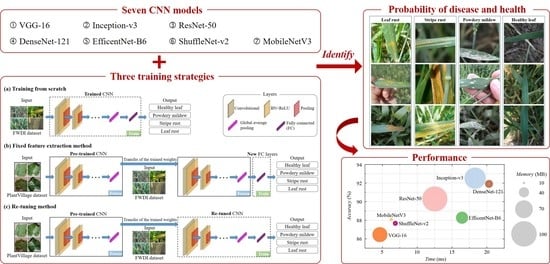
Evaluation of Diverse Convolutional Neural Networks and Training Strategies for Wheat Leaf Disease Identification with Field-Acquired Photographs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. PlantVillage Dataset
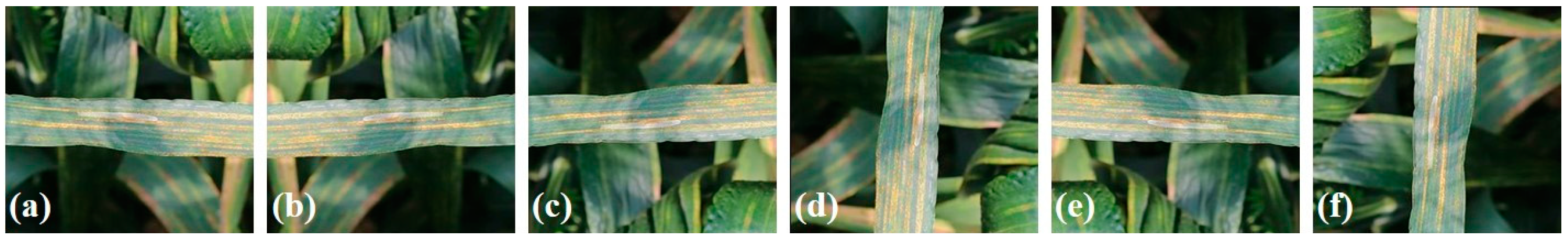
2.1.2. Field-Based Wheat Diseases Images (FWDI) Dataset
2.2. Convolutional Neural Networks (CNNs)
2.2.1. VGG-16
2.2.2. Inception-v3
2.2.3. ResNet-50
2.2.4. DenseNet-121
2.2.5. EfficentNet-B6
2.2.6. ShuffleNet-v2
2.2.7. MobileNetV3
2.3. Comparison and Evaluation
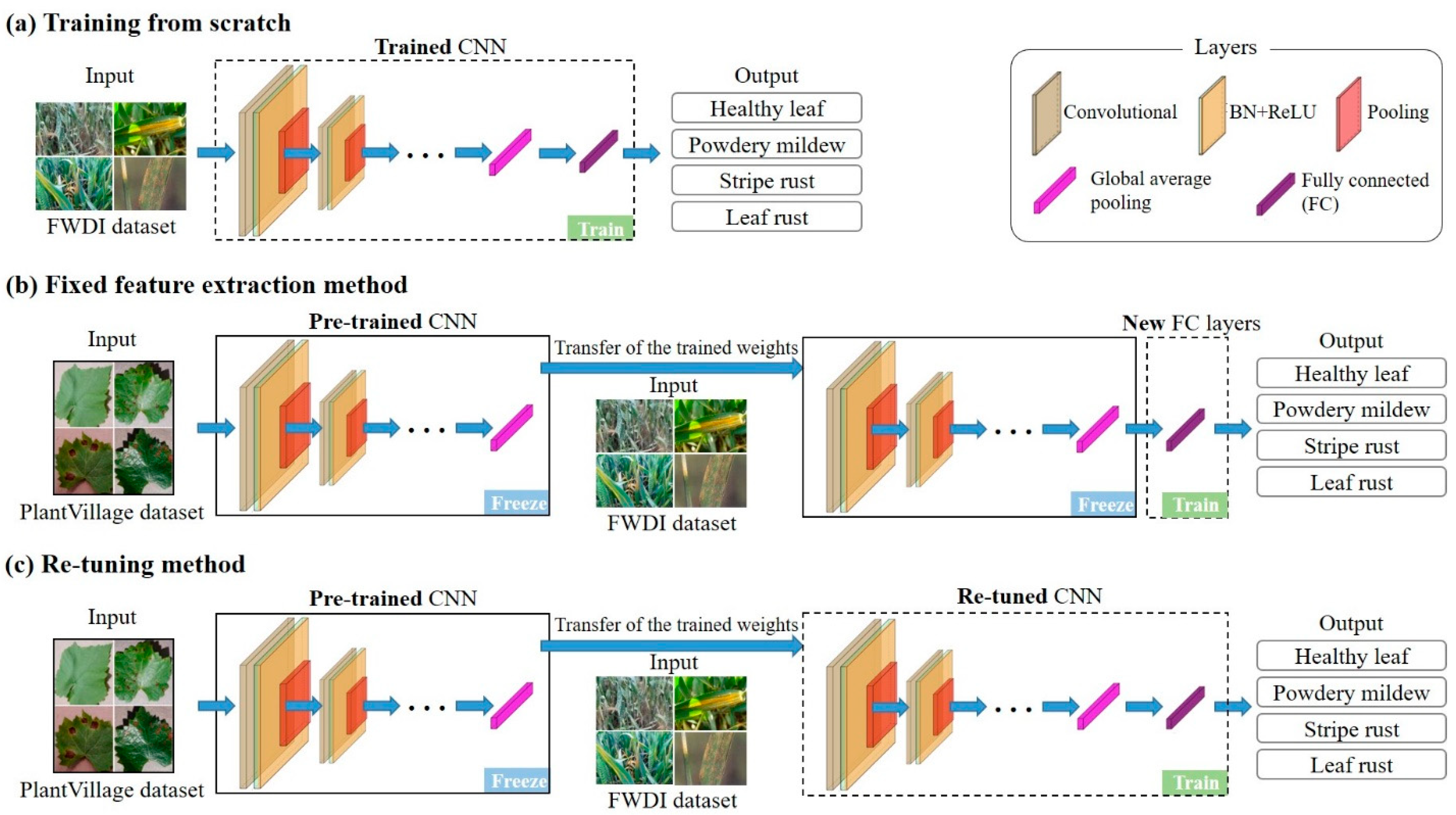
2.3.1. Training Strategies of CNNs
2.3.2. Model Assessment
3. Results
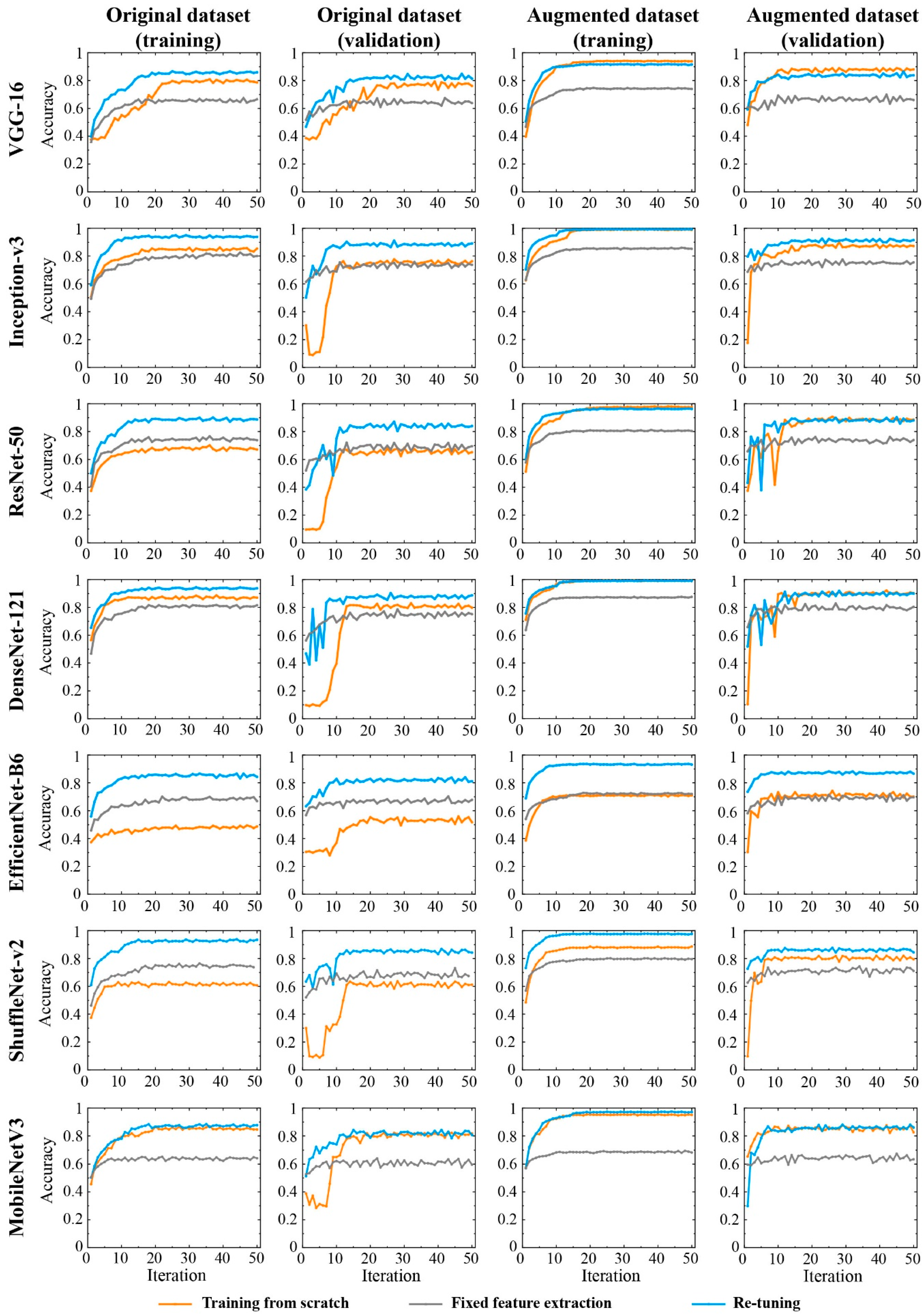
3.1. Accuracy of CNNs Trained by Different Training Sets and Strategies
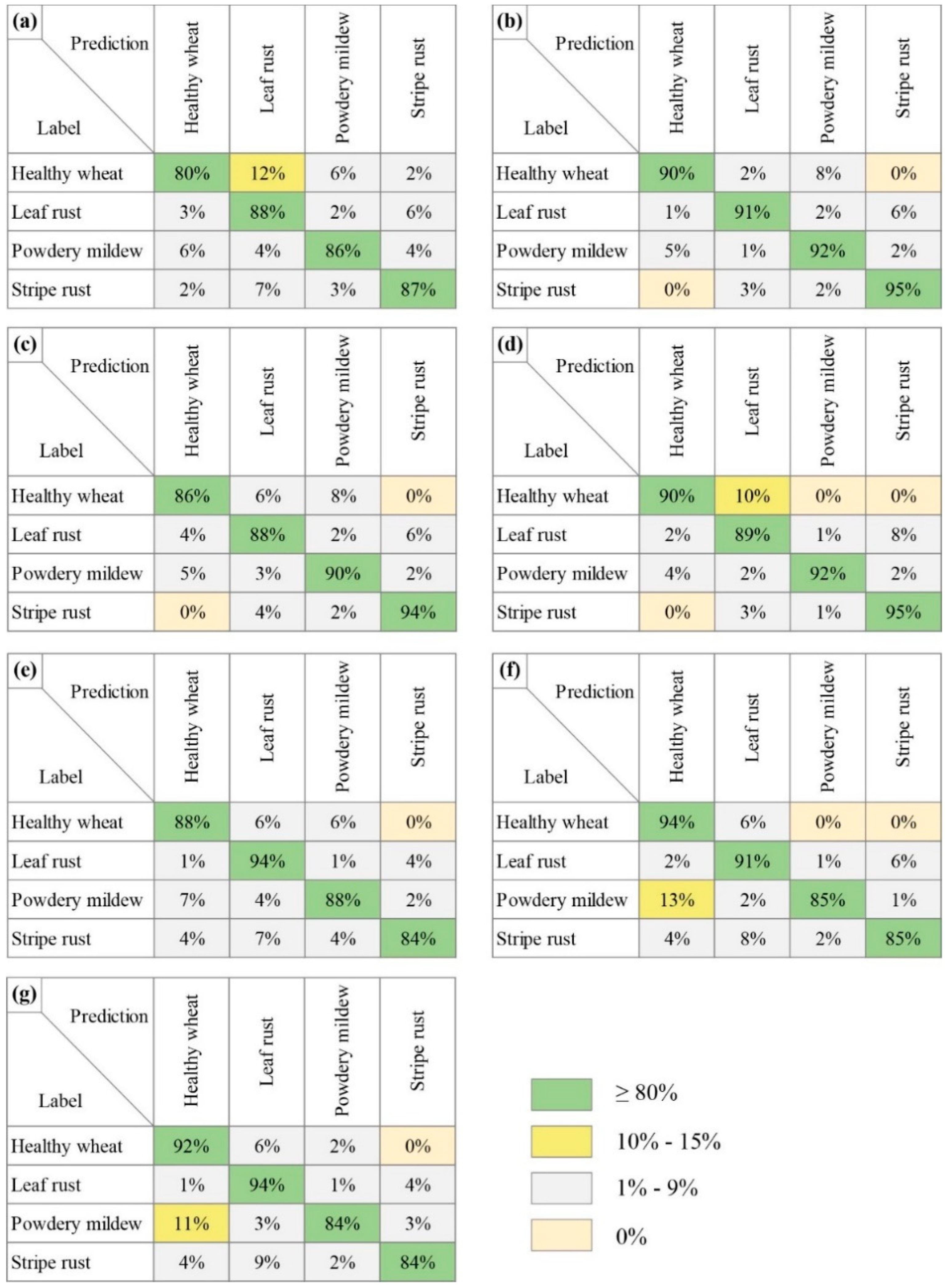
3.2. Wheat Disease Diagnosis
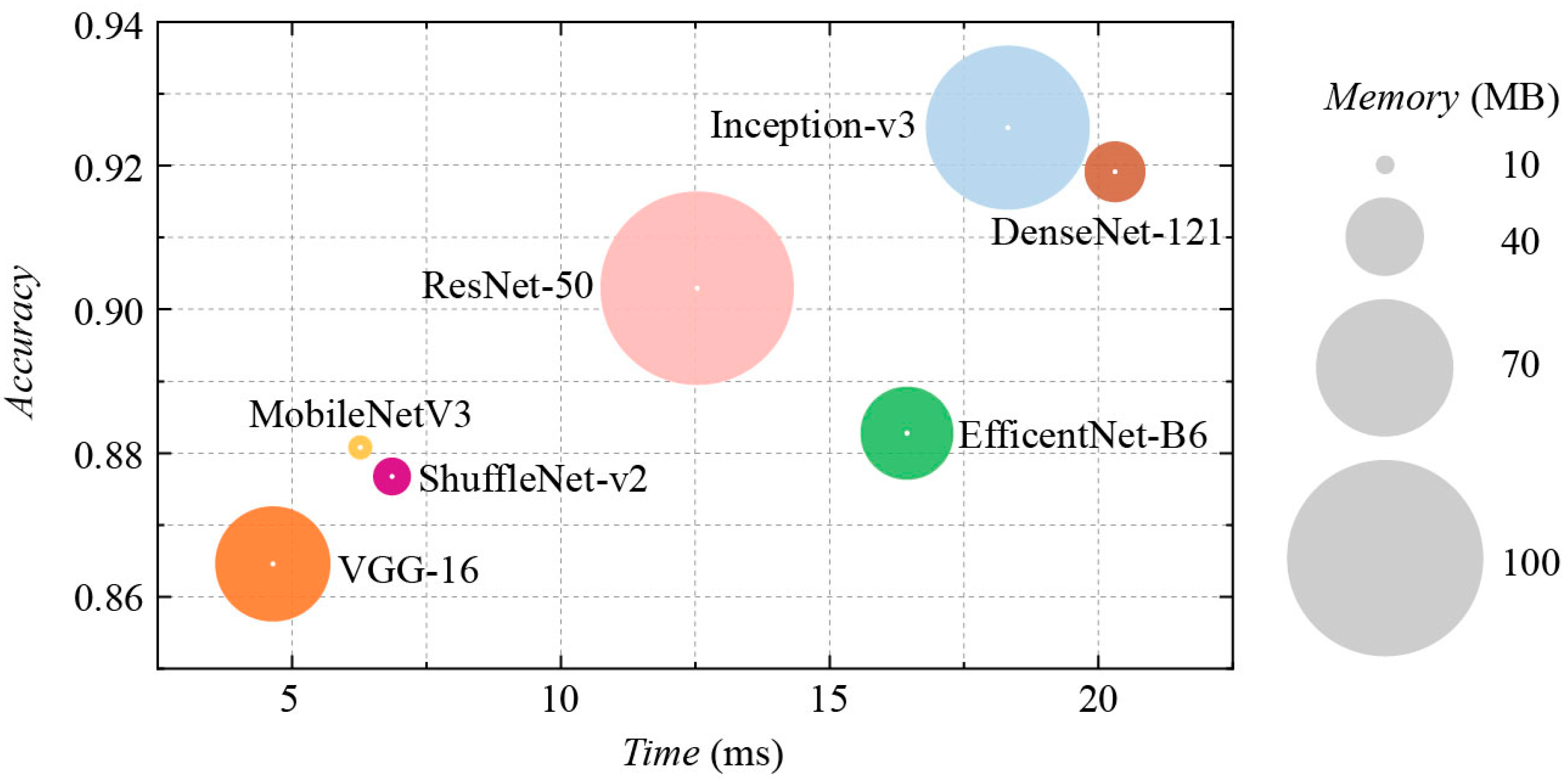
3.3. Comparative Evaluation of CNNs
4. Discussion
4.1. Influencing Factors of CNNs Applied in Crop Diseases Diagnosis
4.2. Limitations and Prospects for Precision Agriculture
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Curtis, B.; Rajaram, S.; Macpherson, H. Bread Wheat: Improvement and Production; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2002. [Google Scholar]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases-a field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Huang, D.S. A Novel Density-Based Clustering Framework by Using Level Set Method. IEEE Trans. Knowl. Data Eng. 2009, 21, 1515–1531. [Google Scholar] [CrossRef]
- Khosrokhani, M.; Nasr, A.H. Applications of the Remote Sensing Technology to Detect and Monitor the Rust Disease in the Wheat–a Literature Review. Geocarto Int. 2022, 1–27. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Processing Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Krizhevsky, A. One weird trick for parallelizing convolutional neural networks. arXiv 2014, arXiv:1404.5997. [Google Scholar] [CrossRef]
- Sermanet, P.; Eigen, D.; Zhang, X.; Mathieu, M.; Fergus, R.; Lecun, Y. OverFeat: Integrated Recognition, Localization and Detection using Convolutional Networks. arXiv 2013, arXiv:1312.6229. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Hu, J.; Zhao, G.; Mei, F.; Zhang, C. An in-field automatic wheat disease diagnosis system. Comput. Electron. Agric. 2017, 142, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Picon, A.; Alvarez-Gila, A.; Seitz, M.; Ortiz-Barredo, A.; Echazarra, J.; Johannes, A. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput. Electron. Agric. 2019, 161, 280–290. [Google Scholar] [CrossRef]
- Bao, W.; Yang, X.; Liang, D.; Hu, G.; Yang, X. Lightweight convolutional neural network model for field wheat ear disease identification. Comput. Electron. Agric. 2021, 189, 106367. [Google Scholar] [CrossRef]
- Panigrahi, S.; Nanda, A.; Swarnkar, T. A Survey on Transfer Learning. In Proceedings of the Intelligent and Cloud Computing; Springer: Singapore, 2021; pp. 781–789. [Google Scholar]
- Pan, S.J.; Qiang, Y. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Ramcharan, A.; Baranowski, K.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep Learning for Image-Based Cassava Disease Detection. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Dong, Z.; Jiang, W.; Yang, Y. Recognition of rice leaf diseases and wheat leaf diseases based on multi-task deep transfer learning. Comput. Electron. Agric. 2021, 186, 106184. [Google Scholar] [CrossRef]
- Amara, J.; Bouaziz, B.; Algergawy, A. A deep learning-based approach for banana leaf diseases classification. In Datenbanksysteme für Business, Technologie und Web (BTW 2017)-Workshopband; Gesellschaft für Informatik e.V.: Stuttgart, Germany, 2017. [Google Scholar]
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A. Deep learning for tomato diseases: Classification and symptoms visualization. Appl. Artif. Intell. 2017, 31, 299–315. [Google Scholar] [CrossRef]
- Cowger, C.; Miranda, L.; Griffey, C.; Hall, M.; Murphy, J.; Maxwell, J.; Sharma, I. Wheat Powdery Mildew; CABI: Oxfordshire, UK, 2012; pp. 84–119. [Google Scholar]
- Wan, A.M.; Chen, X.M.; He, Z. Wheat stripe rust in China. Aust. J. Agric. Res. 2007, 58, 605–619. [Google Scholar] [CrossRef]
- Samborski, D. Wheat leaf rust. In Diseases, Distribution, Epidemiology, and Control; Elsevier: Amsterdam, The Netherlands, 1985; pp. 39–59. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.-C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V. Searching for mobilenetv3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar] [CrossRef]
- Zoph, B.; Le, Q.V. Neural architecture search with reinforcement learning. arXiv 2016, arXiv:1611.01578. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Chollet, F. Keras: The Python Deep Learning Library. Astrophys. Source Code Libr. 2018, ascl:1806.1022. Available online: https://ui.adsabs.harvard.edu/abs/2018ascl.soft06022C/abstract (accessed on 16 June 2022).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Wang, G.; Sun, Y.; Wang, J. Automatic Image-Based Plant Disease Severity Estimation Using Deep Learning. Comput. Intell. Neurosci. 2017, 2017, 2917536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuentes, A.F.; Yoon, S.; Lee, J.; Park, D.S. High-Performance Deep Neural Network-Based Tomato Plant Diseases and Pests Diagnosis System With Refinement Filter Bank. Front. Plant Sci. 2018, 9, 1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbedo, J.G.A. Factors influencing the use of deep learning for plant disease recognition. Biosyst. Eng. 2018, 172, 84–91. [Google Scholar] [CrossRef]
- Ma, N.; Zhang, X.; Zheng, H.T.; Sun, J. ShuffleNet V2: Practical Guidelines for Efficient CNN Architecture Design; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Mustafa, G.; Zheng, H.; Khan, I.H.; Tian, L.; Jia, H.; Li, G.; Cheng, T.; Tian, Y.; Cao, W.; Zhu, Y. Hyperspectral Reflectance Proxies to Diagnose In-Field Fusarium Head Blight in Wheat with Machine Learning. Remote Sens. 2022, 14, 2784. [Google Scholar] [CrossRef]
- Su, J.; Liu, C.; Chen, W.-H. UAV Multispectral Remote Sensing for Yellow Rust Mapping: Opportunities and Challenges. Unmanned Aer. Syst. Precis. Agric. 2022, 107–122. [Google Scholar] [CrossRef]
- León-Rueda, W.A.; León, C.; Caro, S.G.; Ramírez-Gil, J.G. Identification of diseases and physiological disorders in potato via multispectral drone imagery using machine learning tools. Trop. Plant Pathol. 2022, 47, 152–167. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Ba, Y.; Lyu, X.; Zhang, M.; Li, M. Banana Fusarium Wilt Disease Detection by Supervised and Unsupervised Methods from UAV-Based Multispectral Imagery. Remote Sens. 2022, 14, 1231. [Google Scholar] [CrossRef]
- Khaled, A.Y.; Abd Aziz, S.; Bejo, S.K.; Nawi, N.M.; Seman, I.A.; Onwude, D.I. Early detection of diseases in plant tissue using spectroscopy – applications and limitations. Appl. Spectrosc. Rev. 2018, 53, 36–64. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant Methods 2020, 16, 83. [Google Scholar] [CrossRef] [PubMed]


| Dataset | Disease Type | Original Images | Original Training Set | Augmented Images | Augmented Training Set | Test Set |
|---|---|---|---|---|---|---|
| PlantVillage | 26 types | 37,721 | 32,739 | - | - | 4982 |
| FWDI | Powdery mildew | 561 | 449 | 2806 | 2694 | 112 |
| Leaf rust | 808 | 647 | 4043 | 3882 | 161 | |
| Stripe rust | 1015 | 812 | 5075 | 4872 | 203 | |
| Healthy wheat | 259 | 208 | 1299 | 1248 | 51 | |
| Total | 2643 | 2116 | 13,223 | 12,696 | 527 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Liu, H.; Zhao, C.; He, C.; Ma, J.; Cheng, T.; Zhu, Y.; Cao, W.; Yao, X. Evaluation of Diverse Convolutional Neural Networks and Training Strategies for Wheat Leaf Disease Identification with Field-Acquired Photographs. Remote Sens. 2022, 14, 3446. https://doi.org/10.3390/rs14143446
Jiang J, Liu H, Zhao C, He C, Ma J, Cheng T, Zhu Y, Cao W, Yao X. Evaluation of Diverse Convolutional Neural Networks and Training Strategies for Wheat Leaf Disease Identification with Field-Acquired Photographs. Remote Sensing. 2022; 14(14):3446. https://doi.org/10.3390/rs14143446
Chicago/Turabian StyleJiang, Jiale, Haiyan Liu, Chen Zhao, Can He, Jifeng Ma, Tao Cheng, Yan Zhu, Weixing Cao, and Xia Yao. 2022. "Evaluation of Diverse Convolutional Neural Networks and Training Strategies for Wheat Leaf Disease Identification with Field-Acquired Photographs" Remote Sensing 14, no. 14: 3446. https://doi.org/10.3390/rs14143446
APA StyleJiang, J., Liu, H., Zhao, C., He, C., Ma, J., Cheng, T., Zhu, Y., Cao, W., & Yao, X. (2022). Evaluation of Diverse Convolutional Neural Networks and Training Strategies for Wheat Leaf Disease Identification with Field-Acquired Photographs. Remote Sensing, 14(14), 3446. https://doi.org/10.3390/rs14143446