1. Introduction
Canola (
Brassica napus L.) is an oilseed crop produced for its edible oil, high protein meal, and biofuel [
1]. Canada is the leading producer of canola with 18.7 million tons produced in 2020 [
2]. The crop is primarily produced in western Canada, with Saskatchewan accounting for 55 percent of total production. Increased consumer demand necessitates increased canola production and enhanced genetic and agronomic approaches are indispensable for achieving high crop yields.
Crop yield forecasts assist farmers and stakeholders in making timely decisions concerning imports and exports, management decisions, and financial aspects [
3]. Genotype and environmental factors and their interactions make yield prediction extremely difficult due to the intricacy of the yield components. Traditionally, the experts forecasted yield via within-season crop measurements and using statistical approaches such as regression [
4]. With advancements in technology, the acquisition of non-destructive, accurate phenotypic data through remote sensing approaches substantially improved the data quality necessary for reliable yield estimations.
Canola has an indeterminate growth habit with overlapping growth stages. However, each stage can be easily distinguished due to prominent phenological changes. Canola flowers are bright yellow, and hence the reproductive stage can be easily distinguished from the vegetative stage. Flowering intensity is considered a direct contributor to seed yield, as each flower determines the potential number of pods [
5]. Therefore, quantifying flowers through high-throughput methods as an indirect approach to determine the yield is pivotal for plant breeding and crop production [
6]. Furthermore, quantifying the conspicuous canola flower number using unpiloted aerial vehicle (UAV)-based high-resolution RGB imagery is less labor intensive and time consuming than manually counting flowers, especially in breeding trials with hundreds of plots.
Remote sensing techniques extract canopy spectral information, which can study biophysical parameters such as biomass, chlorophyll content, leaf area, and flowering intensity [
7]. Building empirical relationships between the remotely sensed spectral information and the biophysical variables via vegetation indices is quick and straightforward [
8]. Despite the inability of the relationship to be applied in an environment outside the representativeness of the calibration dataset, the use of vegetation indices for remote sensing studies keeps increasing.
Canola flowers reflect more green and red light and absorb more blue light when compared to green vegetation [
9]. These prominent spectral characteristics have led to the development of new vegetation indices [
10,
11,
12,
13,
14]. To our knowledge, except for the Normalized Difference Yellowness Index (NDYI) developed by Long and Sulik [
12], other yellowness indices use spectral information from bands outside the visible region of the electromagnetic spectrum. The contrast between the reduced blue reflectance and increased green and red reflectance [
9] in canola flowers was enhanced through multiplying the band differences to develop the HrFI (
Table 1).
Within-canopy shadow pixels where the canopy objects (leaves and flowers) completely block direct light are problematic, especially when quantifying canola flower pixels using high-resolution imagery (<1 cm), making it necessary to address these challenges [
15]. Therefore, it is imperative to develop new vegetation indices that can filter the pixels disturbed by the effects of shadows in RGB imagery. Furthermore, use of visible bands for these vegetation indices will enhance their applicability where hyperspectral or multispectral sensors are scarce and expensive. Moreover, RGB sensors usually have ultra-high spatial resolution compared to hyper- and multi-spectral imagery, allowing users to gather spatially explicit data. Accurate flower pixel segmentation is critical for image-based assessments with high spatial resolution imagery [
16]. However, intense natural illumination and the influence of shadows from the crop canopy make canopy segmentation from flowers challenging because of the high contrast ratio. The segmentation process often confounds background noise with significant spectral information of flowers [
17] and significantly influences its estimates.
The accuracy of image processing steps such as thresholding and segmentation significantly influences its estimates. The confusion matrix, which is at the core of remote sensing accuracy assessment methods, is used to refine the threshold/classification to improve the accuracy of the process [
18].
We attempt to propose a straightforward method using standard index-based thresholding to detect and extract flower area for yield estimation. Furthermore, the exclusion of the noise from the target is extensively evaluated to improve the classification accuracy of canola flowers.
The study hypothesized that digitized flower information with minimum noise from high-resolution RGB imagery will be a strong indicator of canola reproductive potential and could be used to estimate the canola seed yield. The study aimed to (1) compare vegetation indices to quantify canola flowers using very-high-resolution RGB imagery, and (2) develop canola seed yield prediction models using digitized flower area.
4. Discussion
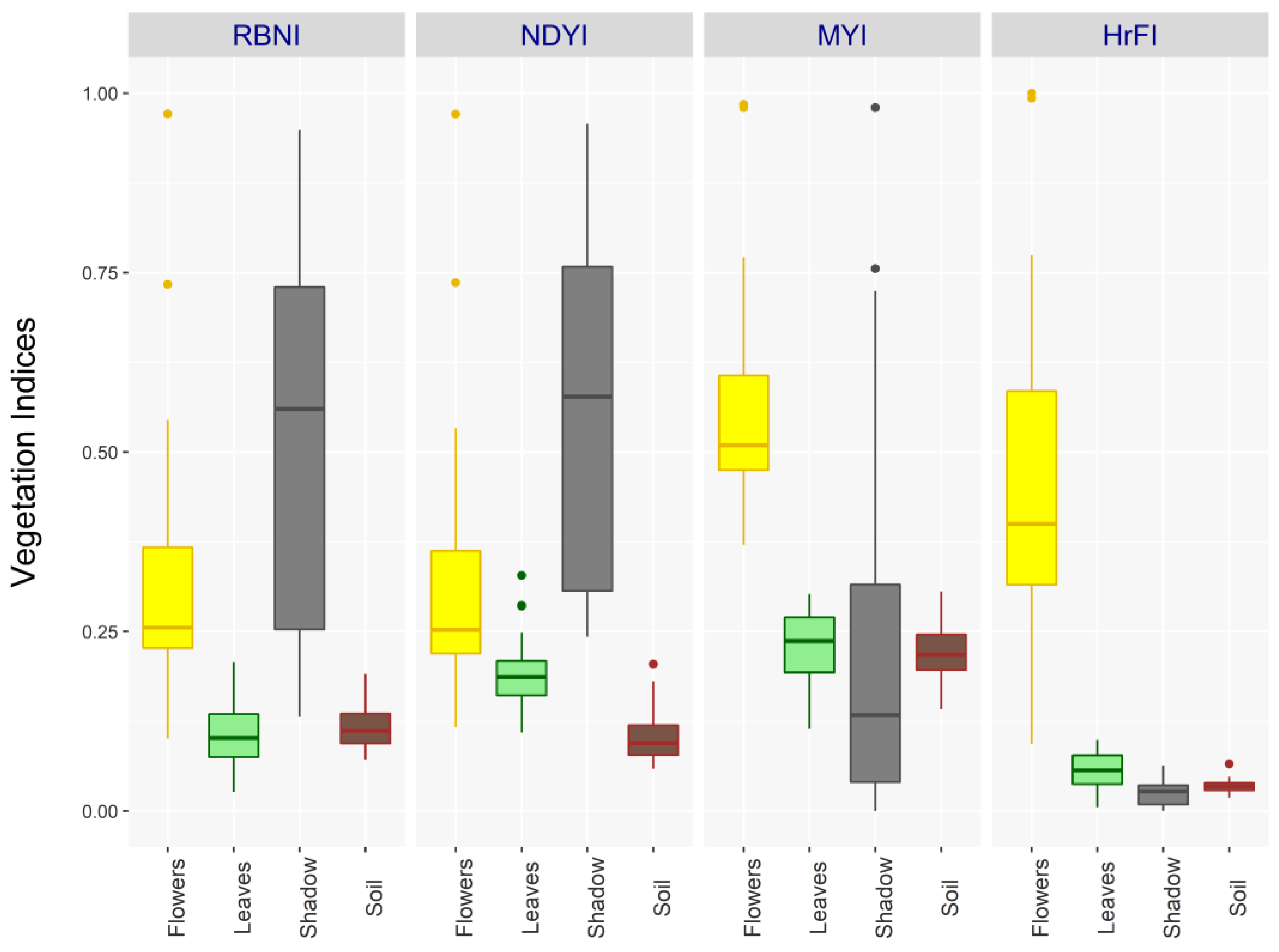
This study aimed to develop a canola seed yield prediction model using high-resolution RGB imagery. The use of vegetation indices to study phenology changes is mathematically less complex, computationally efficient, and cost-effective. The biggest issue with high-resolution images is within-canopy pure shadow pixels that often blend with flower pixels. Canola flowering canopies have different spectral reflectance characteristics than their vegetative canopy, in which flowers reflect more radiation between 500 and 700 nm and absorb slightly more between 400 and 500 nm [
29]. Therefore, the potential of using floral canopy signature for predicting canola seed yield using UAV-based high-resolution RGB imagery was evaluated with a robust dataset from the canola field trial [
13].
In this study, brightness values (DN) were used for processing the imagery. The experimental trial was conducted on fairly uniform land with vegetation ground cover as the material of interest. Furthermore, the three images were acquired under similar environmental conditions at the same time of day. Hence, the effect of atmospheric and illumination effects on uniform land would be less compared to undulating land. However, when mapping a larger landscape with significant changes in topography, use of surface reflectance would be advised, especially when multi-temporal data are used. Property-based methods in which the shadows are identified using the brightness of the shadows are commonly used in the literature [
30]. Vegetative indices are impacted by shadow pixels, and studies have shown that statistical differences exist between shadowed images and sunlit images in soil and vegetation indices [
31]. The pixels completely blocked by any illumination have low reflectance in the red, green, and blue bands. Therefore, due to the inherent nature of their reflectance, when normalized vegetative indices are developed for such pixels, they tend to have higher values (NDYI, RBNI). These pixels have even greater values than flower pixels (
Figure 5), which significantly reduces the accuracy of the extracted features. We also discovered that cast shadow pixels partially occluded by direct light have lower index values than flower pixels and can be automatically masked by a simple background mask. Additionally, a simple multiplication takes advantage of the low reflectance of dark shadow pixels to mask them out. Therefore, the HrFI and MYI introduced in these studies have low values for within-canopy shadow pixels. In most remote sensing studies, shadow pixel restoration is essential [
32]. However, in canola, as the flower canopy is the topmost layer, restored shadow pixels may not significantly contribute to the inferences. The within-canopy shadow pixels may correspond with middle or lower layers of the canola canopy. Hence, the removal of shadow pixels may not have a significant effect on the analysis of the study.
In this study, the within-canopy shadow pixels were considered as image noise. The proposed indices (HrFI) aim to enhance the contrast between area of interest (flower pixels) and within-canopy shadow (noise) (
Figure 5). Spectral ratio transformations enhance noise, especially in pixels with low reflectance (within-canopy shadow pixels in this study). Since the NDYI and RBNI are band ratios, the noise in the images is enhanced [
33]. Therefore, if these ratio indices are to be applied to high-resolution RGB imagery with within-canopy shadows, a bias correction must be undertaken first. The HrFI and MYI overcome the issue of generic enhancement of noise pixels, providing a straightforward method in identifying canola flower pixels.
Plots with high-density planting had a significant amount of pure within-canopy shadows (pure shadow pixels) due to the narrower row spacing than plots with wider row spacing. On the other hand, wider row plots had a higher number of mixed shadow pixels that were partially blocked by direct sunlight, and hence had a higher reflectance than pure shadow pixels. Therefore, removing the effect of within-canopy shadows in variable planting trials is essential. Our analysis of the vegetation indices demonstrated that the HrFI is efficient in capturing the flower canopy reflectance and could distinguish flowers from the soil, shadow, and leaf pixels (
Figure 4,
Figure 5 and
Figure 6). In contrast, other VIs confounded spectral information of floral features, especially when within-canopy shadow pixels are dominant in the imagery. Moreover, we noted that the performance of the MYI closely follows that of the HrFI in thresholding flowering pixels in high-resolution RGB imagery. This could be interpreted as those vegetation indices that enhance the contrast between reduced blue band reflectance, and enhanced red and green band reflectance can efficiently capture the flowering canopy signals of canola.
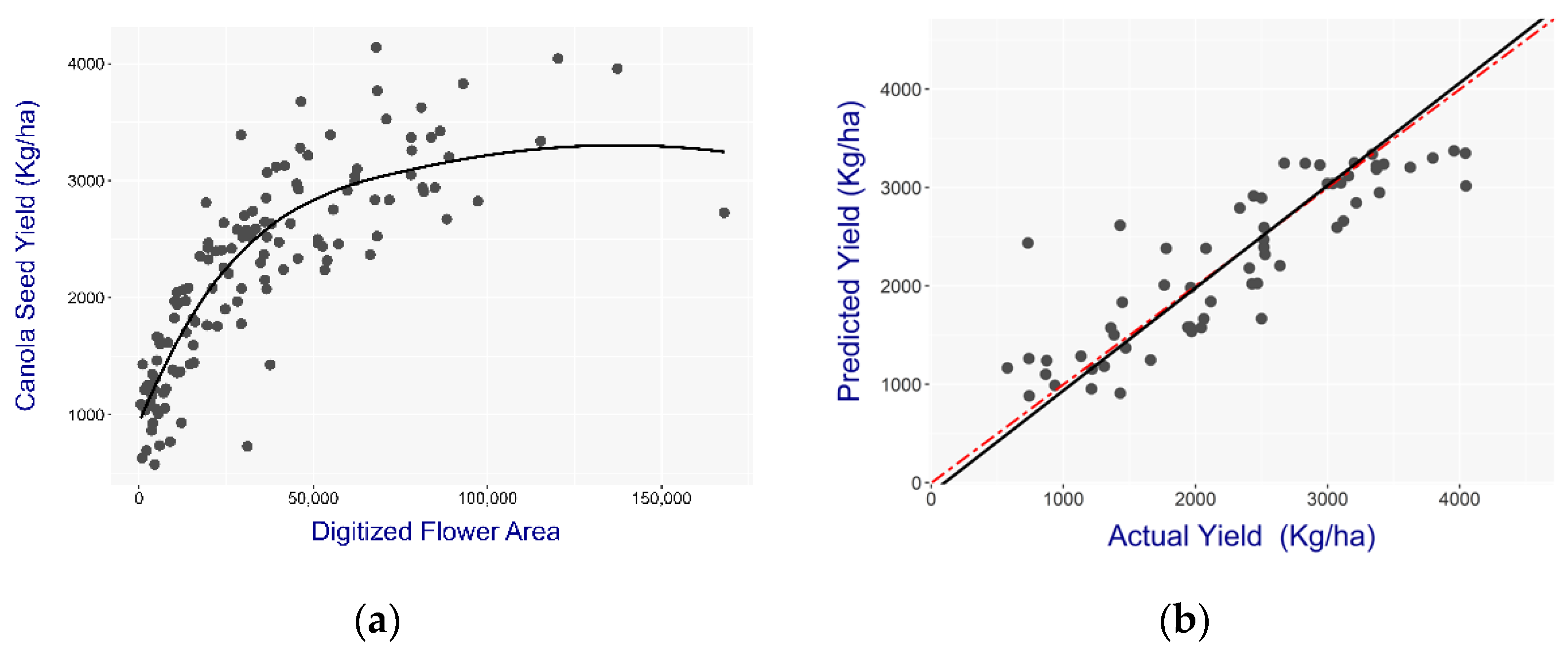
Remote sensing techniques are increasingly used to investigate the relationship between yield and plant growth characteristics. This study found that 82% of the yield variability can be explained using a digitized flower area using a single image date acquired during the peak flowering period. Peak flowering indicates the maximum reproductive potential of the plant and is conducive to estimate the seed yield. Despite canola producing more floral primordials than its photosynthetic capacity, peak flowering has been found to have a strong relationship with the yield [
12]. The early flowering stage can also be used to predict the canola seed yield as the flower area is correlated with the final seed yield at 0.85. The authors of [
5] found that 75% of the pods that are retained until maturity are made from flowers that opened within 11 days from flowering. This could explain the higher predictive power of the early flowering model.
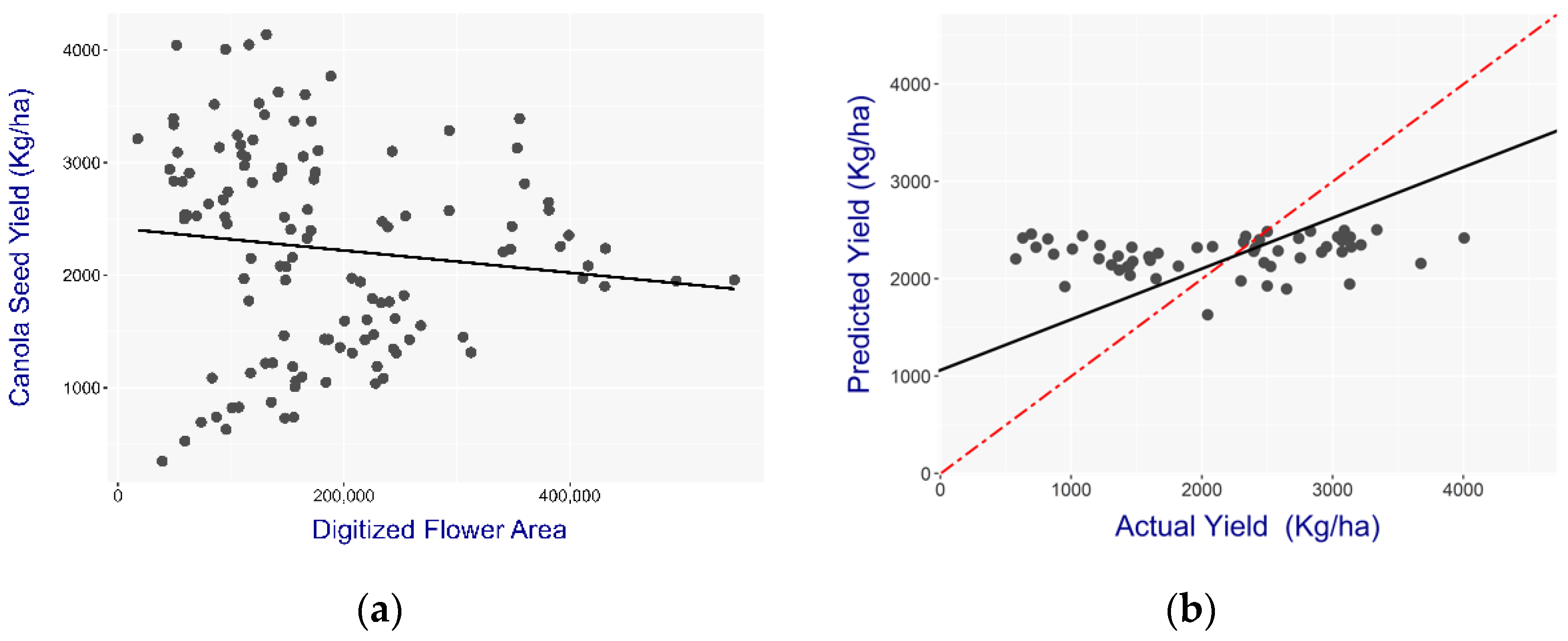
The cumulative flower area explained 75% of the yield potential as a predictor. In contrast, the late flowering model had poor model performance that can be attributed to the late flowering in treatments with low planting density. Plots with high planting density flowered earlier compared to plots with low seeding density [
34]. This effect can be seen in
Figure 10a, where the training model indicates data points arranged in two opposite directions. The negative correlation between the yield and the flower area can be explained by the majority of data points belonging to the high crop density plots, where at that time point (July 23) most flowers have become pods. The minority of data points that indicate a positive trend within the graph flowered later due to the low planting density.
Using a single image date for yield prediction can be less expensive and straightforward than using cumulative flower area. However, the applicability of the single image date data could be sensitive to environmental conditions such as drought and heat stress. Furthermore, it is challenging to identify the exact peak flowering time. High temperatures during the reproductive stage negatively affect seed yield [
35]. Especially when canola is exposed to high heat stress during the late reproductive stage, recovery is low [
36]. Therefore, the flowering information extracted during the peak flowering period before exposure to high environmental stress would not represent the accurate yield potential of the plant. Alternatively, using a time-series change of flower area can capture these environmental effects and could better represent the final seed yield. The authors of [
25] suggested using integrated flowering intensity as a strong indicator of yield potential over a single-date regression approach for medium-resolution UAV-multispectral data. The differences in our study and findings from Zhang et al. (2021) could be attributed to the higher genetic diversity among plots and lower resolution of images in the latter study. The trade-off between the accuracy of the yield prediction and the time expense in using single-date vs. time-series data is to be decided by the user.
This study only used three time points during the reproductive stage. A higher temporal resolution might provide detailed phenology changes to identify the ideal time point necessary to develop an accurate yield model. A major drawback of processing ultra-high-resolution imagery is that it requires high computational power. Furthermore, high-resolution imagery requires a lower flying altitude, necessitating longer flight time. Furthermore, RGB sensors with ultra-high spatial resolution can be expensive. Despite high spatial information, the spectral information of RGB imagery is limited, making it inadequate for certain studies.
The results of this study agree with the hypothesis made in the study. The HrFI developed in this study could successfully segment flower pixels from high-resolution RGB imagery to quantify the flower pixels. The digitized flower area was proven to be a strong predictor of seed yield, both when a single image date and cumulative images over the reproductive period are used. Furthermore, the results of this study accentuate the importance of using high-resolution images in yield prediction.