|---|---|---|
| NDVI_H2 | 0.36 | −0.54 | 0.74 |
| Var_Ph5 | 0.47 | 0.45 | 0.13 |
| Var_Ph3 | 0.43 | 0.54 | 0.07 |
| Var_Ph2 | 0.54 | −0.13 | −0.16 |
| Quartile_3_Ph5 | 0.12 | −0.43 | −0.64 |
| Standard deviation | 1.72 | 1.02 | 0.76 |
| Proportion of Variance | 0.59 | 0.21 | 0.12 |
| Cumulative Proportion | 0.59 | 0.79 | 0.91 |
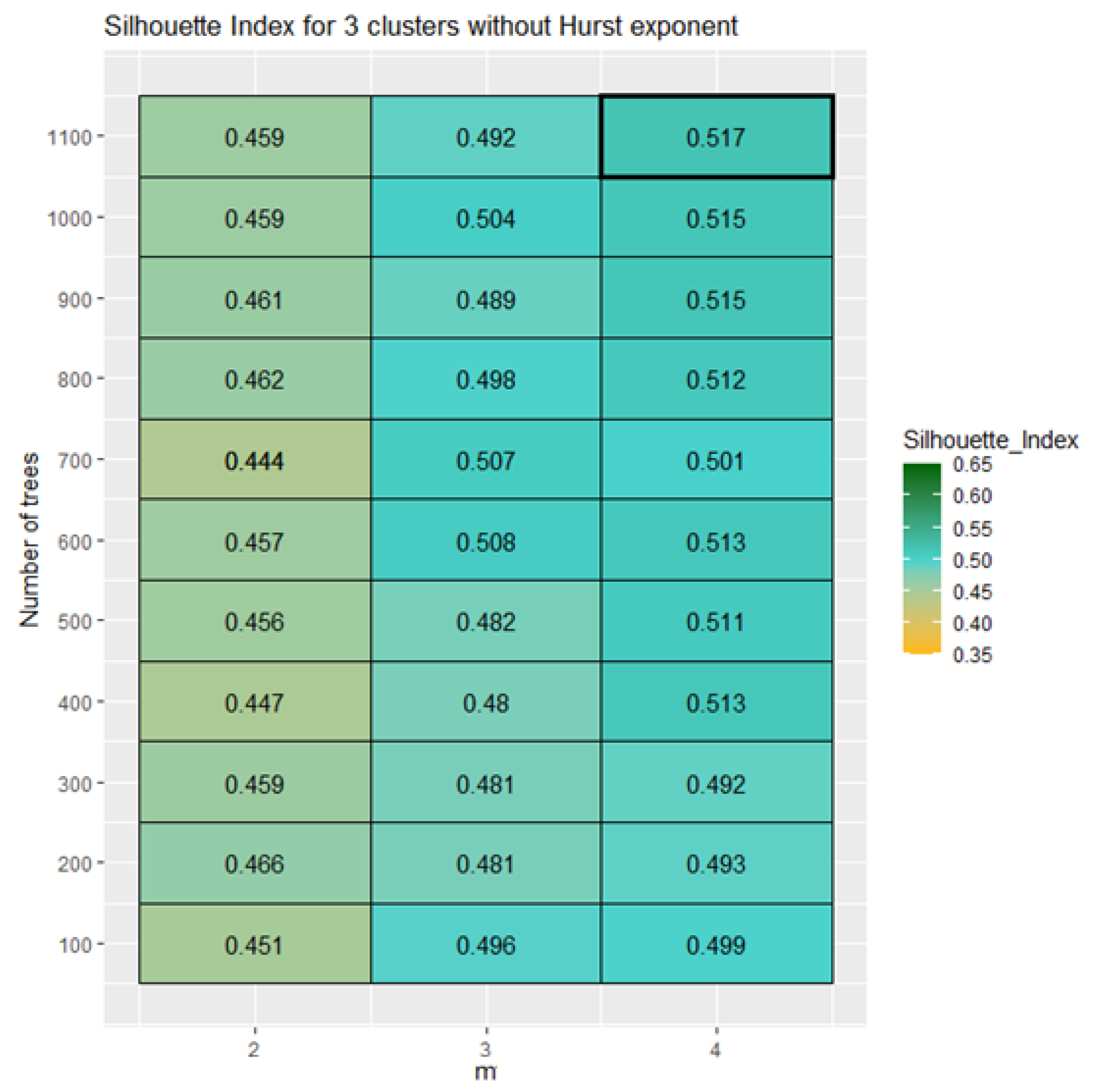
Appendix B. The Silhouette Indexes Calculated for All URF Changing Mtry and Number of Trees for Three Clusters


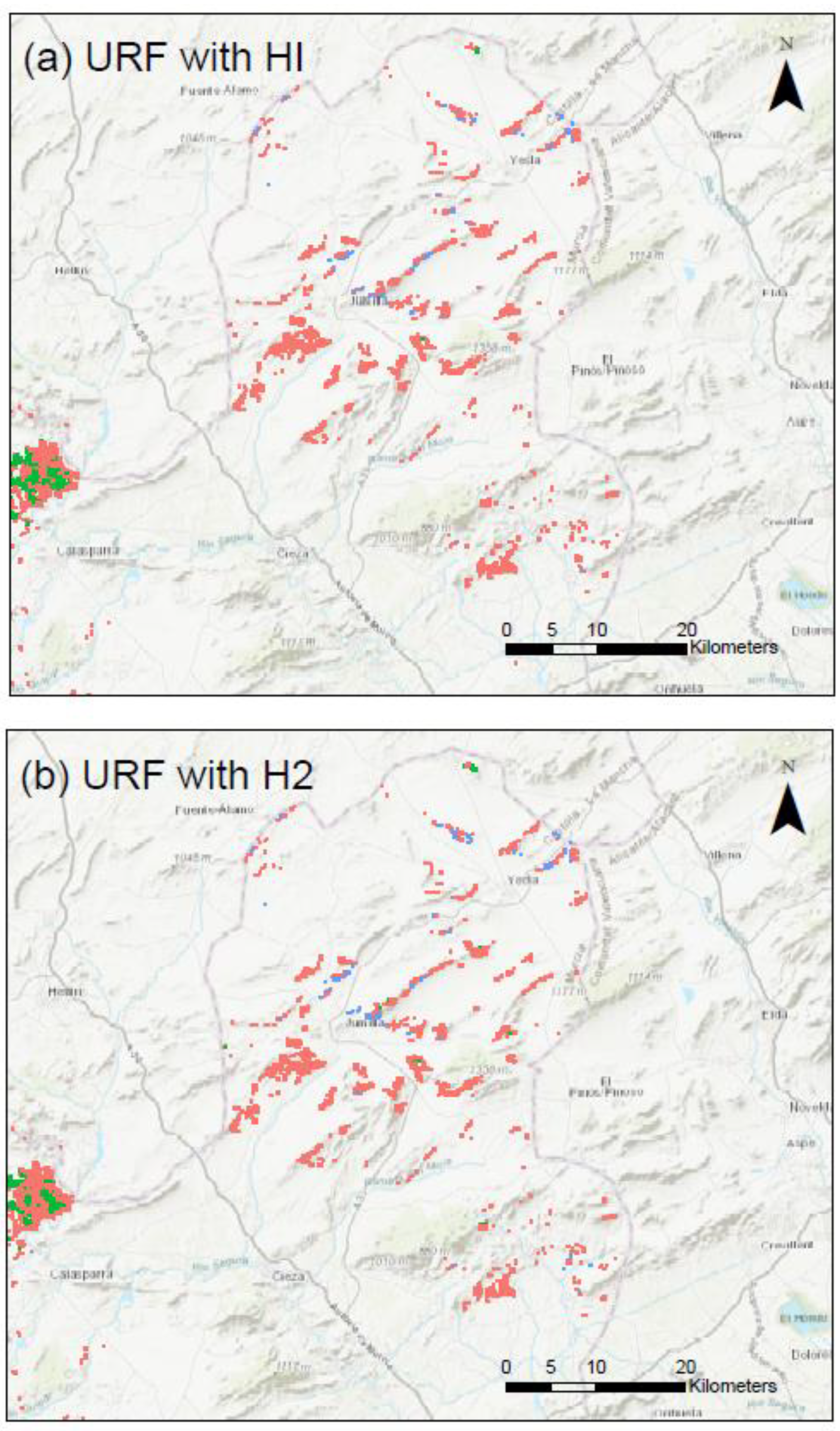
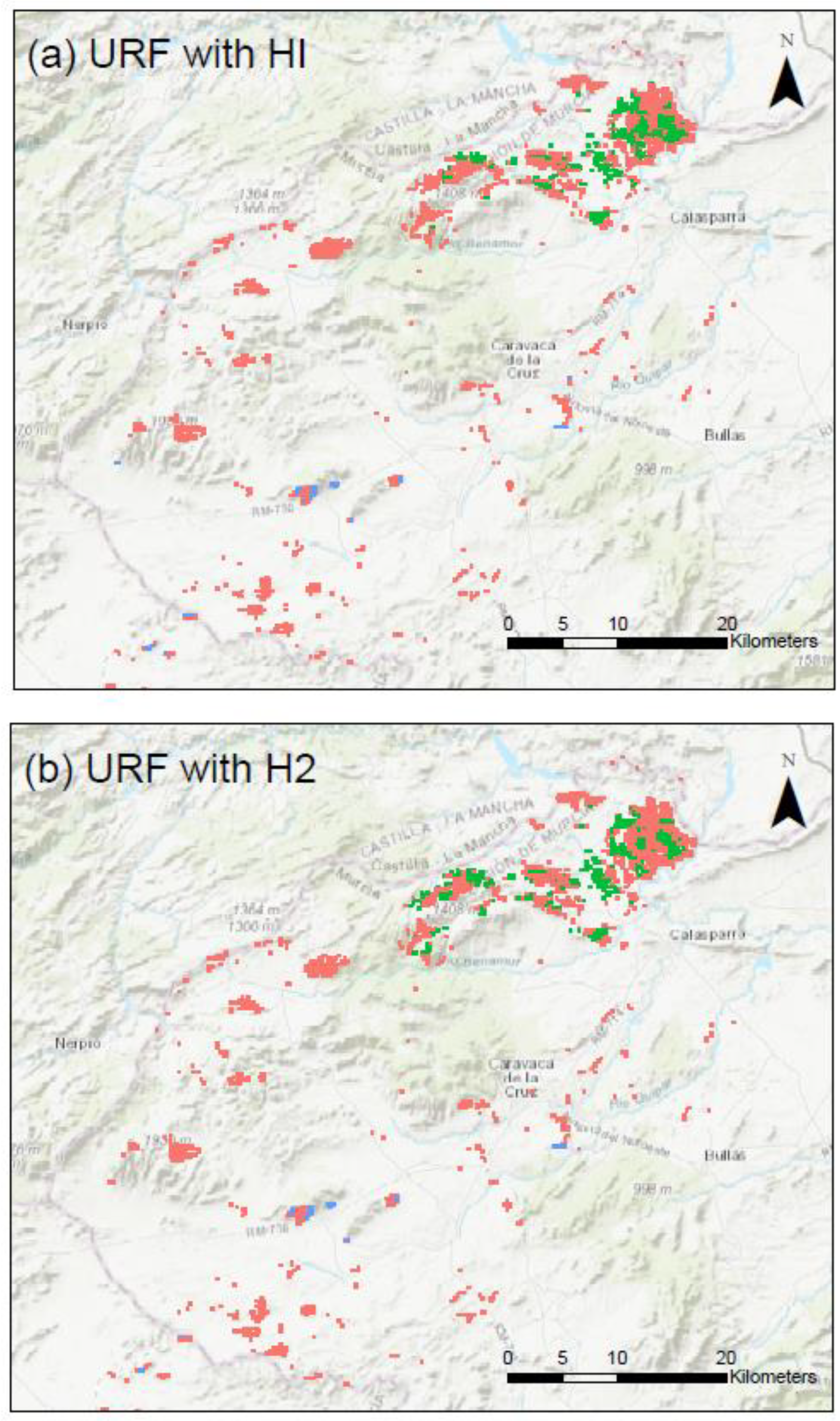
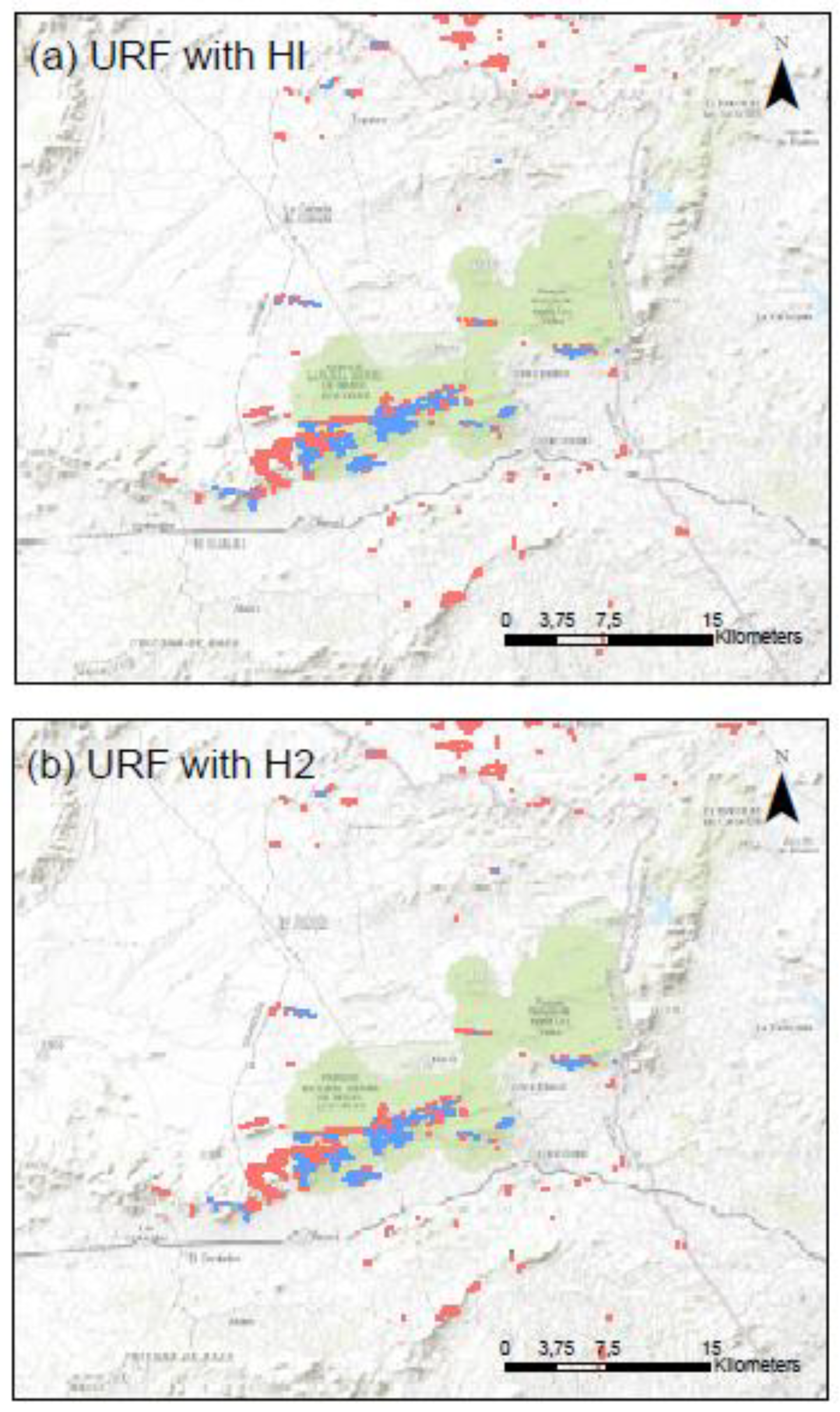
Appendix C. Maps of the Clusters Using URF with HI and H2 for the Three Study Provinces
References
- Levin, S.A. Ecosystems and the Biosphere as Complex Adaptive Systems. Ecosystems 1998, 1, 431–436. [Google Scholar] [CrossRef]
- Levin, S.A. The Problem of Pattern and Scale in Ecology: The Robert H. MacArthur Award Lecture. Ecology 1992, 73, 1943–1967. [Google Scholar] [CrossRef]
- Emanuel, W.R.; Shugart, H.H.; Stevenson, M.P. Climatic Change and the Broad-Scale Distribution of Terrestrial Ecosystem Complexes. Clim. Chang. 1985, 7, 29–43. [Google Scholar] [CrossRef]
- Vetter, S. Rangelands at Equilibrium and Non-Equilibrium: Recent Developments in the Debate. J. Arid Environ. 2005, 62, 321–341. [Google Scholar] [CrossRef]
- Peng, J.; Liu, Z.; Liu, Y.; Wu, J.; Han, Y. Trend Analysis of Vegetation Dynamics in Qinghai–Tibet Plateau Using Hurst Exponent. Ecol. Indic. 2012, 14, 28–39. [Google Scholar] [CrossRef]
- Tong, S.; Zhang, J.; Bao, Y.; Lai, Q.; Lian, X.; Li, N.; Bao, Y. Analyzing Vegetation Dynamic Trend on the Mongolian Plateau Based on the Hurst Exponent and Influencing Factors from 1982–2013. J. Geogr. Sci. 2018, 28, 595–610. [Google Scholar] [CrossRef]
- Almeida-Ñauñay, A.F.; Benito, R.M.; Quemada, M.; Losada, J.C.; Tarquis, A.M. The Vegetation–Climate System Complexity through Recurrence Analysis. Entropy 2021, 23, 559. [Google Scholar] [CrossRef]
- Sanz, E.; Saa-Requejo, A.; Díaz-Ambrona, C.H.; Ruiz-Ramos, M.; Rodríguez, A.; Iglesias, E.; Esteve, P.; Soriano, B.; Tarquis, A.M. Generalized Structure Functions and Multifractal Detrended Fluctuation Analysis Applied to Vegetation Index Time Series: An Arid Rangeland Study. Entropy 2021, 23, 576. [Google Scholar] [CrossRef]
- Bruzzone, O.; Easdale, M.H. Archetypal Temporal Dynamics of Arid and Semi-Arid Rangelands. Remote Sens. Environ. 2021, 254, 112279. [Google Scholar] [CrossRef]
- Rivas-Tabares, D.; Saa-Requejo, A.; Martín-Sotoca, J.J.; Tarquis, A.M. Multiscaling NDVI Series Analysis of Rainfed Cereal in Central Spain. Remote Sens. 2021, 13, 568. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Zhu, X.; Pan, Y.; Liu, Y.; Zhang, D.; Lin, Z. Spatiotemporal Changes in Vegetation Coverage and Its driving Factors in the Three-River Headwaters Region during 2000–2011. J. Geogr. Sci. 2014, 24, 288–302. [Google Scholar] [CrossRef]
- Ndayisaba, F.; Guo, H.; Bao, A.; Guo, H.; Karamage, F.; Kayiranga, A. Understanding the Spatial-Temporal Vegetation Dynamics in Rwanda. Remote Sens. 2016, 8, 129. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. Review of Evidence on Drylands Pastoral Systems and Climate Change: Implications and Opportunities for Mitigation and Adaptation. In Land and Water Discussion Paper 8; Citeseer: Rome, Italy, 2009; p. 38. ISBN 978-92-5-106413-9. [Google Scholar]
- Reid, W.V.; Mooney, H.A.; Cropper, A.; Capistrano, D.; Carpenter, S.R.; Chopra, K.; Dasgupta, P.; Dietz, T.; Duraiappah, A.K.; Hassan, R. Millennium Ecosystem Assessment Synthesis Report; WRI (World Resources Institute): Washington, DC, USA, 2005. [Google Scholar]
- United Nations Conference on Environment and Development. Agenda 21; United Nations Conference on Environment and Development: New York, NY, USA, 1992. [Google Scholar]
- United Nations. Elaboration of an International Convention to Combat Desertification in Countries Experiencing Serious Droughts and/or Desertification Particularly in Africa; UNEP: Geneva, Switzerland, 1994. [Google Scholar]
- Kapalanga, T.S. A Review of Land Degradation Assessment Methods. In Land Restoration Training Programme; Citeseer: Rome, Italy, 2008; Volume 2011, p. 68. [Google Scholar]
- Aide, T.M.; Grau, H.R.; Graesser, J.; Andrade-Nuñez, M.J.; Aráoz, E.; Barros, A.P.; Campos-Cerqueira, M.; Chacon-Moreno, E.; Cuesta, F.; Espinoza, R. Woody Vegetation Dynamics in the Tropical and Subtropical Andes from 2001 to 2014: Satellite Image Interpretation and Expert Validation. Glob. Chang. Biol. 2019, 25, 2112–2126. [Google Scholar] [CrossRef]
- Woodward, F.I.; Lomas, M.R. Vegetation Dynamics–Simulating Responses to Climatic Change. Biol. Rev. 2004, 79, 643–670. [Google Scholar] [CrossRef] [PubMed]
- Warren, A. Land Degradation Is Contextual. Land Degrad. Dev. 2002, 13, 449–459. [Google Scholar] [CrossRef]
- Lambin, E.F.; Turner, B.L.; Geist, H.J.; Agbola, S.B.; Angelsen, A.; Bruce, J.W.; Coomes, O.T.; Dirzo, R.; Fischer, G.; Folke, C.; et al. The Causes of Land-Use and Land-Cover Change: Moving beyond the Myths. Glob. Environ. Chang. 2001, 11, 261–269. [Google Scholar] [CrossRef]
- Gartzia, M.; Fillat, F.; Pérez-Cabello, F.; Alados, C.L. Influence of Agropastoral System Components on Mountain Grassland Vulnerability Estimated by Connectivity Loss. PLoS ONE 2016, 11, e0155193. [Google Scholar] [CrossRef]
- Herrera, P.M.; Davies, J. Governance of the Rangelands in a Changing World. In The Governance of Rangelands; Routledge: Abingdon-on-Thames, UK, 2014; pp. 54–66. ISBN 1315768011. [Google Scholar]
- Robinson, N.P.; Allred, B.W.; Naugle, D.E.; Jones, M.O. Patterns of Rangeland Productivity and Land Ownership: Implications for Conservation and Management. Ecol. Appl. 2019, 29, e01862. [Google Scholar] [CrossRef]
- Perrings, C.; Walker, B. Conservation in the Optimal Use of Rangelands. Ecol. Econ. 2004, 49, 119–128. [Google Scholar] [CrossRef]
- Bird, S.B.; Herrick, J.E.; Wander, M.M.; Wright, S.F. Spatial Heterogeneity of Aggregate Sility and Soil Carbon in Semi-Arid Rangeland. Environ. Pollut. 2002, 116, 445–455. [Google Scholar] [CrossRef]
- Evans, J.P.; Geerken, R. Classifying Rangeland Vegetation Type and Coverage Usitabng a Fourier Component Based Similarity Measure. Remote Sens. Environ. 2006, 105, 1–8. [Google Scholar] [CrossRef]
- Ünal, E.; Mermer, A.; Yildiz, H. Assessment of Rangeland Vegetation Condition from Time Series NDVI Data. J. Field Crops Cent. Res. Inst. 2014, 23, 14–21. [Google Scholar] [CrossRef][Green Version]
- Huang, F.; Wang, P. Vegetation Change of Ecotone in West of Northeast China Plain Using Time-Series Remote Sensing Data. Chin. Geogr. Sci. 2010, 20, 167–175. [Google Scholar] [CrossRef]
- Wang, Y.; Zang, S.; Tian, Y. Mapping Paddy Rice with the Random Forest Algorithm Using MODIS and SMAP Time Series. Chaos Solitons Fractals 2020, 140, 110116. [Google Scholar] [CrossRef]
- Mangiarotti, S.; Sharma, A.K.; Corgne, S.; Hubert-Moy, L.; Ruiz, L.; Sekhar, M.; Kerr, Y. Can the Global Modeling Technique Be Used for Crop Classification? Chaos Solitons Fractals 2018, 106, 363–378. [Google Scholar] [CrossRef]
- Fathizad, H.; Tazeh, M.; Kalantari, S.; Shojaei, S. The Investigation of Spatiotemporal Variations of Land Surface Temperature Based on Land Use Changes Using NDVI in Southwest of Iran. J. Afr. Earth Sci. 2017, 134, 249–256. [Google Scholar] [CrossRef]
- Ahmed, K.R.; Akter, S.; Marandi, A.; Schüth, C. A Simple and Robust Wetland Classification Approach by Using Optical Indices, Unsupervised and Supervised Machine Learning Algorithms. Remote Sens. Appl. Soc. Environ. 2021, 23, 100569. [Google Scholar] [CrossRef]
- Triscowati, D.W.; Sartono, B.; Kurnia, A.; Domiri, D.D.; Wijayanto, A.W. Multitemporal Remote Sensing Data for Classification of Food Crops Plant Phase Using Supervised Random Forest. In Proceedings of the Sixth Geoinformation Science Symposium; International Society for Optics and Photonics, Yogyakarta, Indonesia, 26–27 August 2019; Volume 11311, p. 1131102. [Google Scholar]
- Uehara, T.D.T.; Soares, A.R.; Quevedo, R.P.; Körting, T.S.; Fonseca, L.M.G.; Adami, M. Land Cover Classification of an Area Susceptible to Landslides Using Random Forest and NDVI Time Series Data. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 1345–1348. [Google Scholar]
- Breiman, L.; Cutler, A. Random Forests Manual V4; Technical Report; UC Berkeley: Berkeley, NJ, USA, 2003. [Google Scholar]
- Peerbhay, K.; Mutanga, O.; Lottering, R.; Ismail, R. Mapping Solanum Mauritianum Plant Invasions Using WorldView-2 Imagery and Unsupervised Random Forests. Remote Sens. Environ. 2016, 182, 39–48. [Google Scholar] [CrossRef]
- Lopez, C.; Tucker, S.; Salameh, T.; Tucker, C. An Unsupervised Machine Learning Method for Discovering Patient Clusters Based on Genetic Signatures. J. Biomed. Inform. 2018, 85, 30–39. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Bezdek, J.C.; Pal, N.R. Some New Indexes of Cluster Validity. IEEE Trans. Syst. Man Cybern. Part B 1998, 28, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Frey, B.J.; Dueck, D. Clustering by Passing Messages between Data Points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.E. Long-Term Storage Capacity of Reservoirs. Trans. Am. Soc. Civ. Eng. 1951, 116, 770–799. [Google Scholar] [CrossRef]
- Kantelhardt, J.W.; Zschiegner, S.A.; Koscielny-Bunde, E.; Havlin, S.; Bunde, A.; Stanley, H.E. Multifractal Detrended Fluctuation Analysis of Nonstationary Time Series. Phys. A Stat. Mech. Its Appl. 2002, 316, 87–114. [Google Scholar] [CrossRef]
- Igbawua, T.; Zhang, J.; Yao, F.; Ali, S. Long Range Correlation in Vegetation Over West Africa From 1982 to 2011. IEEE Access 2019, 7, 119151–119165. [Google Scholar] [CrossRef]
- Kalisa, W.; Igbawua, T.; Henchiri, M.; Ali, S.; Zhang, S.; Bai, Y.; Zhang, J. Assessment of Climate Impact on Vegetation Dynamics over East Africa from 1982 to 2015. Sci. Rep. 2019, 9, 16865. [Google Scholar] [CrossRef] [PubMed]
- Ba, R.; Song, W.; Lovallo, M.; Lo, S.; Telesca, L. Analysis of Multifractal and Organization/Order Structure in Suomi-NPP VIIRS Normalized Difference Vegetation Index Series of Wildfire Affected and Unaffected Sites by Using the Multifractal Detrended Fluctuation Analysis and the Fisher-Shannon Analysis. Entropy 2020, 22, 415. [Google Scholar] [CrossRef]
- Emamian, A.; Rashki, A.; Kaskaoutis, D.G.; Gholami, A.; Opp, C.; Middleton, N. Assessing Vegetation Restoration Potential under Different Land Uses and Climatic Classes in Northeast Iran. Ecol. Indic. 2021, 122, 107325. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, E.; Yue, H. Dynamic Monitoring and Trend Analysis of Vegetation Change in Shendong Mining Area Based on MODIS. Remote Sens. Land Resour. 2017, 132–137. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, R.; Gan, F.; Wang, W.; Ding, L.; Yan, B. Evaluation of Spatial-Temporal Variation of Vegetation Restoration in Dexing Copper Mine Area Using Remote Sensing Data. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 2013–2016. [Google Scholar]
- Guo, X.; Zhang, H.; Yuan, T.; Zhao, J.; Xue, Z. Detecting the Temporal Scaling Behavior of the Normalized Difference Vegetation Index Time Series in China Using a Detrended Fluctuation Analysis. Remote Sens. 2015, 7, 12942–12960. [Google Scholar] [CrossRef]
- Martinez-Mena, M.; Castillo, V.; Albaladejo, J. Hydrological and Erosional Response to Natural Rainfall in a Semi-arid Area of South-east Spain. Hydrol. Process. 2001, 15, 557–571. [Google Scholar] [CrossRef]
- Dargie, T.C.D. On the Integrated Interpretation of Indirect Site Ordinations: A Case Study Using Semi-Arid Vegetation in Southeastern Spain. Vegetatio 1984, 55, 37–55. [Google Scholar] [CrossRef]
- Barceló, A.M.; Nunes, L.F. Atlas Climático Ibérico—Iberian Climate Atlas 1971–2000. AEMET: Madrid, Spain, 2009; ISBN 9788478370795. [Google Scholar]
- Fondo Español de Garantía Agraria Visor SigPac (FEGA). Madrid. Spain. Available online: http://www.fega (accessed on 20 January 2022).
- Team, A. Application for Extracting and Exploring Analysis Ready Samples (AppEEARS). NASA EOSDIS Land Processes Distributed Active Archive Center (LP DAAC), USGS/Earth Resources Observation and Science (EROS) Center: Sioux Falls, SD, USA. 2020. Available online: https://lpdaacsvc.cr.usgs.gov/appeears/ (accessed on 2 June 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- EU-DEM v1.1. Available online: https://land.copernicus.eu/imagery-in-situ/eu-dem/eu-dem-v1.1?tab=download (accessed on 4 March 2022).
- ESRI ArcGIS Desktop. Release 10.8.1; ESRI: Redlands, CA, USA, 2020. [Google Scholar]
- Borchers, H.W.; Borchers, M.H.W. Package ‘Pracma’. 2019. Available online: https://CRAN.R-project.org/package=pracma (accessed on 4 March 2022).
- Zhou, Z.; Ding, Y.; Shi, H.; Cai, H.; Fu, Q.; Liu, S.; Li, T. Analysis and Prediction of Vegetation Dynamic Changes in China: Past, Present and Future. Ecol. Indic. 2020, 117, 106642. [Google Scholar] [CrossRef]
- Kendall, M.G. Rank Correlation Methods; Charles Griffin & Co.: London, UK, 1975. [Google Scholar]
- Li, X.; Lanorte, A.; Lasaponara, R.; Lovallo, M.; Song, W.; Telesca, L. Fisher–Shannon and Detrended Fluctuation Analysis of MODIS Normalized Difference Vegetation Index (NDVI) Time Series of Fire-Affected and Fire-Unaffected Pixels. Geomat. Nat. Hazards Risk 2017, 8, 1342–1357. [Google Scholar] [CrossRef]
- Sanz, E.; Saa-Requejo, A.; Díaz-Ambrona, C.H.; Ruiz-Ramos, M.; Rodríguez, A.; Iglesias, E.; Esteve, P.; Soriano, B.; Tarquis, A.M. Normalized Difference Vegetation Index Temporal Responses to Temperature and Precipitation in Arid Rangelands. Remote Sens. 2021, 13, 840. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing Partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Vavrek, M.J. Fossil: Palaeoecological and Palaeogeographical Analysis Tools. Palaeontol. Electron. 2011, 14, 16. [Google Scholar]
- Lloyd, S. Least Squares Quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Oakland, CA, USA, 1 January 1967; Volume 1, pp. 281–297. [Google Scholar]
- MacKay, D. An Example Inference Task: Clustering. In Information Theory, Inference and Learning Algorithms; Cambridge University Press: Cambridge, UK, 2003; Volume 20, pp. 284–292. [Google Scholar]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-Means Clustering Algorithm. J. R. Stat. Soc. Ser. Appl. Stat. 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Breiman, L. Bagging Predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Breiman, L.; Cutler, A. Random Forests-Classification Description. Available online: http://stat-www.berkeley.edu/users/breiman/RandomForests/cc_home.htm (accessed on 21 January 2022).
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Ng, A. Clustering with the K-Means Algorithm. Mach. Learn. 2012.
- Ludwig, J.A.; Tongway, D.J. Viewing Rangelands as Landscape Systems. In Rangeland Desertification; Springer: Amsterdam, The Netherlands, 2000; pp. 39–52. [Google Scholar]
- Tongway, D.J.; Ludwig, J.A. The Nature of Landscape Dysfunction in Rangelands. In Landscape Ecology: Function and Management: Principles from Australia’s Rangelands; CSIRO Publishing: Melbourne, Australia, 1997. [Google Scholar]
- Coronado, A.V.; Carpena, P. Size Effects on Correlation Measures. J. Biol. Phys. 2005, 31, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Schwinning, S.; Parsons, A.J. The Stability of Grazing Systems Revisited: Spatial Models and the Role of Heterogeneity. Funct. Ecol. 1999, 13, 737–747. [Google Scholar] [CrossRef]
- Martens, S.N.; Breshears, D.D.; Meyer, C.W. Spatial Distributions of Understory Light along the Grassland/Forest Continuum: Effects of Cover, Height, and Spatial Pattern of Tree Canopies. Ecol. Model. 2000, 126, 79–93. [Google Scholar] [CrossRef]
- Stavi, I.; Ungar, E.D.; Lavee, H.; Sarah, P. Grazing-Induced Spatial Variability of Soil Bulk Density and Content of Moisture, Organic Carbon and Calcium Carbonate in a Semi-Arid Rangeland. Catena 2008, 75, 288–296. [Google Scholar] [CrossRef]
- Le Houérou, H.N.; Bingham, R.L.; Skerbek, W. Relationship between the Variability of Primary Production and the Variability of Annual Precipitation in World Arid Lands. J. Arid Environ. 1988, 15, 1–18. [Google Scholar] [CrossRef]
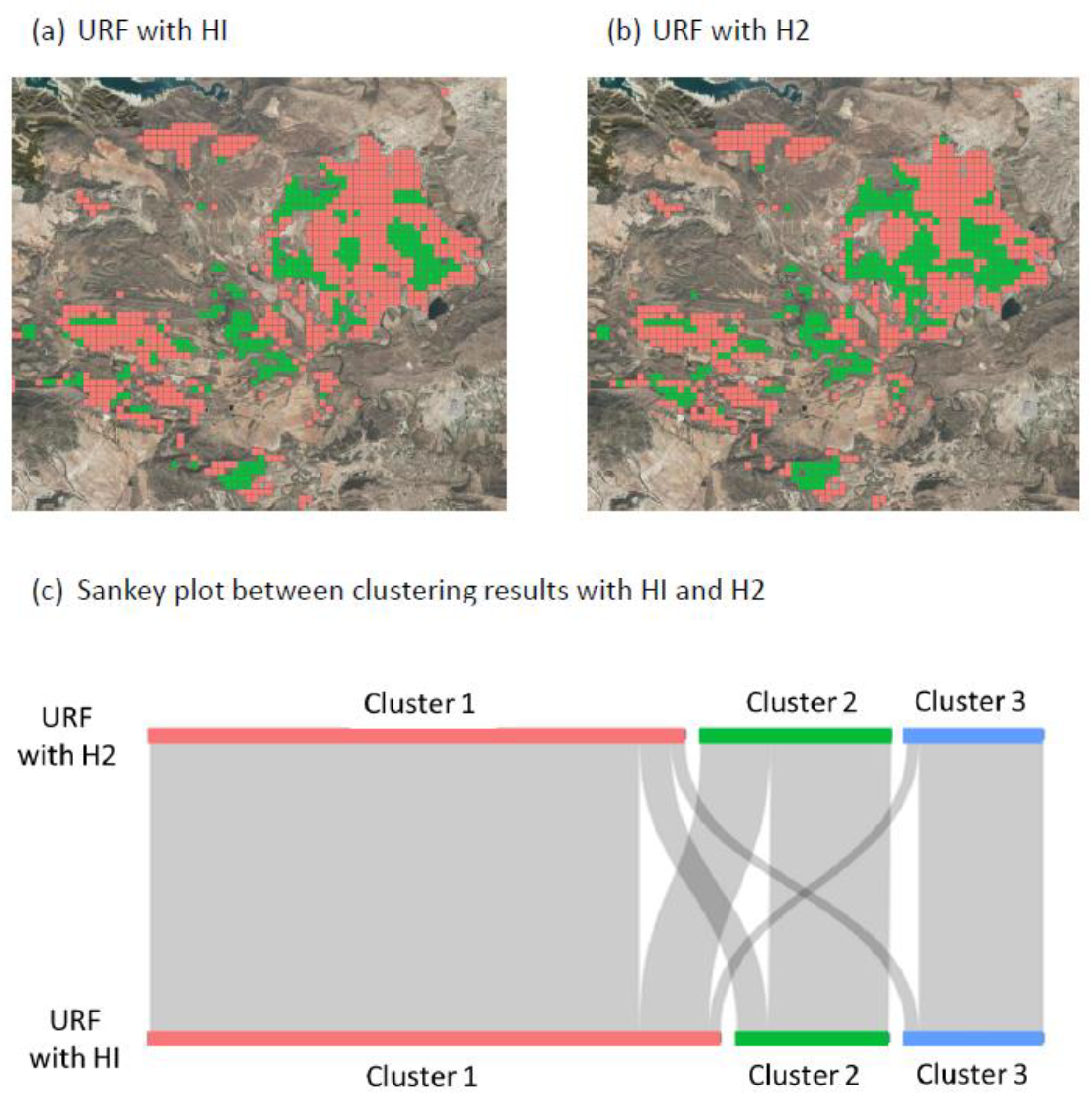
| Analysis | K-Means | Unsupervised Random Forest | ||
|---|---|---|---|---|
| 3 Clusters | 4 Clusters | 3 Clusters | 4 Clusters | |
| Without H2/HI | 0.33 | 0.34 | 0.51 | 0.49 |
| With H2 | 0.33 | 0.34 | 0.62 | 0.47 |
| With HI | 0.33 | 0.34 | 0.50 | 0.45 |
| Hurst Exponent | Elevation | Slope | Var_Ph5 | Var_Ph2 |
|---|---|---|---|---|
| H2 | −0.81 | −0.53 | 0.54 | 0.29 |
| HI | −0.25 | −0.07 | 0.05 | 0.21 |
| Cluster | Woodland | Shrubland | Grassland |
|---|---|---|---|
| 1 | 48.0 | 7.7 | 44.3 |
| 2 | 99.7 | 0.3 | 0.0 |
| 3 | 15.5 | 4.4 | 80.1 |
| Significance | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|
| Significant decrease | 6.2% | 0 % | 2.2% |
| Not significant | 26.5% | 10% | 34.3% |
| Significant increase | 67.3% | 90% | 63.5% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanz, E.; Sotoca, J.J.M.; Saa-Requejo, A.; Díaz-Ambrona, C.H.; Ruiz-Ramos, M.; Rodríguez, A.; Tarquis, A.M. Clustering Arid Rangelands Based on NDVI Annual Patterns and Their Persistence. Remote Sens. 2022, 14, 4949. https://doi.org/10.3390/rs14194949
Sanz E, Sotoca JJM, Saa-Requejo A, Díaz-Ambrona CH, Ruiz-Ramos M, Rodríguez A, Tarquis AM. Clustering Arid Rangelands Based on NDVI Annual Patterns and Their Persistence. Remote Sensing. 2022; 14(19):4949. https://doi.org/10.3390/rs14194949
Chicago/Turabian StyleSanz, Ernesto, Juan José Martín Sotoca, Antonio Saa-Requejo, Carlos H. Díaz-Ambrona, Margarita Ruiz-Ramos, Alfredo Rodríguez, and Ana M. Tarquis. 2022. "Clustering Arid Rangelands Based on NDVI Annual Patterns and Their Persistence" Remote Sensing 14, no. 19: 4949. https://doi.org/10.3390/rs14194949
APA StyleSanz, E., Sotoca, J. J. M., Saa-Requejo, A., Díaz-Ambrona, C. H., Ruiz-Ramos, M., Rodríguez, A., & Tarquis, A. M. (2022). Clustering Arid Rangelands Based on NDVI Annual Patterns and Their Persistence. Remote Sensing, 14(19), 4949. https://doi.org/10.3390/rs14194949










