SC-CAN: Spectral Convolution and Channel Attention Network for Wheat Stress Classification
Abstract
:1. Introduction
2. Related Works
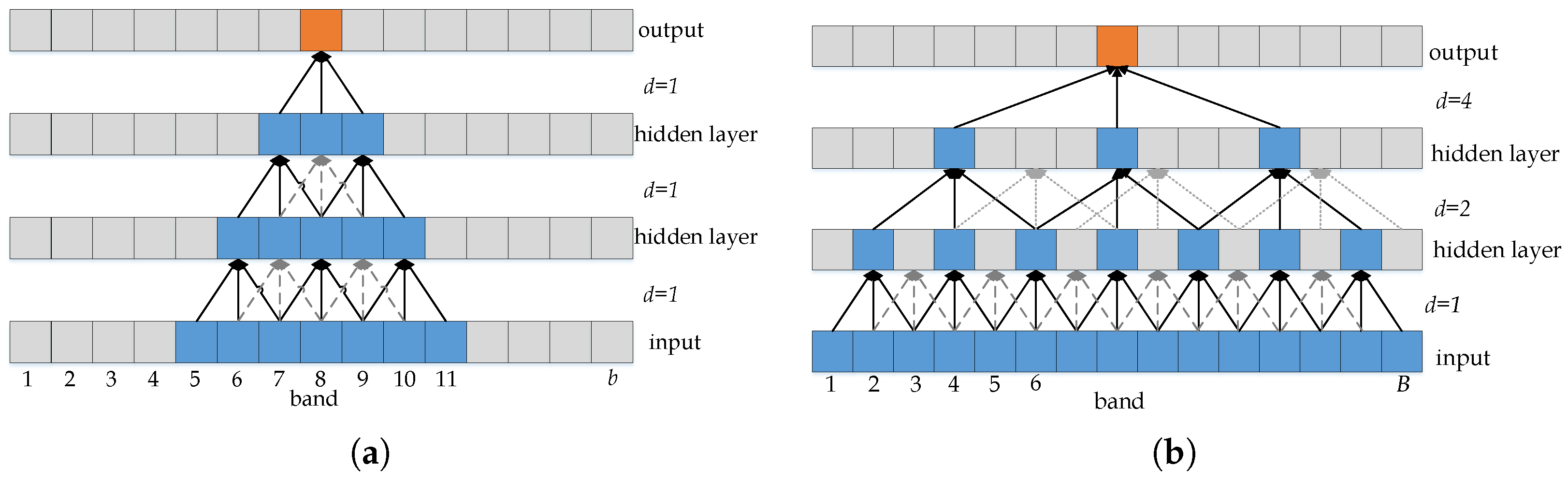
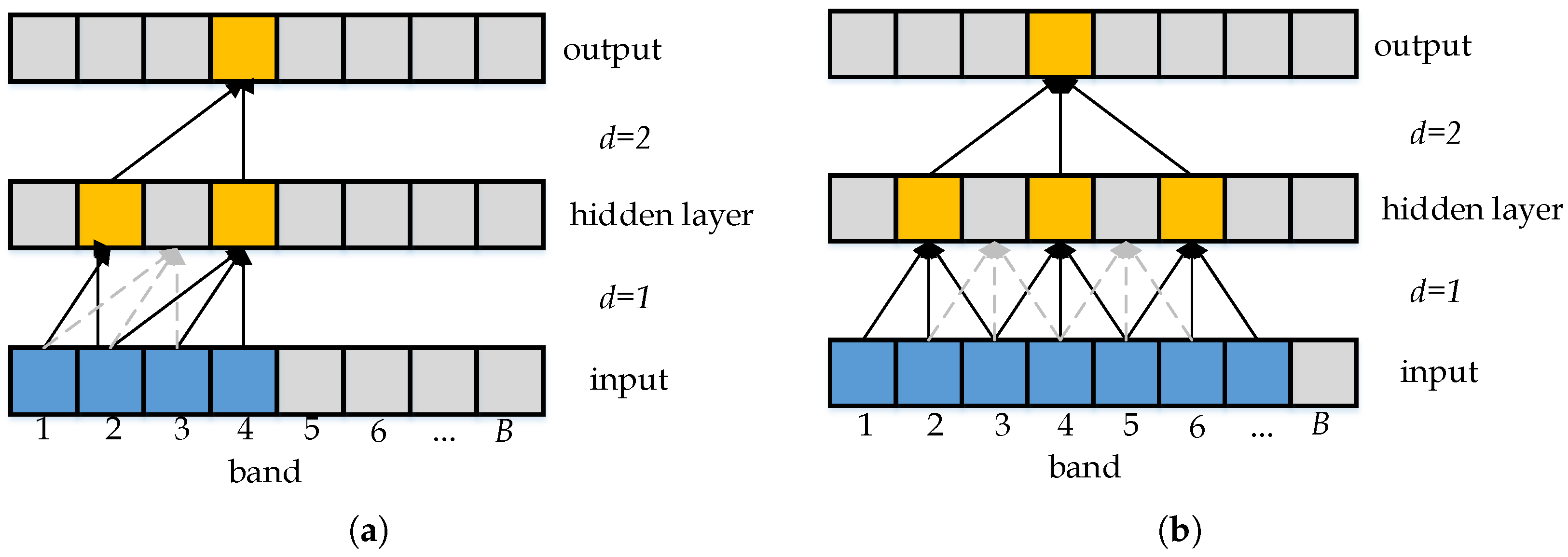
2.1. Dilated Convolution
2.2. Attention Module
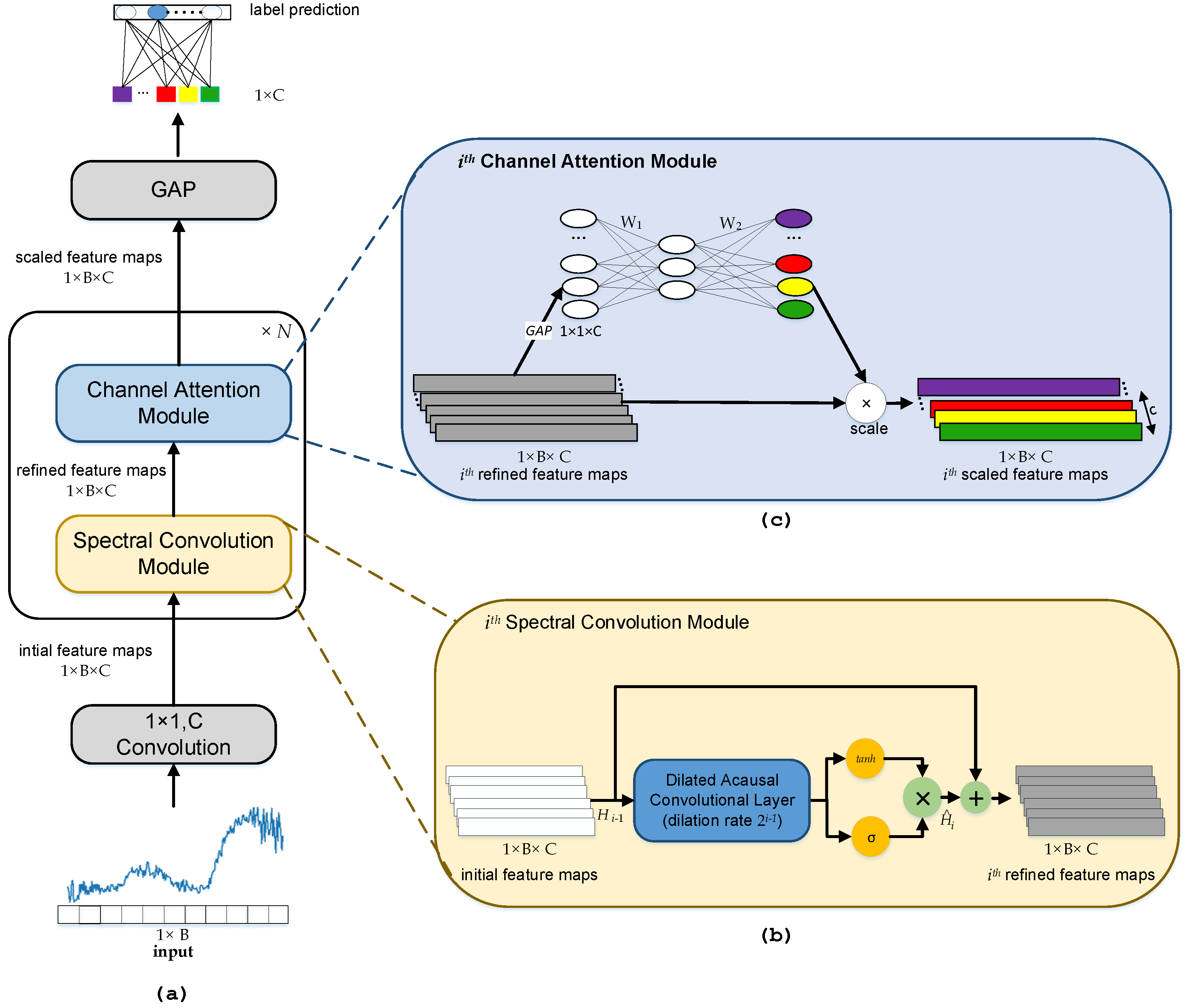
3. Proposed Methodology
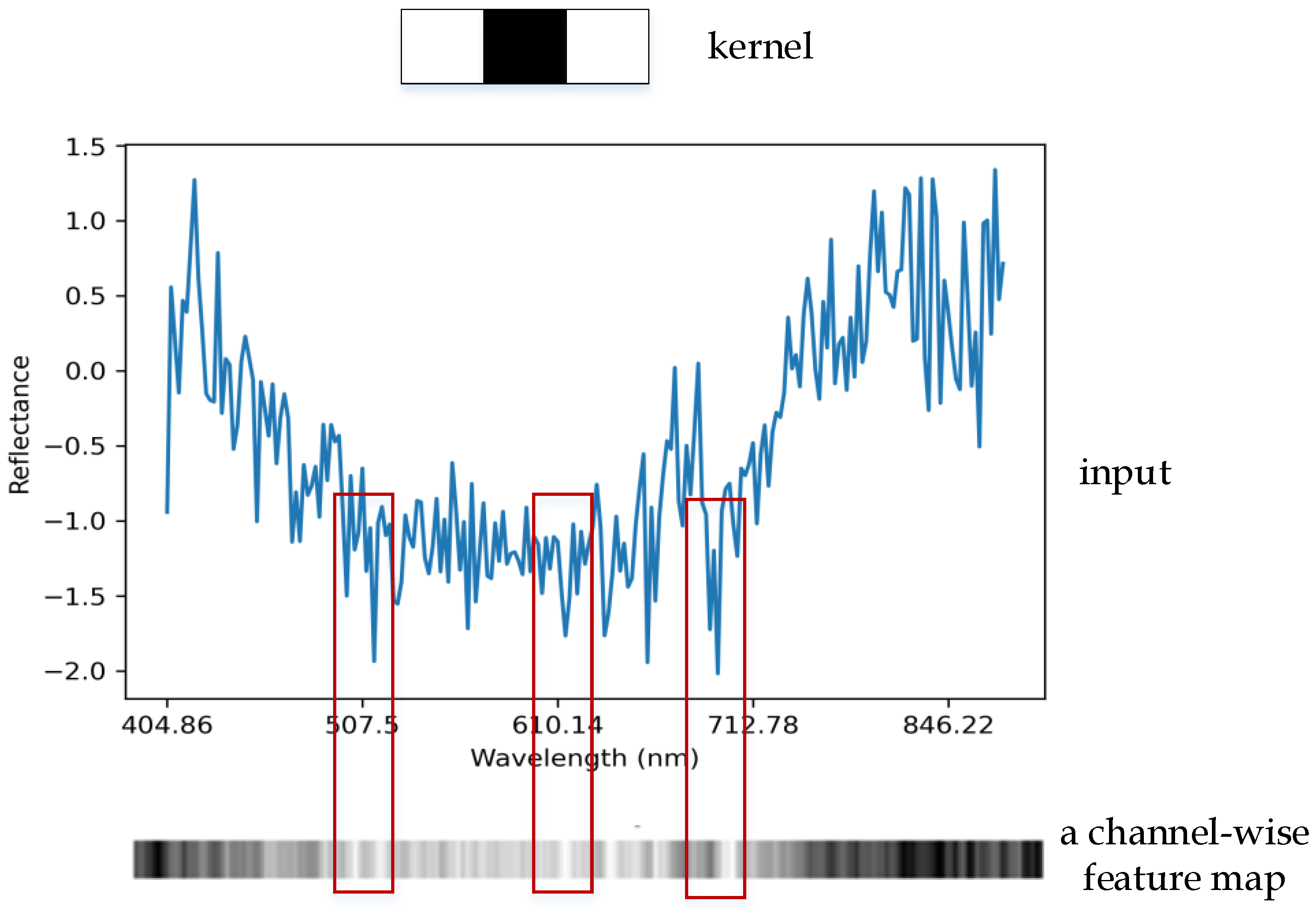
3.1. Spectral Convolution Module
3.2. Channel Attention Module
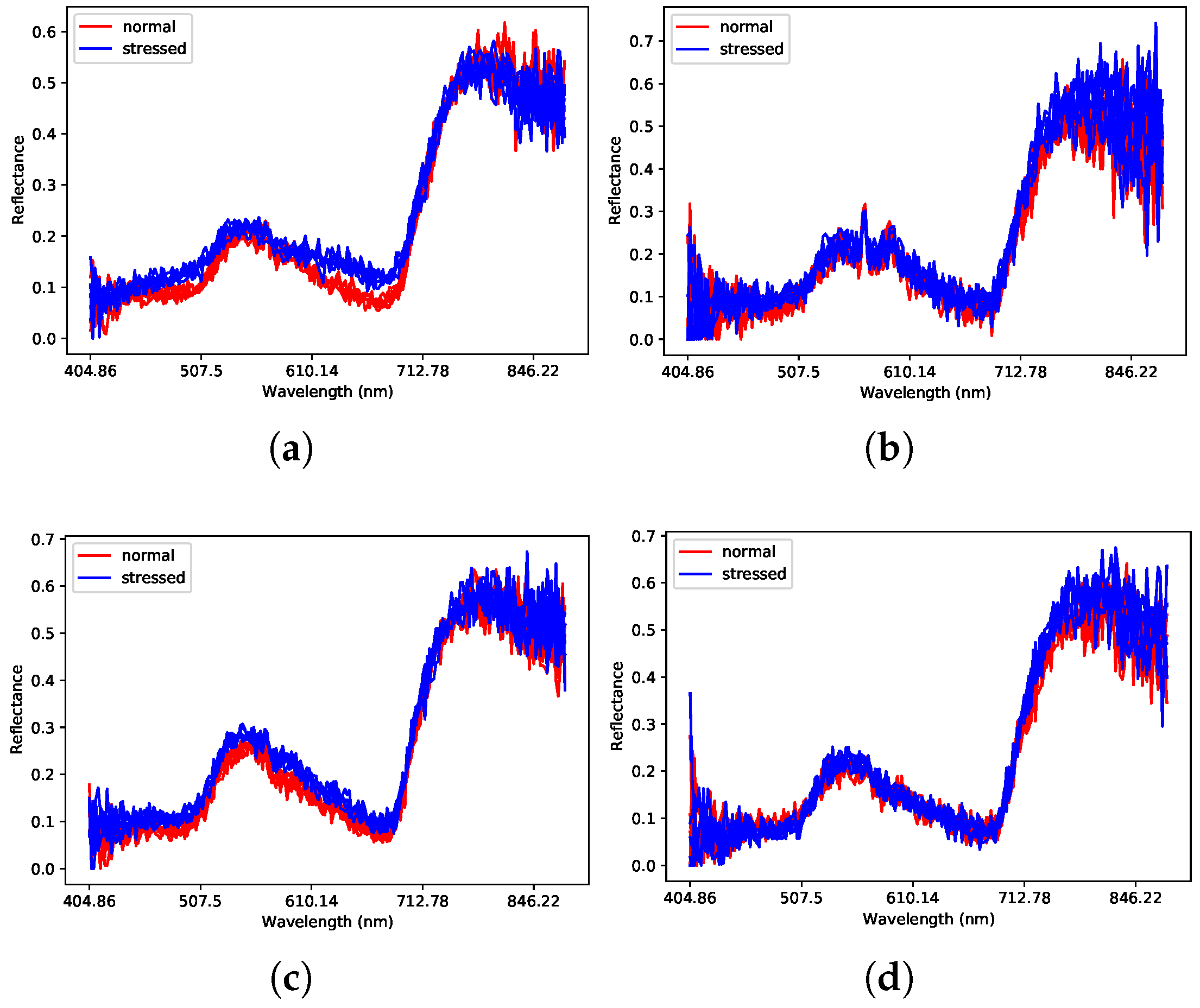
4. Experiments and Analysis
4.1. Experimental Settings
- For the experiments with CS, co(CS), sp(CS), and Kharchia datasets, alike [10], we used 70% data as training samples and 30% data as testing samples. In each experiment, we applied 5-fold cross-validation and reported mean and standard deviation. As preprocessing, we utilized a standardization technique to rescale data to have a mean of 0 and a standard deviation of 1. For training, we used Adam optimizer with a learning rate of 0.0003, the batch size was 256. The number of output channel (C) was 196, and the number of iterations was 200. For evaluation, we computed the F1 measure of control (F1C0) and stressed salt (F1C1) classes, Overall Accuracy (OA) and Average Accuracy (AA), to evaluate the proposed method’s performance.
- For the Fusarium dataset experiments, the total number of samples is 809,200. We randomly selected 227,484 samples and used the remaining samples for testing. However, since around 200,000 samples have zero value in all their bands, we discarded these samples. Then, we used the Synthetic Minority Oversampling technique (SMOTE) to oversample the minority class to overcome the class imbalance problem. In each experiment, we applied 5-fold cross-validation. The training settings were the same as those of the CS dataset, except for a batch size of 128 and a learning rate of 0.0002. For evaluation, we computed the F1 measure of background (F1background), healthy (F1healthy), and disease (F1disease) classes, OA and AA, to evaluate the proposed method’s performance with the Fusarium dataset.
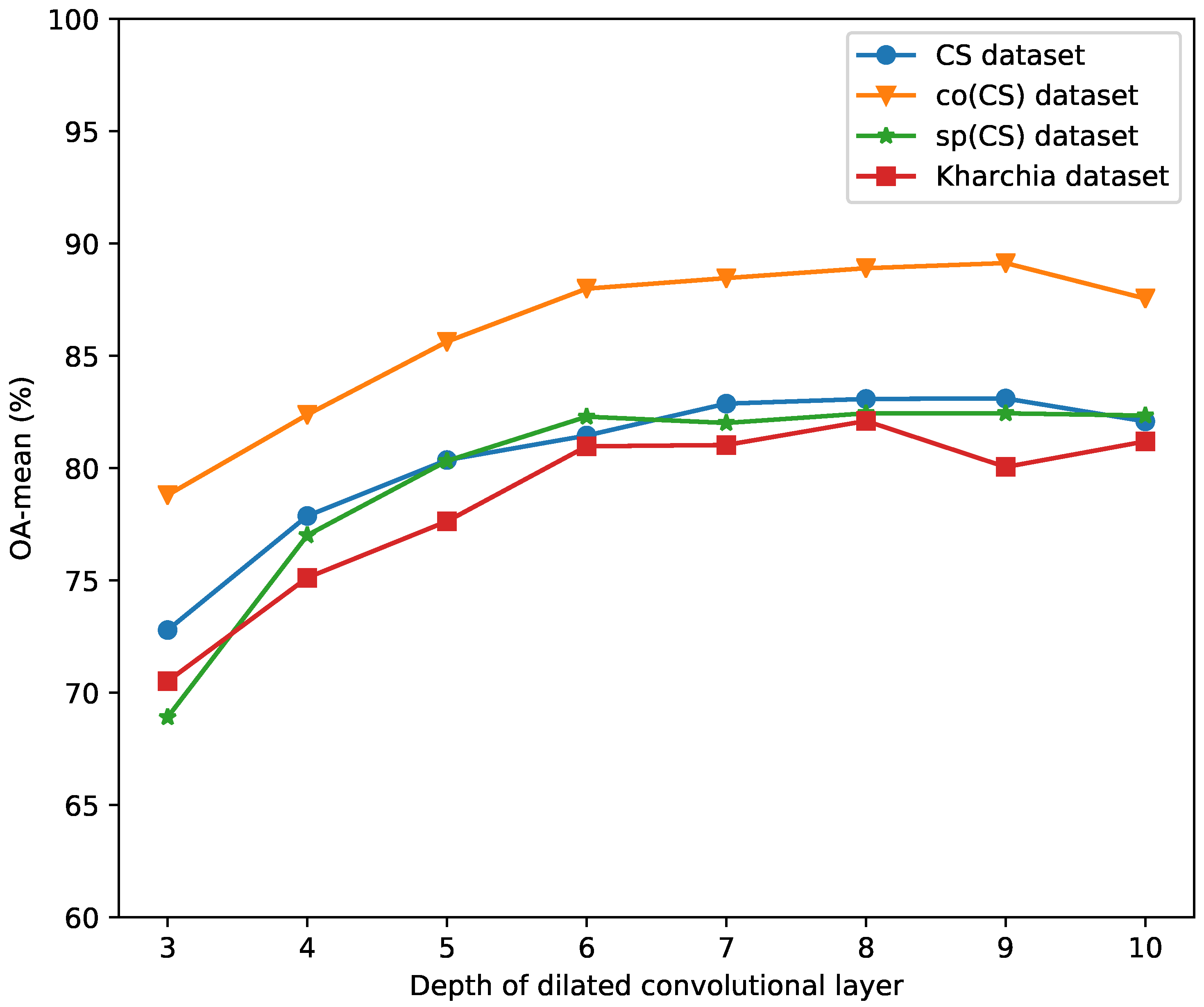
4.2. Impact of the Number of Dilated Convolution Layers (Number of N)
4.3. Ablation Analysis
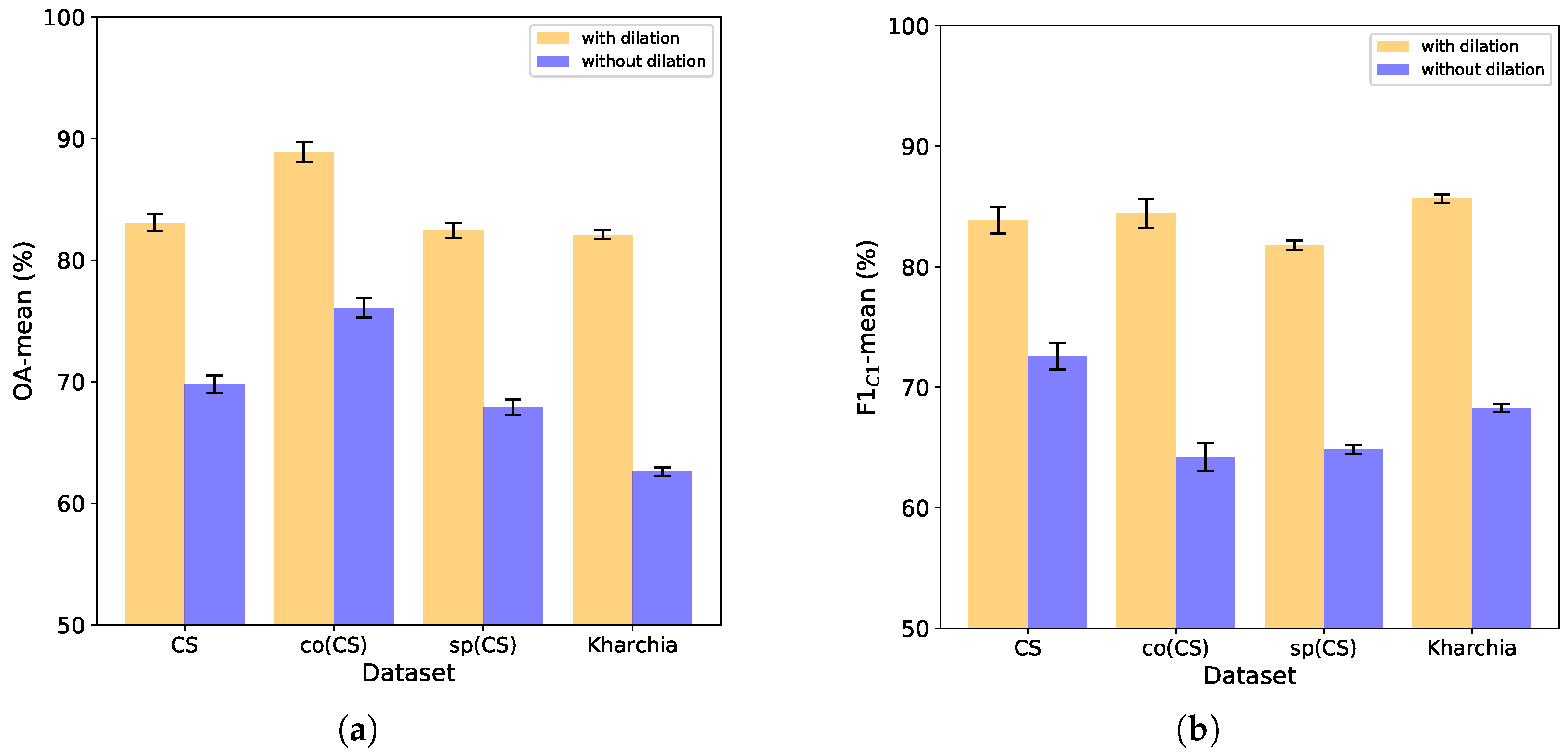
4.3.1. Impact of Dilation on Performance
4.3.2. Impact of Channel Attention Module on Performance
4.4. Comparison with Existing Methods
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sarwat, M.; Ahmad, A.; Abdin, M.Z.; Ibrahim, M.M. Stress Signaling in Plants: Genomics and Proteomics Perspective; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–350. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Peng, Z. Rice diseases detection and classification using attention based neural network and bayesian optimization. Expert Syst. Appl. 2021, 178, 114770. [Google Scholar] [CrossRef]
- Chandel, N.S.; Chakraborty, S.K.; Rajwade, Y.A.; Dubey, K.; Tiwari, M.K.; Jat, D. Identifying crop water stress using deep learning models. Neural Comput. Appl. 2020, 33, 5353–5367. [Google Scholar] [CrossRef]
- Mahlein, A.K. Plant Disease Detection by Imaging Sensors–Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Li, S.; Wu, H.; Wan, D.; Zhu, J. An effective feature selection method for hyperspectral image classification based on genetic algorithm and support vector machine. Knowl.-Based Syst. 2011, 24, 40–48. [Google Scholar] [CrossRef]
- Audebert, N.; Le Saux, B.; Lefevre, S. Deep Learning for Classification of Hyperspectral Data: A Comparative Review. IEEE Geosci. Remote Sens. Mag. 2019, 7, 159–173. [Google Scholar] [CrossRef]
- Huerta, E.B.; Duval, B.; Hao, J.K. Fuzzy Logic for Elimination of Redundant Information of Microarray Data. Genom. Proteom. Bioinform. 2008, 6, 61–73. [Google Scholar] [CrossRef]
- Moghimi, A.; Yang, C.; Miller, M.E.; Kianian, S.F.; Marchetto, P.M. A Novel Approach to Assess Salt Stress Tolerance in Wheat Using Hyperspectral Imaging. Front. Plant Sci. 2018, 9, 1182. [Google Scholar] [CrossRef]
- Moghimi, A.; Yang, C.; Marchetto, P.M. Ensemble Feature Selection for Plant Phenotyping: A Journey from Hyperspectral to Multispectral Imaging. IEEE Access 2018, 6, 56870–56884. [Google Scholar] [CrossRef]
- Li, S.; Song, W.; Fang, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Deep Learning for Hyperspectral Image Classification: An Overview. IEEE Trans. Geosci. Remote Sens. 2019, 57, 6690–6709. [Google Scholar] [CrossRef]
- Shah, S.A.A.; Bennamoun, M.; Boussaïd, F. Iterative deep learning for image set based face and object recognition. Neurocomputing 2016, 174, 866–874. [Google Scholar] [CrossRef]
- Graves, A.; Mohamed, A.R.; Hinton, G. Speech recognition with deep recurrent neural networks. In Proceedings of the ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 6645–6649. [Google Scholar]
- Chow, V. Predicting auction price of vehicle license plate with deep recurrent neural network. Expert Syst. Appl. 2020, 142, 113008. [Google Scholar] [CrossRef]
- Mou, L.; Ghamisi, P.; Zhu, X.X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef]
- Zhou, F.; Hang, R.; Liu, Q.; Yuan, X. Hyperspectral image classification using spectral-spatial LSTMs. Neurocomputing 2019, 328, 39–47. [Google Scholar] [CrossRef]
- Hochreiter, S.; Bengio, Y.; Frasconi, P.; Schmidhuber, J. A field guide to dynamical recurrent neural networks. In Gradient Flow in Recurrent Nets: The Difficulty of Learning Long-Term Dependencies; Wiley-IEEE Press: Hoboken, NJ, USA, 2001; pp. 237–243. [Google Scholar]
- Lipton, Z.C.; Berkowitz, J.; Elkan, C. A Critical Review of Recurrent Neural Networks for Sequence Learning. arXiv 2015, arXiv:1506.00019. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, F.; Hang, R.; Yuan, X. Bidirectional-Convolutional LSTM Based Spectral-Spatial Feature Learning for Hyperspectral Image Classification. Remote Sens. 2017, 9, 1330. [Google Scholar] [CrossRef]
- Lea, C.; Flynn, M.D.; Vidal, R.; Reiter, A.; Hager, G.D. Temporal Convolutional Networks for Action Segmentation and Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 156–165. [Google Scholar]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep Convolutional Neural Networks for Hyperspectral Image Classification. J. Sens. 2015, 2015, 1–12. [Google Scholar] [CrossRef]
- Peng, Z.; Huang, W.; Gu, S.; Xie, L.; Wang, Y.; Jiao, J.; Ye, Q. Conformer: Local Features Coupling Global Representations for Visual Recognition. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021; pp. 367–376. [Google Scholar]
- Jin, X.; Jie, L.; Wang, S.; Qi, H.; Li, S. Classifying Wheat Hyperspectral Pixels of Healthy Heads and Fusarium Head Blight Disease Using a Deep Neural Network in the Wild Field. Remote Sens. 2018, 10, 395. [Google Scholar] [CrossRef]
- Van Den Oord, A.; Dieleman, S.; Zen, H.; Simonyan, K.; Vinyals, O.; Graves, A.; Kalchbrenner, N.; Senior, A.W.; Kavukcuoglu, K. WaveNet: A generative model for raw audio. SSW 2016, 125, 2. [Google Scholar]
- Zhu, L.; Li, C.; Wang, B.; Yuan, K.; Yang, Z. DCGSA: A global self-attention network with dilated convolution for crowd density map generating. Neurocomputing 2020, 378, 455–466. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Chen, L.; Zhang, H.; Xiao, J.; Nie, L.; Shao, J.; Liu, W.; Chua, T.S. SCA-CNN: Spatial and Channel-Wise Attention in Convolutional Networks for Image Captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Wang, J.; Jiang, T.; Cui, Z.; Cao, Z. Filter pruning with a feature map entropy importance criterion for convolution neural networks compressing. Neurocomputing 2021, 461, 41–54. [Google Scholar] [CrossRef]
- Karlsson, I.; Friberg, H.; Kolseth, A.K.; Steinberg, C.; Persson, P. Agricultural factors affecting Fusarium communities in wheat kernels. Int. J. Food Microbiol. 2017, 252, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Peiris, K.H.S.; Dong, Y.; Davis, M.A.; Bockus, W.W.; Dowell, F.E. Estimation of the Deoxynivalenol and Moisture Contents of Bulk Wheat Grain Samples by FT-NIR Spectroscopy. Cereal Chem. J. 2017, 94, 677–682. [Google Scholar] [CrossRef]
- Iliev, I.; Krezhova, D.; Yanev, T.; Kirova, E.; Alexieva, V. Response of chlorophyll fluorescence to salinity stress on the early growth stage of the soybean plants (Glycine max L.). In Proceedings of the RAST 2009—Proceedings of 4th International Conference on Recent Advances Space Technologies, Istanbul, Turkey, 11–13 June 2009; pp. 403–407. [Google Scholar] [CrossRef]
- Hernández, E.I.; Melendez-Pastor, I.; Navarro-Pedreño, J.; Gómez, I. Spectral indices for the detection of salinity effects in melon plants. Sci. Agric. 2014, 71, 324–330. [Google Scholar] [CrossRef]
- Hamzeh, S.; Naseri, A.A.; AlaviPanah, S.K.; Bartholomeus, H.; Herold, M. Assessing the accuracy of hyperspectral and multispectral satellite imagery for categorical and Quantitative mapping of salinity stress in sugarcane fields. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 412–421. [Google Scholar] [CrossRef]
- Cao, F.; Guo, W. Deep hybrid dilated residual networks for hyperspectral image classification. Neurocomputing 2020, 384, 170–181. [Google Scholar] [CrossRef]
- Pan, B.; Xu, X.; Shi, Z.; Zhang, N.; Luo, H.; Lan, X. DSSNet: A Simple Dilated Semantic Segmentation Network for Hyperspectral Imagery Classification. IEEE Geosci. Remote Sens. Lett. 2020, 17, 1968–1972. [Google Scholar] [CrossRef]
- Pooja, K.; Nidamanuri, R.R.; Mishra, D. Multi-Scale Dilated Residual Convolutional Neural Network for Hyperspectral Image Classification. In Proceedings of the Workshop on Hyperspectral Image and Signal Processing, Evolution in Remote Sensing, Amsterdam, The Netherlands, 14–16 January 2019; Volume 2019, pp. 1–5. [Google Scholar]
- Hamaguchi, R.; Fujita, A.; Nemoto, K.; Imaizumi, T.; Hikosaka, S. Effective Use of Dilated Convolutions for Segmenting Small Object Instances in Remote Sensing Imagery. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision WACV, Lake Tahoe, NV, USA, 12–15 March 2018; Volume 2018, pp. 1442–1450. [Google Scholar]
- Cotrozzi, L. Spectroscopic detection of forest diseases: A review (1970–2020). J. For. Res. 2022, 33, 21–38. [Google Scholar] [CrossRef]
- Hou, J.; Wang, G.; Chen, X.; Xue, J.H.; Zhu, R.; Yang, H. Spatial-Temporal Attention Res-TCN for Skeleton-based Dynamic Hand Gesture Recognition. In Proceedings of the European Conference on Computer Vision (ECCV) Workshops, Munich, Germany, 8–14 September 2018. [Google Scholar]
- Zagoruyko, S.; Komodakis, N. Paying more attention to attention: Improving the performance of convolutional neural networks via attention transfer. In Proceedings of the 5th International Conference on Learning Representations, ICLR 2017—Conference Track Proceedings, Toulon, France, 24–26 April 2017. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5999–6009. [Google Scholar]
- Cheng, J.; Dong, L.; Lapata, M. Long Short-Term Memory-Networks for Machine Reading. In Proceedings of the EMNLP 2016—Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 551–561. [Google Scholar]
- Lin, Z.; Feng, M.; dos Santos, C.N.; Yu, M.; Xiang, B.; Zhou, B.; Bengio, Y. A Structured Self-attentive Sentence Embedding. arXiv 2017, arXiv:1703.03130. [Google Scholar]
- Parikh, A.P.; Täckström, O.; Das, D.; Uszkoreit, J. A Decomposable Attention Model for Natural Language Inference. In Proceedings of the EMNLP 2016—Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 2249–2255. [Google Scholar]
- Mou, L.; Zhu, X.X. Learning to Pay Attention on Spectral Domain: A Spectral Attention Module-Based Convolutional Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 110–122. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Z.; Shuai, S.; Sun, Q. Spectral group attention networks for hyperspectral image classification with spectral separability analysis. Infrared Phys. Technol. 2020, 108, 103340. [Google Scholar] [CrossRef]
- Ribalta Lorenzo, P.; Tulczyjew, L.; Marcinkiewicz, M.; Nalepa, J. Hyperspectral Band Selection Using Attention-Based Convolutional Neural Networks. IEEE Access 2020, 8, 42384–42403. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Guo, W.; Ye, H.; Cao, F. Feature-Grouped Network with Spectral-Spatial Connected Attention for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5500413. [Google Scholar] [CrossRef]
- Zhu, X.; Cheng, D.; Zhang, Z.; Lin, S.; Dai, J. An Empirical Study of Spatial Attention Mechanisms in Deep Networks. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019. [Google Scholar]
- Farha, Y.A.; Gall, J. MS-TCN: Multi-stage temporal convolutional network for action segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- van den Oord, A.; Kalchbrenner, N.; Vinyals, O.; Espeholt, L.; Graves, A.; Kavukcuoglu, K. Conditional Image Generation with PixelCNN Decoders. Adv. Neural Inf. Process. Syst. 2016, 29, 4797–4805. [Google Scholar]
- Khotimah, W.N.; Bennamoun, M.; Boussaid, F.; Sohel, F.; Edwards, D. A high-performance spectral-spatial residual network for hyperspectral image classification with small training data. Remote Sens. 2020, 12, 3137. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, L.; Du, B.; Zhang, F. Spectral-Spatial Unified Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5893–5909. [Google Scholar] [CrossRef]
- Hong, D.; Han, Z.; Yao, J.; Gao, L.; Zhang, B.; Plaza, A.; Chanussot, J. SpectralFormer: Rethinking Hyperspectral Image Classification with Transformers. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5518615. [Google Scholar] [CrossRef]
| Dataset | Performance | With Attention | Without Attention |
|---|---|---|---|
| CS | OA | 83.08 ± 0.70 | 81.02 ± 2.79 |
| AA | 83.15 ± 0.43 | 82.32 ± 1.61 | |
| F1C0 | 82.21 ± 0.30 | 78.24 ± 5.96 | |
| F1C1 | 83.86 ± 1.09 | 82.78 ± 1.94 | |
| co(CS) | OA | 88.90 ± 0.81 | 85.24 ± 0.91 |
| AA | 88.07 ± 0.88 | 84.01 ± 1.01 | |
| F1C0 | 91.38 ± 0.62 | 88.11 ± 1.05 | |
| F1C1 | 84.41 ± 1.17 | 80.46 ± 0.97 | |
| sp(CS) | OA | 82.44 ± 0.62 | 79.73 ± 1.04 |
| AA | 82.52 ± 0.52 | 80.20 ± 1.07 | |
| F1C0 | 83.03 ± 1.09 | 80.93 ± 1.35 | |
| F1C1 | 83.03 ± 1.09 | 78.22 ± 2.10 | |
| Kharchia | OA | 82.10 ± 0.36 | 78.80 ± 2.09 |
| AA | 81.25 ± 0.43 | 78.60 ± 2.00 | |
| F1C0 | 76.23 ± 0.34 | 73.58 ± 1.22 | |
| F1C1 | 85.65 ± 0.35 | 82.07 ± 3.15 |
| Method | F1C0 | F1C1 | F1-mean | OA | AA |
|---|---|---|---|---|---|
| CS | |||||
| 1DCNN | 71.40 ± 1.64 | 77.21 ± 0.46 | 74.31 ± 0.93 | 74.65 ± 0.79 | 74.50 ± 0.74 |
| RNN | 76.82 ± 2.68 | 80.02 ± 1.49 | 78.42 ± 1.96 | 78.57 ± 1.86 | 78.52 ± 1.89 |
| LSTM | 77.16 ± 0.88 | 81.27 ± 0.37 | 79.21 ± 0.60 | 79.42 ± 0.56 | 79.25 ± 0.54 |
| sRN | 79.78 ± 0.57 | 82.14 ± 0.95 | 80.97 ± 0.66 | 81.05 ± 0.69 | 80.98 ± 0.57 |
| spectralFormer | 77.55 ± 1.78 | 80.60 ± 0.96 | 79.08 ± 1.31 | 79.20 ± 1.26 | 79.09 ± 1.32 |
| SFS_Forward | 78.87 | 76.55 | 77.71 | - | - |
| SC-CAN | 82.21 ± 0.30 | 83.86 ± 1.09 | 83.03 ± 0.66 | 83.08 ± 0.70 | 83.15 ± 0.43 |
| co(CS) | |||||
| 1DCNN | 79.40 ± 0.51 | 61.84 ± 1.03 | 70.62 ± 0.73 | 73.25 ± 0.65 | 70.89 ± 0.72 |
| RNN | 82.20 ± 1.20 | 66.70 ± 2.75 | 74.45 ± 1.76 | 76.82 ± 1.47 | 74.98 ± 1.58 |
| LSTM | 84.36 ± 0.42 | 70.88 ± 0.98 | 77.62 ± 0.64 | 79.65 ± 0.54 | 78.05 ± 0.60 |
| sRN | 85.03 ± 0.65 | 70.74 ± 1.75 | 77.89 ± 1.05 | 80.20 ± 0.81 | 79.00 ± 0.97 |
| spectralFormer | 86.09 ± 1.03 | 73.88 ± 3.45 | 79.99 ± 2.21 | 81.86 ± 1.67 | 80.57 ± 1.52 |
| SC-CAN | 91.38 ± 0.62 | 84.41 ± 1.17 | 87.89 ± 0.89 | 88.90 ± 0.81 | 88.07 ± 0.88 |
| sp(CS) | |||||
| 1DCNN | 68.42 ± 0.63 | 65.32 ± 0.66 | 66.87 ± 0.57 | 66.95 ± 0.58 | 66.89 ± 0.58 |
| RNN | 79.31 ± 0.57 | 74.36 ± 1.38 | 76.83 ± 0.97 | 77.10 ± 0.89 | 77.57 ± 0.73 |
| LSTM | 76.07 ± 0.96 | 73.07 ± 1.26 | 74.57 ± 0.95 | 74.67 ± 0.94 | 74.70 ± 0.97 |
| sRN | 77.88 ± 0.53 | 74.70 ± 0.76 | 76.29 ± 0.47 | 76.41 ± 0.45 | 76.47 ± 0.44 |
| spectralFormer | 77.84 ± 1.45 | 75.21 ± 1.53 | 76.52 ± 1.22 | 76.62 ± 1.24 | 76.73 ± 1.33 |
| SC-CAN | 83.03 ± 1.09 | 81.78 ± 0.39 | 82.40 ± 0.60 | 82.44 ± 0.62 | 82.52 ± 0.52 |
| Kharchia | |||||
| 1DCNN | 53.46 ± 0.66 | 71.49 ± 0.59 | 62.47 ± 0.51 | 64.64 ± 0.54 | 62.55 ± 0.53 |
| RNN | 61.71 ± 4.56 | 80.44 ± 1.06 | 71.07 ± 2.38 | 74.18 ± 1.45 | 73.72 ± 1.66 |
| LSTM | 66.97 ± 0.98 | 79.91 ± 0.62 | 73.44 ± 0.74 | 75.02 ± 0.71 | 73.63 ± 0.75 |
| sRN | 69.71 ± 1.54 | 82.50 ± 0.73 | 76.11 ± 0.97 | 77.83 ± 0.85 | 76.87 ± 0.98 |
| spectralFormer | 67.57 ± 1.06 | 81.04 ± 1.22 | 74.30 ± 0.99 | 76.08 ± 1.13 | 74.93 ± 1.30 |
| SC-CAN | 76.23 ± 0.34 | 85.65 ± 0.35 | 80.94 ± 0.33 | 82.10 ± 0.36 | 81.25 ± 0.43 |
| Method | F1disease | F1healthy | F1background | OA | AA |
|---|---|---|---|---|---|
| 1D CNN | 52.71 ± 1.38 | 76.50 ± 0.29 | 79.21 ± 0.69 | 61.37 ± 33.89 | 62.58 ± 34.55 |
| RNN | 51.59 ± 8.29 | 83.27 ± 4.14 | 80.51 ± 1.77 | 79.79 ± 1.13 | 72.33 ± 3.46 |
| LSTM | 51.15 ± 4.57 | 77.35 ± 0.71 | 83.03 ± 1.54 | 78.36 ± 1.24 | 82.86 ± 0.65 |
| sRN | 39.78 ± 18.70 | 73.97 ± 14.35 | 77.56 ± 6.78 | 72.31 ± 11.68 | 76.09 ± 9.85 |
| spectralFormer | 62.99 ± 4.45 | 81.91 ± 0.61 | 84.18 ± 0.63 | 82.00 ± 0.89 | 72.59 ± 1.88 |
| 2D-CNN-BidGRU 1 | 52 | 71 | 88 | 74.30 | - |
| 2D-CNN-BidGRU 2 | 30.19 ± 0.85 | 62.30 ± 0.42 | 77.01 ± 0.26 | 66.70 ± 0.48 | 70.47 ± 0.19 |
| SC-CAN | 70.38 ± 3.10 | 83.25 ± 0.62 | 83.42 ± 1.68 | 82.78 ± 0.97 | 83.83 ± 1.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khotimah, W.N.; Boussaid, F.; Sohel, F.; Xu, L.; Edwards, D.; Jin, X.; Bennamoun, M. SC-CAN: Spectral Convolution and Channel Attention Network for Wheat Stress Classification. Remote Sens. 2022, 14, 4288. https://doi.org/10.3390/rs14174288
Khotimah WN, Boussaid F, Sohel F, Xu L, Edwards D, Jin X, Bennamoun M. SC-CAN: Spectral Convolution and Channel Attention Network for Wheat Stress Classification. Remote Sensing. 2022; 14(17):4288. https://doi.org/10.3390/rs14174288
Chicago/Turabian StyleKhotimah, Wijayanti Nurul, Farid Boussaid, Ferdous Sohel, Lian Xu, David Edwards, Xiu Jin, and Mohammed Bennamoun. 2022. "SC-CAN: Spectral Convolution and Channel Attention Network for Wheat Stress Classification" Remote Sensing 14, no. 17: 4288. https://doi.org/10.3390/rs14174288
APA StyleKhotimah, W. N., Boussaid, F., Sohel, F., Xu, L., Edwards, D., Jin, X., & Bennamoun, M. (2022). SC-CAN: Spectral Convolution and Channel Attention Network for Wheat Stress Classification. Remote Sensing, 14(17), 4288. https://doi.org/10.3390/rs14174288









