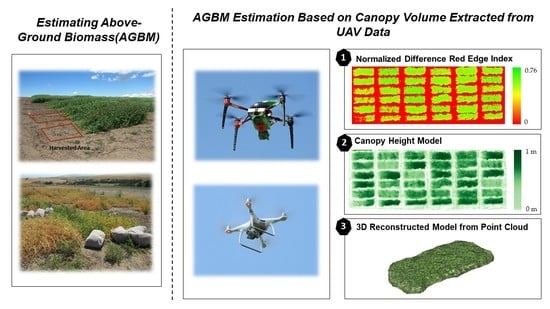
Optimization of UAV-Based Imaging and Image Processing Orthomosaic and Point Cloud Approaches for Estimating Biomass in a Forage Crop
Abstract
:1. Introduction
2. Materials and Methods
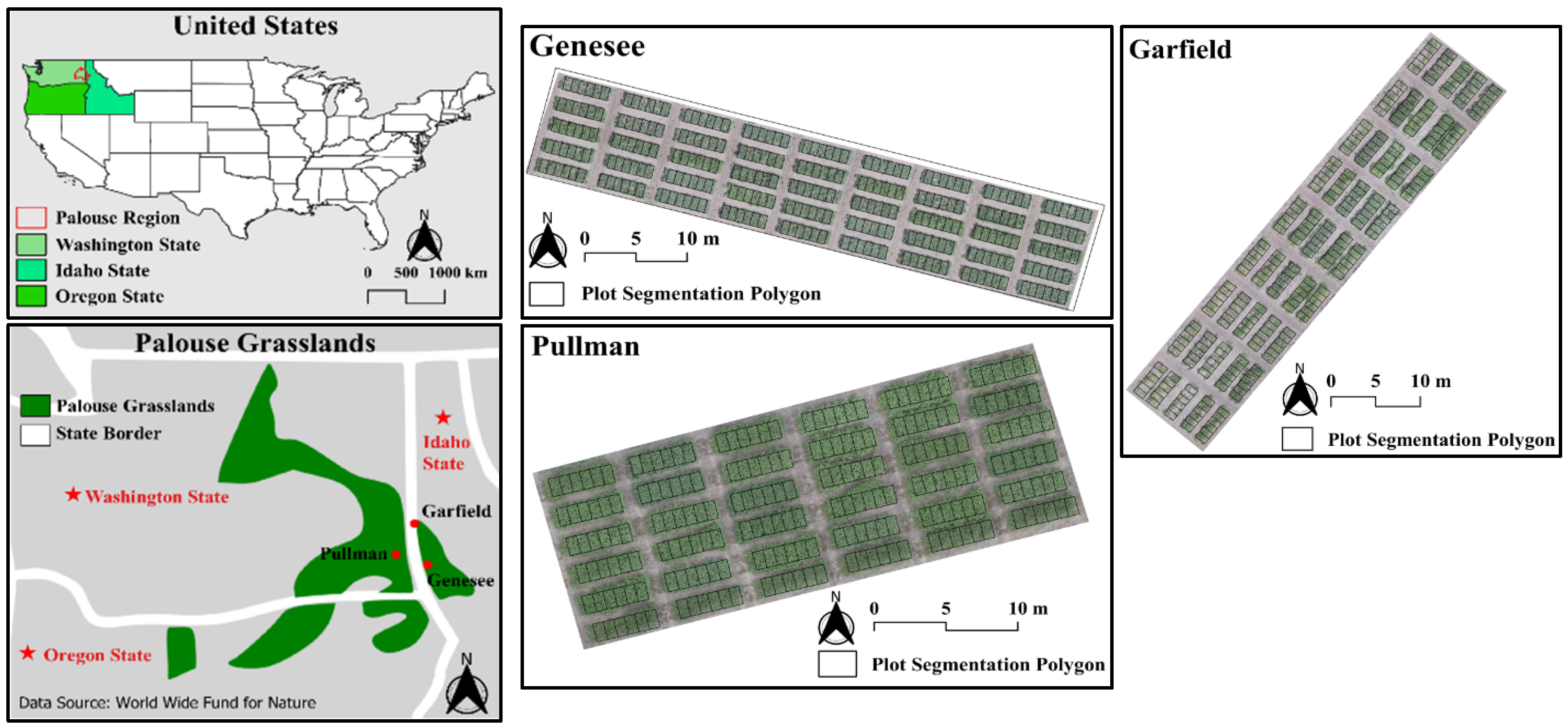
2.1. Study Area
2.2. Data Acquisition
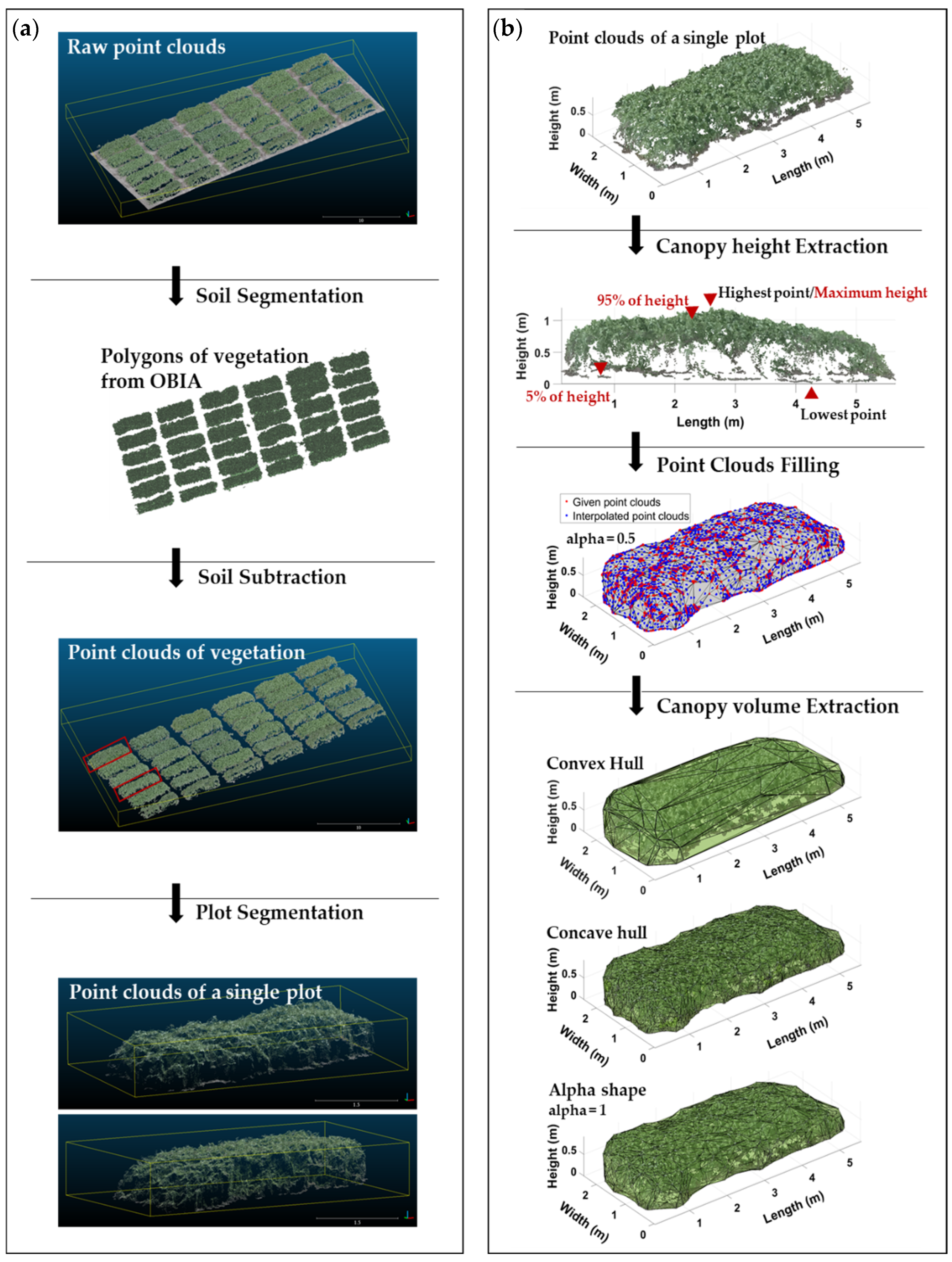
2.3. Image Processing
2.3.1. Vegetation Indices
| Vegetation Index | Formulation | Reference |
|---|---|---|
| CIgr: Chlorophyll Index Green | [49] | |
| CIre: Chlorophyll Index Red Edge | [49] | |
| EVI2: Enhanced Vegetation Index 2 | [50] | |
| GNDVI: Green Normalized Difference Vegetation Index | [51] | |
| MCARI2: Modified Chlorophyll Absorption Ratio Index 2 | [52] | |
| MTVI2: Modified Triangular Vegetation Index 2 | [52] | |
| NDRE: Normalized Difference Red Edge | [53] | |
| NDVI: Normalized Difference Vegetation Index | [54] | |
| NDWI: Normalized Difference Water Index | [55] | |
| OSAVI: Optimized Soil-Adjusted Vegetation Index | [56] | |
| RDVI: Renormalized Difference Vegetation Index | [57] | |
| RGBVI: Red–Green–Blue Vegetation Index | [58] |
2.3.2. Canopy Height Model from Digital Surface Model
2.3.3. Three-Dimensional Reconstruction Model of Point Clouds
2.4. Data Analysis
3. Result and Discussion
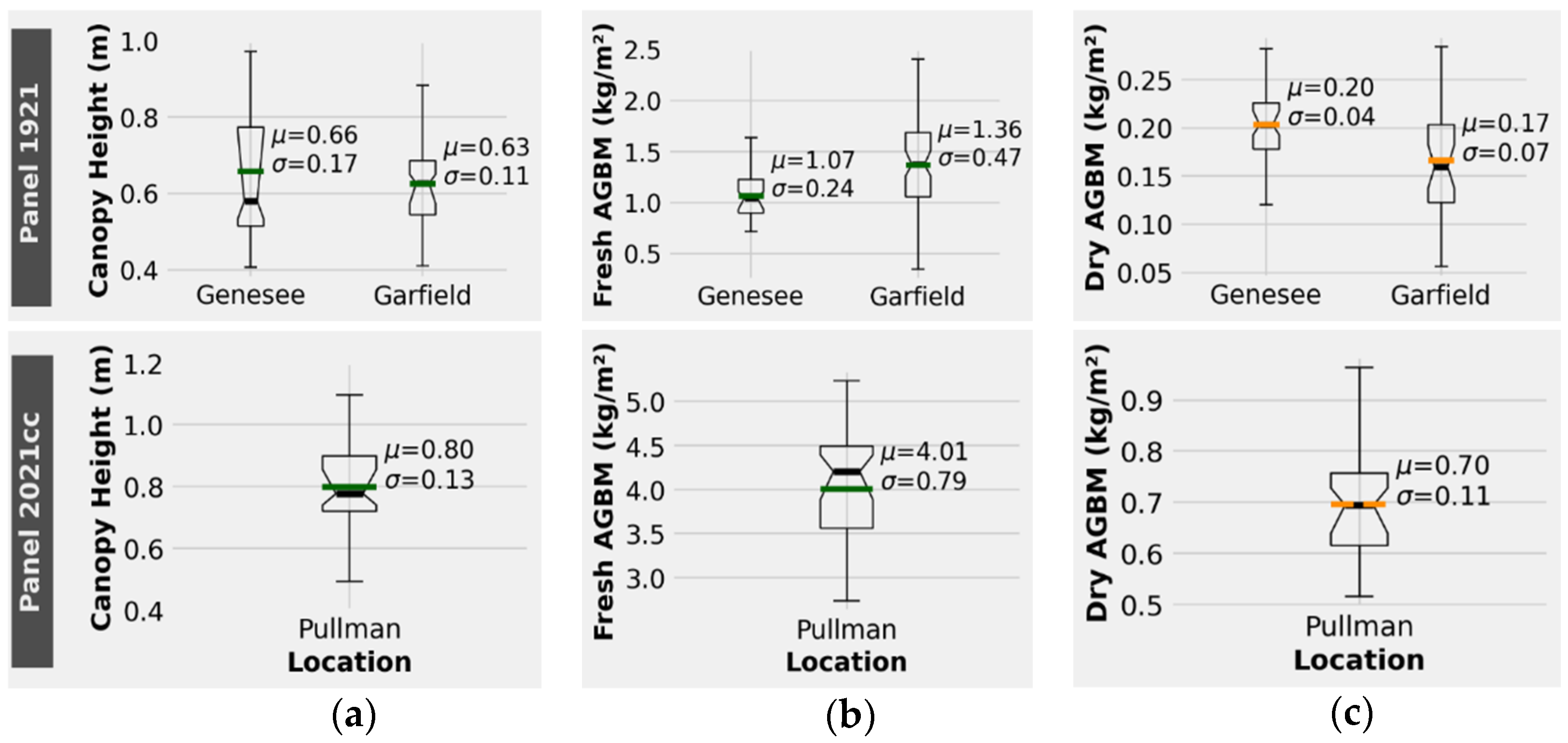
3.1. Ground Reference Data
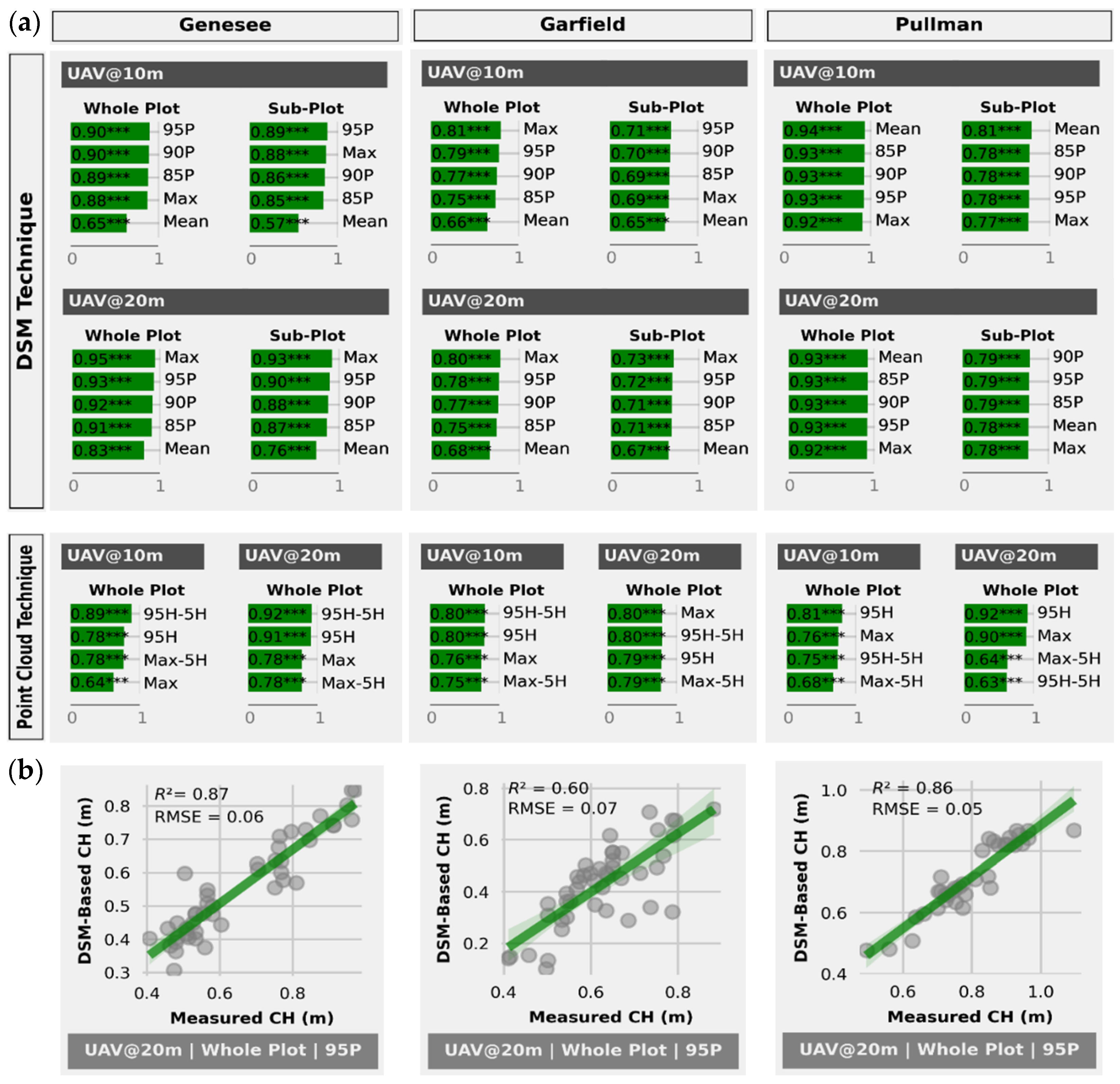
3.2. Canopy Height Estimation
3.2.1. DSM-Based Technique for Canopy Height Estimation
3.2.2. Point-Cloud-Based Technique for Canopy Height Estimation
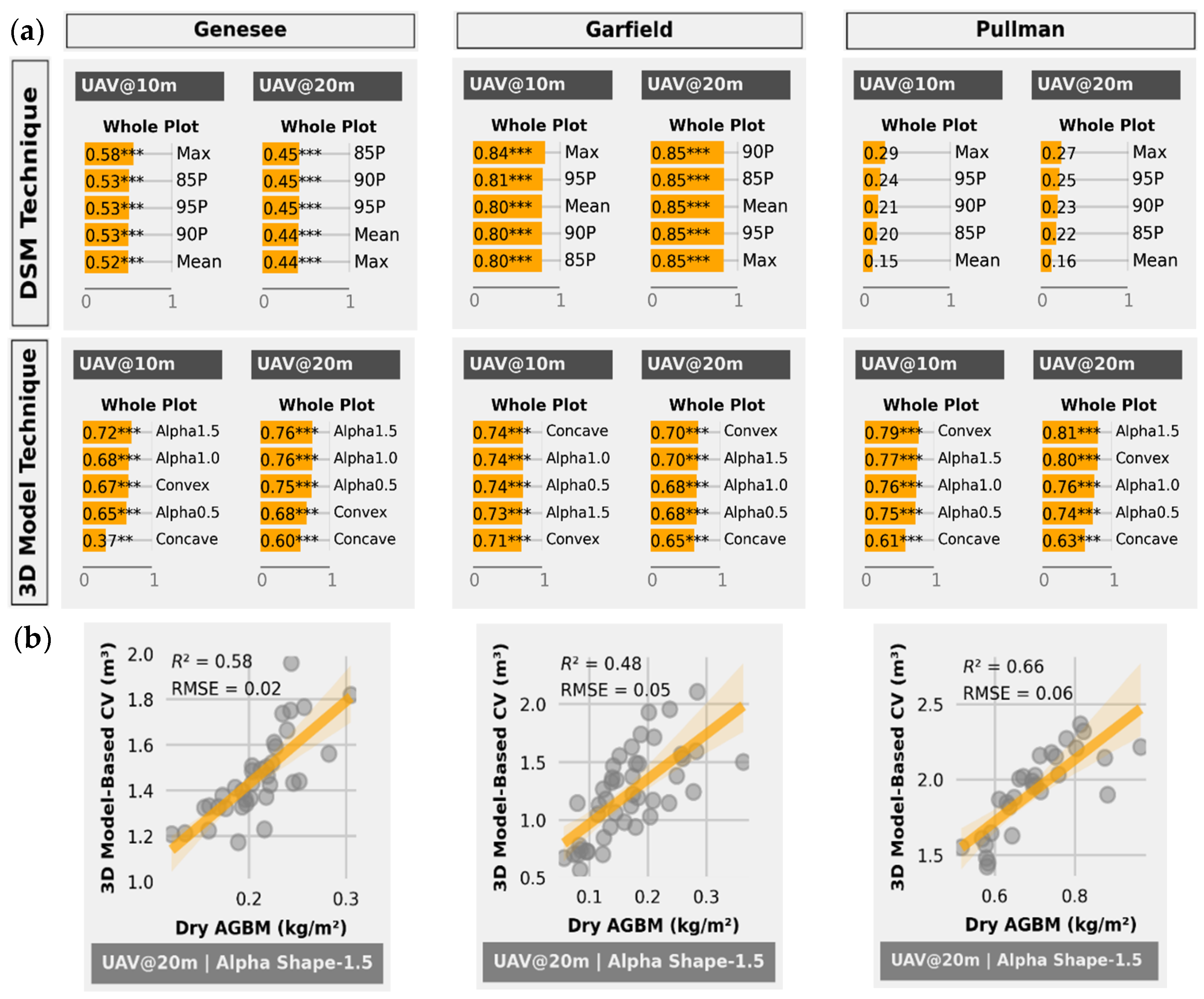
3.3. Fresh AGBM Estimation
3.3.1. Vegetation-Index-Based Technique
3.3.2. DSM-Based Technique
3.3.3. Three-Dimensional Model-Based Technique
3.4. Dry AGBM Estimation
4. Summary and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fraser, M.D.; Fychan, R.; Jones, R. The effect of harvest date and inoculation on the yield, fermentation characteristics and feeding value of forage pea and field bean silages. Grass Forage Sci. 2001, 56, 218–230. [Google Scholar] [CrossRef]
- Chen, C.; Miller, P.; Muehlbauer, F.; Neill, K.; Wichman, D.; McPhee, K. Winter pea and lentil response to seeding date and micro-and macro-environments. Agron. J. 2006, 98, 1655–1663. [Google Scholar] [CrossRef] [Green Version]
- Clark, A. Managing Cover Crops Profitably, 3rd ed.; Sustainable Agriculture Network: Beltsville, MD, USA, 2008. [Google Scholar]
- Tulbek, M.C.; Lam, R.S.H.; Wang, Y.C.; Asavajaru, P.; Lam, A. Pea: A Sustainable Vegetable Protein Crop. In Sustainable Protein Sources; Nadathur, S.R., Wanasundara, J.P.D., Scanlin, L., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 145–164. [Google Scholar] [CrossRef]
- Steinfeld, H.; Gerder, P.; Wassenaar, T.D.; Castel, V.; Rosales, M.; de Haan, C. Livestock’s Long Shadow: Environmental Issues and Options; Food and Agriculture Organization: Rome, Italy, 2006. [Google Scholar]
- Gerber, P.J.; Steinfeld, H.; Henderson, B.; Mottet, A.; Opio, C.; Dijkman, J.; Falcucci, A.; Tempio, G. Tackling Climate Change through Livestock: A Global Assessment of Emissions and Mitigation Opportunities; Food and Agriculture Organization: Rome, Italy, 2013. [Google Scholar]
- Annicchiarico, P.; Russi, L.; Romani, M.; Pecetti, L.; Nazzicari, N. Farmer-participatory vs. conventional market-oriented breeding of inbred crops using phenotypic and genome-enabled approaches: A pea case study. Field Crops Res. 2019, 232, 30–39. [Google Scholar] [CrossRef]
- Insua, J.R.; Utsumi, S.A.; Basso, B. Estimation of spatial and temporal variability of pasture growth and digestibility in grazing rotations coupling unmanned aerial vehicle (UAV) with crop simulation models. PLoS ONE 2019, 14, e0212773. [Google Scholar] [CrossRef] [Green Version]
- Ligoski, B.; Gonçalves, L.F.; Claudio, F.L.; Alves, E.M.; Krüger, A.M.; Bizzuti, B.E.; Lima, P.D.M.T.; Abdalla, A.L.; Paim, T.D.P. Silage of intercropping corn, palisade grass, and pigeon pea increases protein content and reduces in vitro methane production. Agronomy 2020, 10, 1784. [Google Scholar] [CrossRef]
- Quirós Vargas, J.J.; Zhang, C.; Smitchger, J.A.; McGee, R.J.; Sankaran, S. Phenotyping of plant biomass and performance traits using remote sensing techniques in pea (Pisum sativum, L.). Sensors 2019, 19, 2031. [Google Scholar] [CrossRef] [Green Version]
- Furbank, R.T.; Tester, M. Phenomics–technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef] [Green Version]
- Maesano, M.; Khoury, S.; Nakhle, F.; Firrincieli, A.; Gay, A.; Tauro, F.; Harfouche, A. UAV-based LiDAR for high-throughput determination of plant height and above-ground biomass of the bioenergy grass arundo donax. Remote Sens. 2020, 12, 3464. [Google Scholar] [CrossRef]
- Jung, J.; Maeda, M.; Chang, A.; Bhandari, M.; Ashapure, A.; Landivar-Bowles, J. The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr. Opin. Biotechnol. 2021, 70, 15–22. [Google Scholar] [CrossRef]
- Li, D.; Quan, C.; Song, Z.; Li, X.; Yu, G.; Li, C.; Muhammad, A. High-throughput plant phenotyping platform (HT3P) as a novel tool for estimating agronomic traits from the lab to the field. Front. Bioeng. Biotechnol. 2021, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.V.; Sangjan, W.; Selvaraj, M.G.; McGee, R.J.; Sankaran, S. Effect of the solar zenith angles at different latitudes on estimated crop vegetation indices. Drones 2021, 5, 80. [Google Scholar] [CrossRef]
- Zhang, C.; Serra, S.; Quirós Vargas, J.; Sangjan, W.; Musacchi, S.; Sankaran, S. Non-invasive sensing techniques to phenotype multiple apple tree architectures. Inf. Process. Agric. 2021; in press. [Google Scholar] [CrossRef]
- De Jesus Colwell, F.; Souter, J.; Bryan, G.J.; Compton, L.J.; Boonham, N.; Prashar, A. Development and validation of methodology for estimating potato canopy structure for field crop phenotyping and improved breeding. Front. Plant Sci. 2021, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Carroll, M.E.; Singh, A.; Swetnam, T.L.; Merchant, N.; Sarkar, S.; Singh, A.K.; Ganapathysubramanian, B. UAS-based plant phenotyping for research and breeding applications. Plant Phenom. 2021, 2021, 9840192. [Google Scholar] [CrossRef]
- Sangjan, W.; Sankaran, S. Phenotyping architecture traits of tree species using remote sensing techniques. Trans. ASABE 2021, 64, 1611–1624. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Tian, T.; Yin, J. A review of unmanned aerial vehicle low-altitude remote sensing (UAV-LARS) use in agricultural monitoring in China. Remote Sens. 2021, 13, 1221. [Google Scholar] [CrossRef]
- Tilly, N.; Aasen, H.; Bareth, G. Fusion of plant height and vegetation indices for the estimation of barley biomass. Remote Sens. 2015, 7, 11449–11480. [Google Scholar] [CrossRef] [Green Version]
- Wengert, M.; Piepho, H.P.; Astor, T.; Graß, R.; Wijesingha, J.; Wachendorf, M. Assessing spatial variability of barley whole crop biomass yield and leaf area index in silvoarable agroforestry systems using UAV-borne remote sensing. Remote Sens. 2021, 13, 2751. [Google Scholar] [CrossRef]
- Sankaran, S.; Zhou, J.; Khot, L.R.; Trapp, J.J.; Mndolwa, E.; Miklas, P.N. High-throughput field phenotyping in dry bean using small unmanned aerial vehicle based multispectral imagery. Comput. Electron. Agric. 2018, 151, 84–92. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Zhou, M.; Li, D.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Improved estimation of rice aboveground biomass combining textural and spectral analysis of UAV imagery. Precis. Agric. 2019, 20, 611–629. [Google Scholar] [CrossRef]
- Wan, L.; Zhang, J.; Dong, X.; Du, X.; Zhu, J.; Sun, D.; Liu, Y.; He, Y.; Cen, H. Unmanned aerial vehicle-based field phenotyping of crop biomass using growth traits retrieved from PROSAIL model. Comput. Electron. Agric. 2021, 187, 106304. [Google Scholar] [CrossRef]
- Yue, J.; Yang, G.; Tian, Q.; Feng, H.; Xu, K.; Zhou, C. Estimate of winter-wheat above-ground biomass based on UAV ultrahigh-ground-resolution image textures and vegetation indices. ISPRS J. Photogramm. Remote Sens. 2019, 150, 226–244. [Google Scholar] [CrossRef]
- Roy Choudhury, M.; Das, S.; Christopher, J.; Apan, A.; Chapman, S.; Menzies, N.W.; Dang, Y.P. Improving biomass and grain yield prediction of wheat genotypes on sodic soil using integrated high-resolution multispectral, hyperspectral, 3D point cloud, and machine learning techniques. Remote Sens. 2021, 13, 3482. [Google Scholar] [CrossRef]
- Schönberger, J.L.; Frahm, J. Structure-from-Motion revisited. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 4104–4113. [Google Scholar]
- Thompson, A.L.; Thorp, K.R.; Conley, M.M.; Elshikha, D.M.; French, A.N.; Andrade-Sanchez, P.; Pauli, D. Comparing nadir and multi-angle view sensor technologies for measuring in-field plant height of upland cotton. Remote Sens. 2019, 11, 700. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Zhang, L.; Zhang, H.; Han, W.; Peng, X. Estimating above-ground biomass of maize using features derived from UAV-based RGB imagery. Remote Sens. 2019, 11, 1261. [Google Scholar] [CrossRef] [Green Version]
- Gilliot, J.M.; Michelin, J.; Hadjard, D.; Houot, S. An accurate method for predicting spatial variability of maize yield from UAV-based plant height estimation: A tool for monitoring agronomic field experiments. Precis. Agric. 2021, 22, 897–921. [Google Scholar] [CrossRef]
- Acorsi, M.G.; das Dores Abati Miranda, F.; Martello, M.; Smaniotto, D.A.; Sartor, L.R. Estimating biomass of black oat using UAV-based RGB imaging. Agronomy 2019, 9, 344. [Google Scholar] [CrossRef] [Green Version]
- Peprah, C.O.; Yamashita, M.; Yamaguchi, T.; Sekino, R.; Takano, K.; Katsura, K. Spatio-temporal estimation of biomass growth in rice using canopy surface model from unmanned aerial vehicle images. Remote Sens. 2021, 13, 2388. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Maimaitiyiming, M.; Hartling, S.; Peterson, K.T.; Maw, M.J.W.; Shakoor, N.; Mockler, T.; Fritschi, F.B. Vegetation index weighted canopy volume model (CVMVI) for soybean biomass estimation from unmanned aerial system-based RGB imagery. ISPRS J. Photogramm. Remote Sens. 2019, 151, 27–41. [Google Scholar] [CrossRef]
- Toda, Y.; Kaga, A.; Kajiya-Kanegae, H.; Hattori, T.; Yamaoka, S.; Okamoto, M.; Tsujimoto, H.; Iwata, H. Genomic prediction modeling of soybean biomass using UAV-based remote sensing and longitudinal model parameters. Plant Genome 2021, 14, e20157. [Google Scholar] [CrossRef]
- Yue, J.; Yang, G.; Li, C.; Li, Z.; Wang, Y.; Feng, H.; Xu, B. Estimation of winter wheat above-ground biomass using unmanned aerial vehicle-based snapshot hyperspectral sensor and crop height improved models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, B.P.; Spangenberg, G.; Kant, S. Fusion of spectral and structural information from aerial images for improved biomass estimation. Remote Sens. 2020, 12, 3164. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Matese, A. Evaluation of novel precision viticulture tool for canopy biomass estimation and missing plant detection based on 2.5D and 3D approaches using RGB images acquired by UAV platform. Plant Methods 2020, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Reddersen, B.; Fricke, T.; Wachendorf, M. A multi-sensor approach for predicting biomass of extensively managed grassland. Comput. Electron. Agric. 2014, 109, 247–260. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating biomass of barley using crop surface models (CSMs) derived from UAV-based RGB imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef] [Green Version]
- Rogers, S.R.; Manning, I.; Livingstone, W. Comparing the spatial accuracy of digital surface models from four unoccupied aerial systems: Photogrammetry versus LiDAR. Remote Sens. 2020, 12, 2806. [Google Scholar] [CrossRef]
- Dong, X.; Kim, W.Y.; Lee, K.H. Drone-based three-dimensional photogrammetry and concave hull by slices algorithm for apple tree volume mapping. J. Biosyst. Eng. 2021, 46, 474–484. [Google Scholar] [CrossRef]
- Kothawade, G.S.; Chandel, A.K.; Schrader, M.J.; Rathnayake, A.P.; Khot, L.R. High throughput canopy characterization of a commercial apple orchard using aerial RGB imagery. In Proceedings of the 2021 IEEE International Workshop on Metrology for Agriculture and Forestry (MetroAgriFor), Trento-Bolzano, Italy, 3–5 November 2021; pp. 177–181. [Google Scholar] [CrossRef]
- Qi, Y.; Dong, X.; Chen, P.; Lee, K.H.; Lan, Y.; Lu, X.; Jia, R.; Deng, J.; Zhang, Y. Canopy volume extraction of Citrus reticulate Blanco cv. Shatangju trees using UAV image-based point cloud deep learning. Remote Sens. 2021, 13, 3437. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Jiang, Q.; Fang, S.; Peng, Y.; Gong, Y.; Zhu, R.; Wu, X.; Ma, Y.; Duan, B.; Liu, J. UAV-based biomass estimation for rice-combining spectral, TIN-based structural and meteorological features. Remote Sens. 2019, 11, 890. [Google Scholar] [CrossRef] [Green Version]
- Tefera, A.T.; Banerjee, B.P.; Pandey, B.R.; James, L.; Puri, R.R.; Cooray, O.; Marsh, J.; Richards, M.; Kant, S.; Fitzgerald, G.J.; et al. Estimating early season growth and biomass of field pea for selection of divergent ideotypes using proximal sensing. Field Crops Res. 2022, 277, 108407. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Remote sensing of chlorophyll concentration in higher plant leaves. Adv. Space Res. 1998, 22, 689–692. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Quantitative estimation of chlorophyll-a using reflectance spectra: Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- McFeeters, S.K. The use of the normalized difference water index (NDWI) in the delineation of open water features. Int. J. Remote Sens. 1996, 17, 1425–1432. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Duarte, L.; Silva, P.; Teodoro, A.C. Development of a QGIS plugin to obtain parameters and elements of plantation trees and vineyards with aerial photographs. ISPRS Int. J. Geo-Inf. 2018, 7, 109. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Luo, B.; Hong, H.; Su, X.; Wang, Y.; Liu, J.; Wang, C.; Zhang, J.; Wei, L. Object detection based on global-local saliency constraint in aerial images. Remote Sens. 2020, 12, 1435. [Google Scholar] [CrossRef]
- Jian, M.; Wang, J.; Yu, H.; Wang, G.; Meng, X.; Yang, L.; Dong, J.; Yin, Y. Visual saliency detection by integrating spatial position prior of object with background cues. Expert Syst. Appl. 2021, 168, 114219. [Google Scholar] [CrossRef]
- Huyan, L.; Bai, Y.; Li, Y.; Jiang, D.; Zhang, Y.; Zhou, Q.; Wei, J.; Liu, J.; Zhang, Y.; Cui, T. A Lightweight Object Detection Framework for Remote Sensing Images. Remote Sens. 2021, 13, 683. [Google Scholar] [CrossRef]
- Chakraborty, M.; Khot, L.R.; Sankaran, S.; Jacoby, P.W. Evaluation of mobile 3D light detection and ranging based canopy mapping system for tree fruit crops. Comput. Electron. Agric. 2019, 158, 284–293. [Google Scholar] [CrossRef]
- Saarinen, N.; Calders, K.; Kankare, V.; Yrttimaa, T.; Junttila, S.; Luoma, V.; Huuskonen, S.; Hynynen, J.; Verbeeck, H. Understanding 3D structural complexity of individual Scots pine trees with different management history. Ecol. Evol. 2021, 11, 2561–2572. [Google Scholar] [CrossRef]
- Yan, Z.; Liu, R.; Cheng, L.; Zhou, X.; Ruan, X.; Xiao, Y. A concave hull methodology for calculating the crown volume of individual trees based on vehicle-borne LiDAR data. Remote Sens. 2019, 11, 623. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Hassanzadeh, A.; Kikkert, J.; Pethybridge, S.J.; van Aardt, J. Comparison of UAS-based structure-from-motion and LiDAR for structural characterization of short broadacre crops. Remote Sens. 2021, 13, 3975. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Paterson, A.H.; Sun, S.; Xu, R.; Robertson, J. Quantitative analysis of cotton canopy size in field conditions using a consumer-grade RGB-D camera. Front. Plant Sci. 2018, 8, 2233. [Google Scholar] [CrossRef] [Green Version]
- Fonstad, M.A.; Dietrich, J.T.; Courville, B.C.; Jensen, J.L.; Carbonneau, P.E. Topographic structure from motion: A new development in photogrammetric measurement. Earth Surf. Processes Landf. 2013, 38, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Jensen, J.L.; Mathews, A.J. Assessment of image-based point cloud products to generate a bare earth surface and estimate canopy heights in a woodland ecosystem. Remote Sens. 2016, 8, 50. [Google Scholar] [CrossRef] [Green Version]
- Salach, A.; Bakuła, K.; Pilarska, M.; Ostrowski, W.; Górski, K.; Kurczyński, Z. Accuracy assessment of point clouds from LiDAR and dense image matching acquired using the UAV platform for DTM creation. ISPRS Int. J. Geo-Inf. 2018, 7, 342. [Google Scholar] [CrossRef] [Green Version]
- Kanke, Y.; Tubana, B.; Dalen, M.; Harrell, D. Evaluation of red and red-edge reflectance-based vegetation indices for rice biomass and grain yield prediction models in paddy fields. Precis. Agric. 2016, 17, 507–530. [Google Scholar] [CrossRef]
- Cheng, T.; Song, R.; Li, D.; Zhou, K.; Zheng, H.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Spectroscopic estimation of biomass in canopy components of paddy rice using dry matter and chlorophyll indices. Remote Sens. 2017, 9, 319. [Google Scholar] [CrossRef] [Green Version]
- Peña, J.M.; Torres-Sánchez, J.; Serrano-Pérez, A.; De Castro, A.I.; López-Granados, F. Quantifying efficacy and limits of unmanned aerial vehicle (UAV) technology for weed seedling detection as affected by sensor resolution. Sensors 2015, 15, 5609–5626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, X.; Liu, S.; Baret, F.; Hemerlé, M.; Comar, A. Estimates of plant density of wheat crops at emergence from very low altitude UAV imagery. Remote Sens. Environ. 2017, 198, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Candón, D.; De Castro, A.I.; López-Granados, F. Assessing the accuracy of mosaics from unmanned aerial vehicle (UAV) imagery for precision agriculture purposes in wheat. Precis. Agric. 2014, 15, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Mesas-Carrascosa, F.J.; Torres-Sánchez, J.; Clavero-Rumbao, I.; García-Ferrer, A.; Peña, J.M.; Borra-Serrano, I.; López-Granados, F. Assessing optimal flight parameters for generating accurate multispectral orthomosaicks by UAV to support site-specific crop management. Remote Sens. 2015, 7, 12793–12814. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, C.; Takeda, F.; Kramer, E.A.; Ashrafi, H.; Hunter, J. 3D point cloud data to quantitatively characterize size and shape of shrub crops. Hortic. Res. 2019, 6, 43. [Google Scholar] [CrossRef] [Green Version]
- Kuželka, K.; Slavík, M.; Surový, P. Very high density point clouds from UAV laser scanning for automatic tree stem detection and direct diameter measurement. Remote Sens. 2020, 12, 1236. [Google Scholar] [CrossRef] [Green Version]
- Moreira, B.M.; Goyanes, G.; Pina, P.; Vassilev, O.; Heleno, S. Assessment of the influence of survey design and processing choices on the accuracy of tree diameter at breast height (DBH) measurements using UAV-based photogrammetry. Drones 2021, 5, 43. [Google Scholar] [CrossRef]



| Season | Field Location | GrowthStage | UAV’s Flight—Image Acquisition | Ground Reference Data | ||||
|---|---|---|---|---|---|---|---|---|
| Date | Flight Altitude (m) | Camera | GSD 5 (cm/pixel) | Fresh AGBM 6 | Dry AGBM | |||
| 2019 | Genesee | F50 1 | 18 June | 10 | RGB 3 | 0.21 | 19 June | - |
| 20 | RGB | 0.50 | ||||||
| 20 | MS 4 | 1.34 | ||||||
| PM 2 | 29 July | 10 | RGB | 0.19 | - | 31 July | ||
| 20 | RGB | 0.52 | ||||||
| Garfield | F50 | 24 June | 10 | RGB | 0.22 | 25 June | - | |
| 20 | RGB | 0.51 | ||||||
| 20 | MS | 1.19 | ||||||
| PM | 29 July | 10 | RGB | 0.21 | - | 31 July | ||
| 20 | RGB | 0.52 | ||||||
| 2020 | Pullman | F50 | 10 June | 10 | RGB | 0.25 | 12 June | - |
| 20 | RGB | 0.51 | ||||||
| 20 | MS | 1.26 | ||||||
| PM | 27 July | 10 | RGB | 0.29 | - | 27 July | ||
| 20 | RGB | 0.55 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sangjan, W.; McGee, R.J.; Sankaran, S. Optimization of UAV-Based Imaging and Image Processing Orthomosaic and Point Cloud Approaches for Estimating Biomass in a Forage Crop. Remote Sens. 2022, 14, 2396. https://doi.org/10.3390/rs14102396
Sangjan W, McGee RJ, Sankaran S. Optimization of UAV-Based Imaging and Image Processing Orthomosaic and Point Cloud Approaches for Estimating Biomass in a Forage Crop. Remote Sensing. 2022; 14(10):2396. https://doi.org/10.3390/rs14102396
Chicago/Turabian StyleSangjan, Worasit, Rebecca J. McGee, and Sindhuja Sankaran. 2022. "Optimization of UAV-Based Imaging and Image Processing Orthomosaic and Point Cloud Approaches for Estimating Biomass in a Forage Crop" Remote Sensing 14, no. 10: 2396. https://doi.org/10.3390/rs14102396
APA StyleSangjan, W., McGee, R. J., & Sankaran, S. (2022). Optimization of UAV-Based Imaging and Image Processing Orthomosaic and Point Cloud Approaches for Estimating Biomass in a Forage Crop. Remote Sensing, 14(10), 2396. https://doi.org/10.3390/rs14102396