Evaluating the Effectiveness of Machine Learning and Deep Learning Models Combined Time-Series Satellite Data for Multiple Crop Types Classification over a Large-Scale Region
Abstract
:1. Introduction
2. Materials and Methods
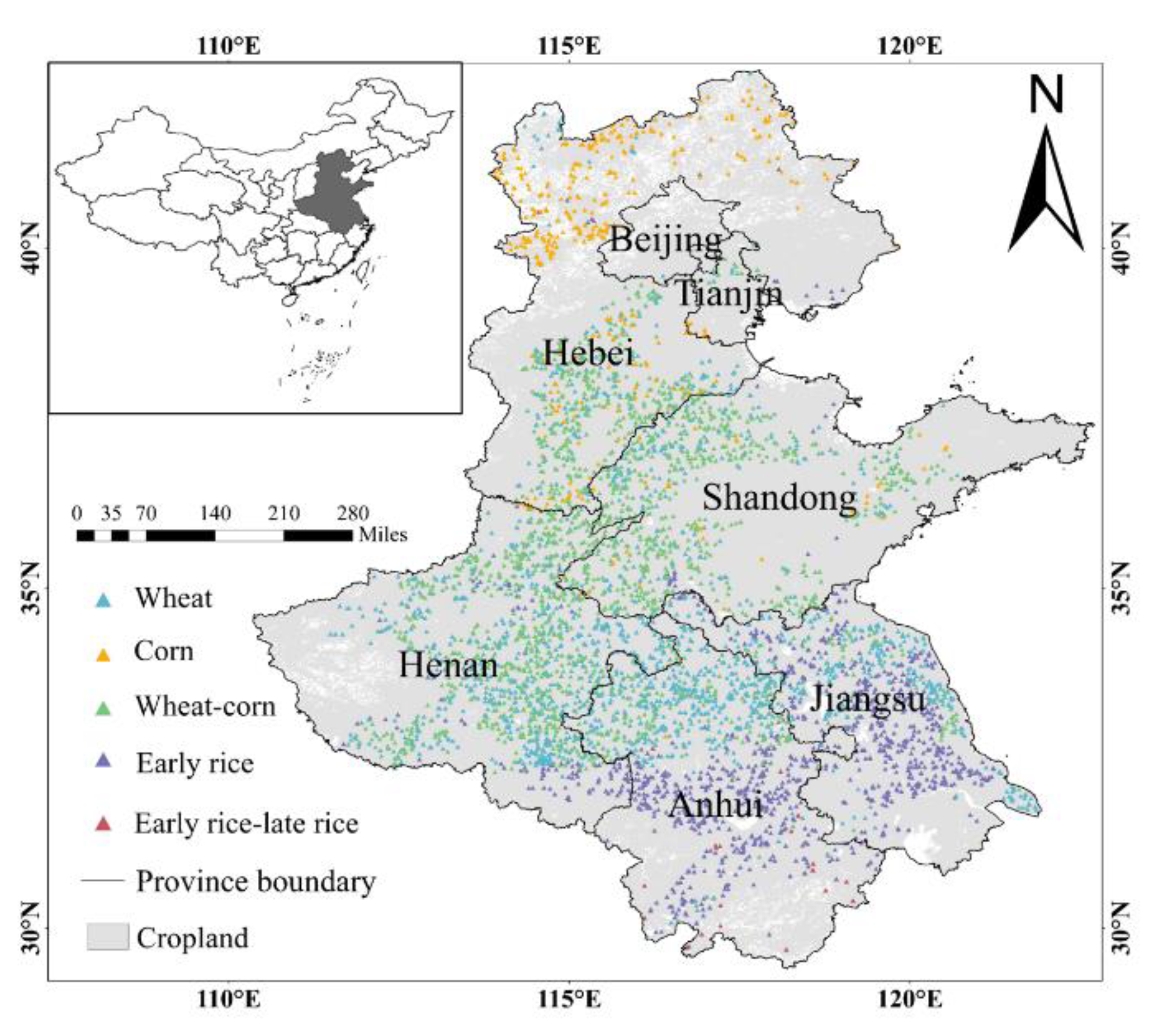
2.1. Study Area
2.2. Data
2.2.1. MODIS Data
2.2.2. Sample Data
2.3. Method
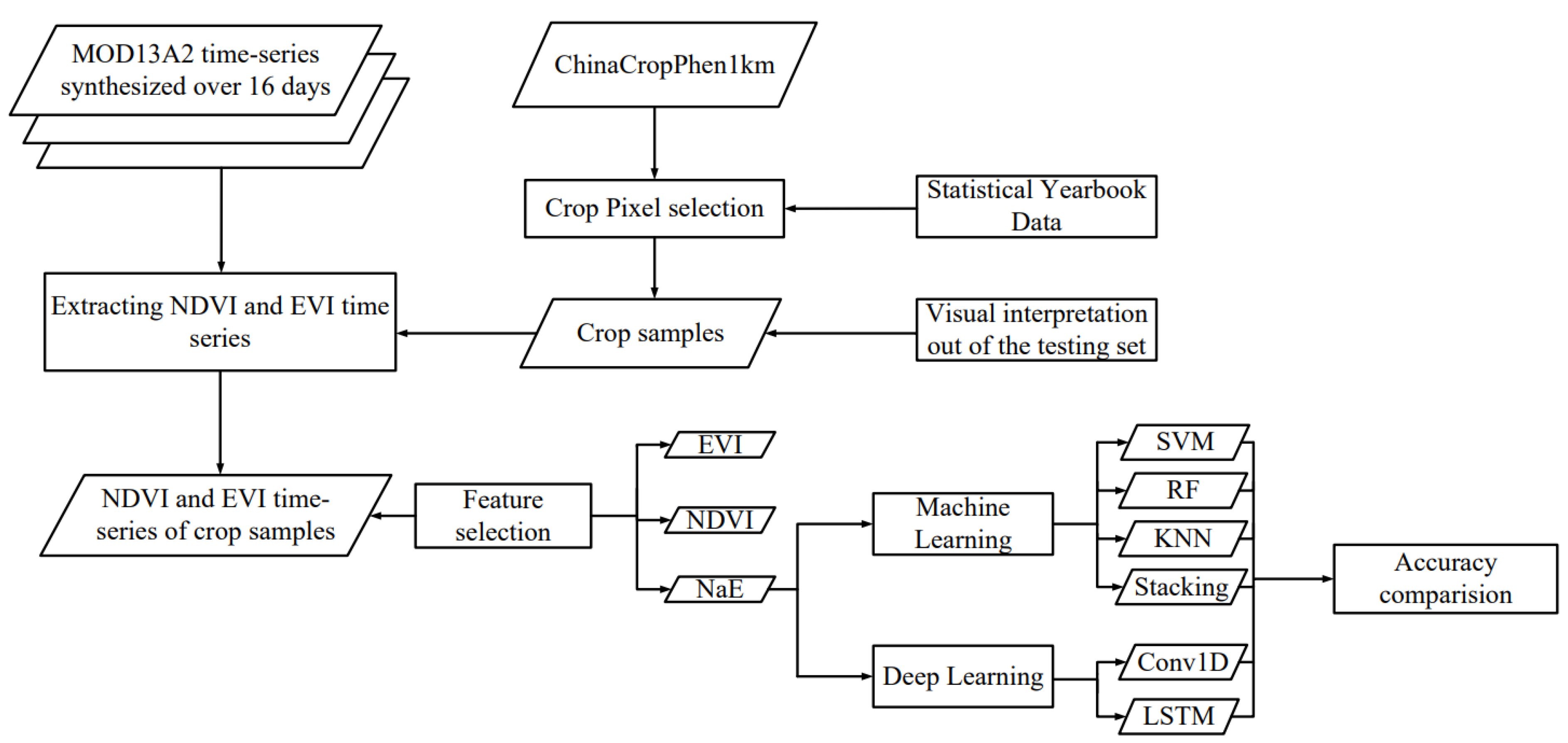
2.3.1. Data Acquisition and Preparation
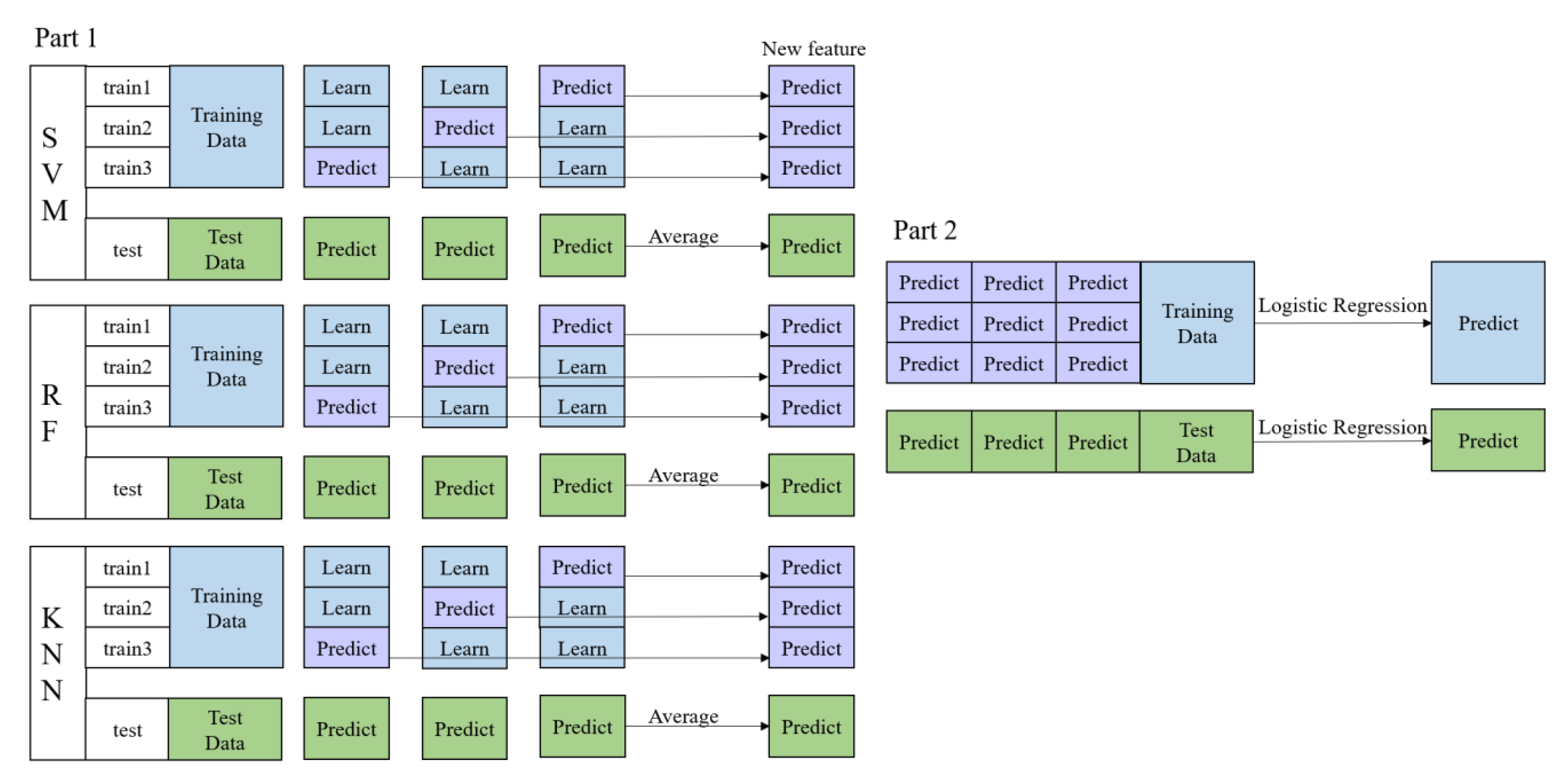
2.3.2. Machine Learning Classifiers
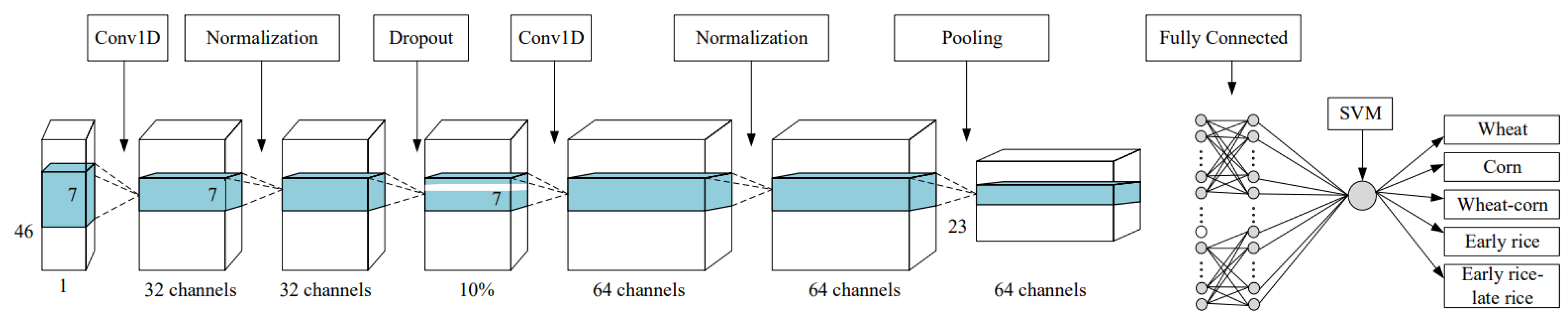
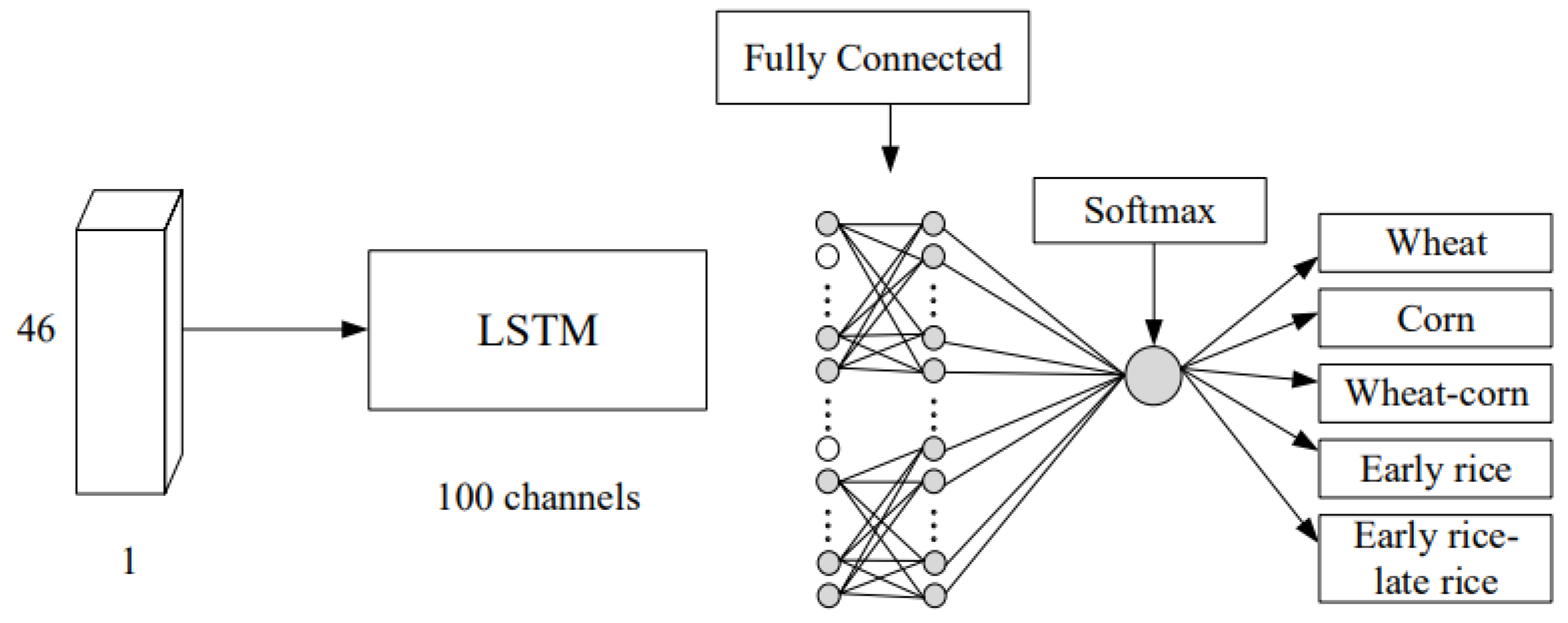
2.3.3. Deep Learning Classifiers
2.4. Accuracy Evaluation
3. Results
3.1. Feature Selection
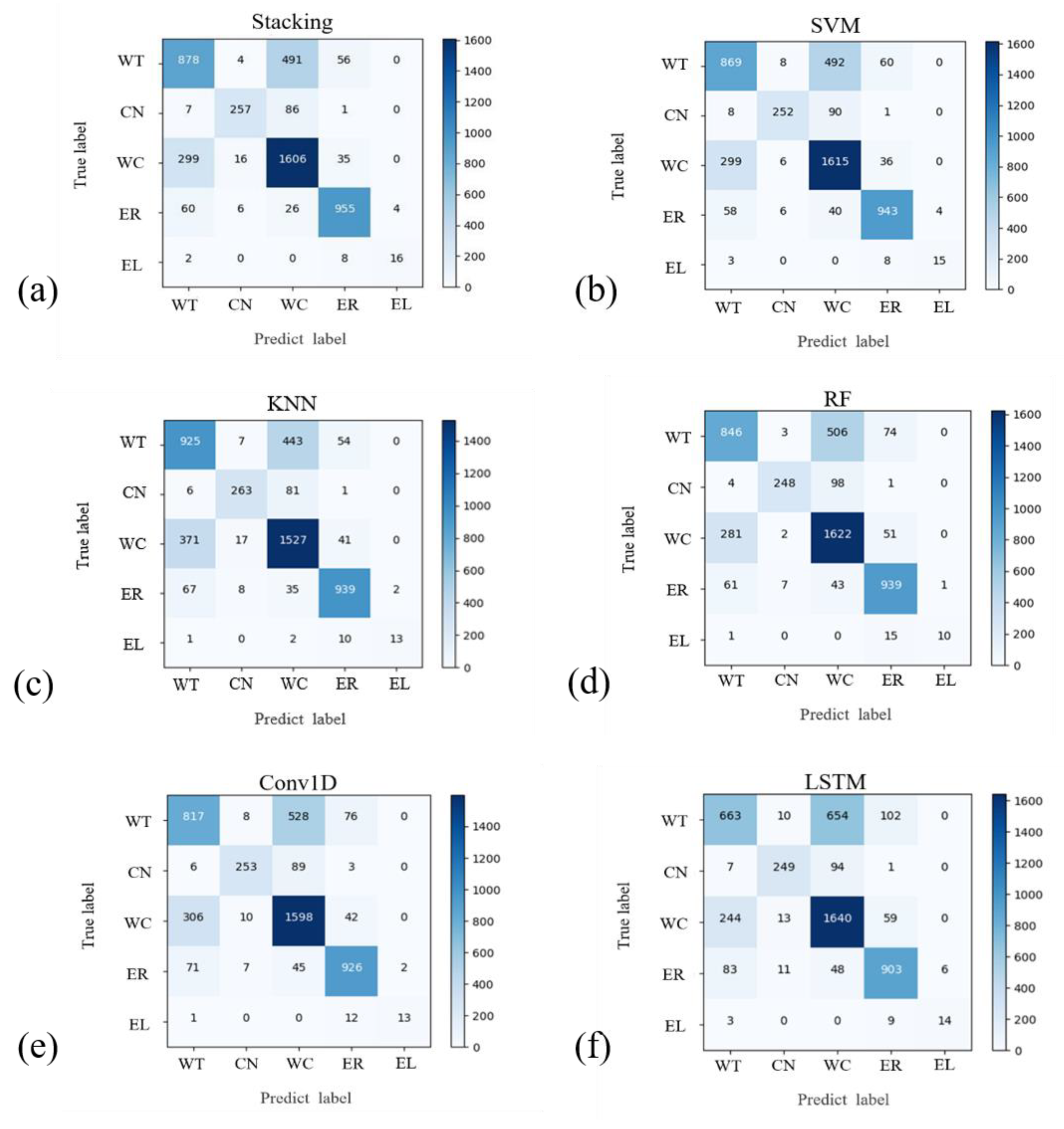
3.2. Machine Learning and Deep Learning Classification Results
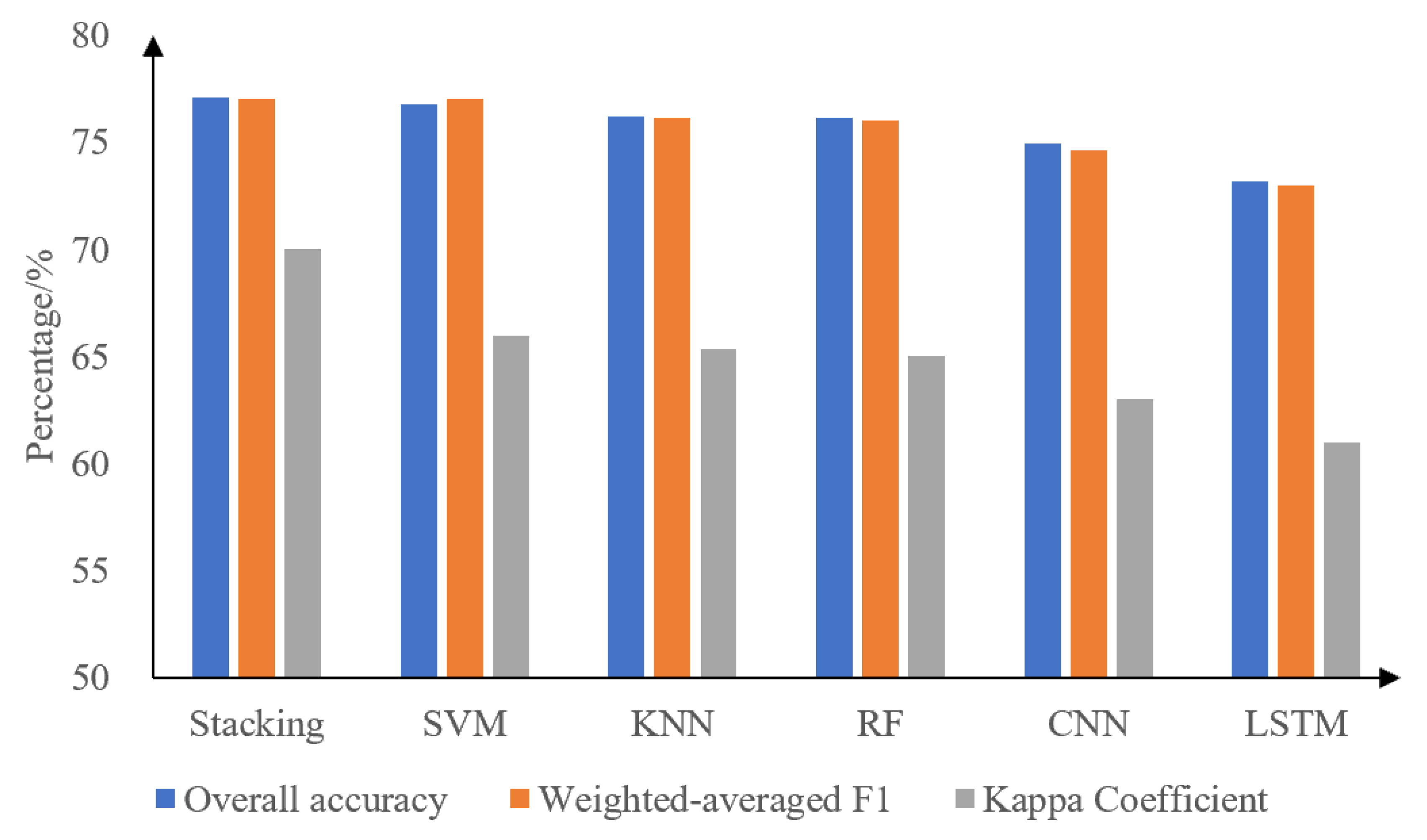
3.3. Comparison of Machine Learning and Deep Learning Results
4. Discussion
4.1. Effectiveness of the Used Models
4.2. Potential Refinements
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Xun, L.; Zhang, J.; Cao, D.; Wang, J.; Zhang, S.; Yao, F. Mapping Cotton Cultivated Area Combining Remote Sensing with a Fused Representation-Based Classification Algorithm. Comput. Electron. Agric. 2021, 181, 105940. [Google Scholar] [CrossRef]
- Xun, L.; Zhang, J.; Cao, D.; Yang, S.; Yao, F. A Novel Cotton Mapping Index Combining Sentinel-1 SAR and Sentinel-2 Multispectral Imagery. ISPRS J. Photogramm. Remote Sens. 2021, 181, 148–166. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, J.; Deng, F.; Zhang, S.; Zhang, D.; Xun, L.; Ji, M.; Feng, Q. Superpixel-Based Regional-Scale Grassland Community Classification Using Genetic Programming with Sentinel-1 SAR and Sentinel-2 Multispectral Images. Remote Sens. 2021, 13, 67. [Google Scholar] [CrossRef]
- Xun, L.; Zhang, J.; Cao, D.; Zhang, S.; Yao, F. Crop Area Identification Based on Time Series EVI2 and Sparse Representation Approach: A Case Study in Shandong Province, China. IEEE Access 2019, 7, 157513–157523. [Google Scholar] [CrossRef]
- Felegari, S.; Sharifi, A.; Moravej, K.; Amin, M.; Golchin, A.; Muzirafuti, A.; Tariq, A.; Zhao, N. Integration of Sentinel 1 and Sentinel 2 Satellite Images for Crop Mapping. Appl. Sci. 2021, 11, 104. [Google Scholar] [CrossRef]
- Valero, S.; Arnaud, L.; Planells, M.; Ceschia, E. Synergy of Sentinel-1 and Sentinel-2 Imagery for Early Seasonal Agricultural Crop Mapping. Remote Sens. 2021, 13, 4891. [Google Scholar] [CrossRef]
- Eerens, H.; Haesen, D.; Rembold, F.; Urbano, F.; Tote, C.; Bydekerke, L. Image Time Series Processing for Agriculture Monitoring. Environ. Model. Softw. 2014, 53, 154–162. [Google Scholar] [CrossRef]
- Zhang, G.; Xiao, X.; Dong, J.; Kou, W.; Jin, C.; Qin, Y.; Zhou, Y.; Wang, J.; Menarguez, M.A.; Biradar, C. Mapping Paddy Rice Planting Areas through Time Series Analysis of MODIS Land Surface Temperature and Vegetation Index Data. ISPRS J. Photogramm. Remote Sens. 2015, 106, 157–171. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, S.; Feng, L.; Zhang, J.; Deng, F. Mapping Maize Cultivated Area Combining MODIS EVI Time Series and the Spatial Variations of Phenology over Huanghuaihai Plain. Appl. Sci. 2020, 10, 2667. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, L.; Yao, F. Improved Maize Cultivated Area Estimation over a Large Scale Combining MODIS-EVI Time Series Data and Crop Phenological Information. ISPRS J. Photogramm. Remote Sens. 2014, 94, 102–113. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, J.; Deng, F.; Zhang, S.; Zhang, D.; Xun, L.; Javed, T.; Liu, G.; Liu, D.; Ji, M. Fusion of Gf and Modis Data for Regional-Scale Grassland Community Classification with Evi2 Time-Series and Phenological Features. Remote Sens. 2021, 13, 835. [Google Scholar] [CrossRef]
- Qiu, B.; Feng, M.; Tang, Z. A Simple Smoother Based on Continuous Wavelet Transform: Comparative Evaluation Based on the Fidelity, Smoothness and Efficiency in Phenological Estimation. Int. J. Appl. Earth Obs. Geoinf. 2016, 47, 91–101. [Google Scholar] [CrossRef]
- Potgieter, A.B.; Apan, A.; Hammer, G.; Dunn, P. Early-Season Crop Area Estimates for Winter Crops in NE Australia Using MODIS Satellite Imagery. ISPRS J. Photogramm. Remote Sens. 2010, 65, 380–387. [Google Scholar] [CrossRef]
- Sakamoto, T.; Yokozawa, M.; Toritani, H.; Shibayama, M.; Ishitsuka, N.; Ohno, H. A Crop Phenology Detection Method Using Time-Series MODIS Data. Remote Sens. Environ. 2005, 96, 366–374. [Google Scholar] [CrossRef]
- Funk, C.; Budde, M.E. Phenologically-Tuned MODIS NDVI-Based Production Anomaly Estimates for Zimbabwe. Remote Sens. Environ. 2009, 113, 115–125. [Google Scholar] [CrossRef]
- Jönsson, P.; Eklundh, L. Seasonality Extraction by Function Fitting to Time-Series of Satellite Sensor Data. IEEE Trans. Geosci. Remote Sens. 2002, 40, 1824–1832. [Google Scholar] [CrossRef]
- Jönsson, P.; Eklundh, L. TIMESAT—A Program for Analyzing Time-Series of Satellite Sensor Data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, T.; Van Nguyen, N.; Ohno, H.; Ishitsuka, N.; Yokozawa, M. Spatio-Temporal Distribution of Rice Phenology and Cropping Systems in the Mekong Delta with Special Reference to the Seasonal Water Flow of the Mekong and Bassac Rivers. Remote Sens. Environ. 2006, 100, 1–16. [Google Scholar] [CrossRef]
- Galford, G.L.; Mustard, J.F.; Melillo, J.; Gendrin, A.; Cerri, C.C.; Cerri, C.E.P. Wavelet Analysis of MODIS Time Series to Detect Expansion and Intensification of Row-Crop Agriculture in Brazil. Remote Sens. Environ. 2008, 112, 576–587. [Google Scholar] [CrossRef]
- Shao, Y.; Lunetta, R.S.; Wheeler, B.; Iiames, J.S.; Campbell, J.B. An Evaluation of Time-Series Smoothing Algorithms for Land-Cover Classifications Using MODIS-NDVI Multi-Temporal Data. Remote Sens. Environ. 2016, 174, 258–265. [Google Scholar] [CrossRef]
- Chen, J.; Jönsson, P.; Tamura, M.; Gu, Z.; Matsushita, B.; Eklundh, L. A Simple Method for Reconstructing a High-Quality NDVI Time-Series Data Set Based on the Savitzky-Golay Filter. Remote Sens. Environ. 2004, 91, 332–344. [Google Scholar] [CrossRef]
- Olsson, L. Fourier Series for Analysis of Temporal Sequences of Satellite Sensor Imagery. Int. J. Remote Sens. 1994, 15, 3735–3741. [Google Scholar] [CrossRef]
- Verhoef, W.; Menenti, M.; Azzali, S. Cover a Colour Composite of NOAA-AVHRR-NDVI Based on Time Series Analysis (1981–1992). Int. J. Remote Sens. 1996, 17, 231–235. [Google Scholar] [CrossRef]
- He, T.; Xie, C.; Liu, Q.; Guan, S.; Liu, G. Evaluation and Comparison of Random Forest and A-LSTM Networks for Large-Scale Winter Wheat Identification. Remote Sens. 2019, 11, 1665. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Hu, L.; Zhou, H. Deep Learning Based Multi-Temporal Crop Classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Guidici, D.; Clark, M.L. One-Dimensional Convolutional Neural Network Land-Cover Classification of Multi-Seasonal Hyperspectral Imagery in the San Francisco Bay Area, California. Remote Sens. 2017, 9, 629. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Shukla, S.; Sharma, K.K.; Singh, K.K.; Akbari, A.S. Classification of Land Cover and Land Use Using Deep Learning. In Machine Vision and Augmented Intelligence—Theory and Applications; Springer: Berlin/Heidelberg, Germany, 2021; pp. 321–327. [Google Scholar] [CrossRef]
- Bermúdez, J.D.; Achanccaray, P.; Sanches, I.D.; Cue, L.; Happ, P.; Feitosa, R.Q. Evaluation of Recurrent Neural Networks for Crop Recognition from Multitemporal Remote Sensing Images. An. Do Congr. Bras. De Cartogr. E XXVI Expo. 2017, 2017, 800–804. [Google Scholar]
- Chen, S.W.; Tao, C.S. Multi-Temporal PolSAR Crops Classification Using Polarimetric-Feature-Driven Deep Convolutional Neural Network. In Proceedings of the RSIP 2017—International Workshop on Remote Sensing with Intelligent Processing, Shanghai, China, 18–21 May 2017. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D Convolutional Neural Networks for Crop Classification with Multi-Temporal Remote Sensing Images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Mou, L.; Bruzzone, L.; Zhu, X.X. Learning Spectral-Spatialoral Features via a Recurrent Convolutional Neural Network for Change Detection in Multispectral Imagery. IEEE Trans. Geosci. Remote Sens. 2019, 57, 924–935. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests, machine learning 45. J. Clin. Microbiol. 2001, 2, 199–228. [Google Scholar]
- Mitchell, T.M. Machine Learning; McGraw-Hill: New York, NY, USA, 2003. [Google Scholar]
- Zhong, L.; Hawkins, T.; Biging, G.; Gong, P. A Phenology-Based Approach to Map Crop Types in the San Joaquin Valley, California. Int. J. Remote Sens. 2011, 32, 7777–7804. [Google Scholar] [CrossRef]
- Lu, H.; He, J.; Liu, L. Discussion on multispectral remote sensing image classification integrating object-oriented image analysis and KNN algorithm. Sci. Technol. Innov. Appl. 2019, 11, 27–30. [Google Scholar]
- Chan, J.C.W.; Paelinckx, D. Evaluation of Random Forest and Adaboost Tree-Based Ensemble Classification and Spectral Band Selection for Ecotope Mapping Using Airborne Hyperspectral Imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Juan, P.; Yang, C.; Song, Y.; Zhai, Z.; Xu, H. Classification of rice phenotypic omics entities based on stacking integrated learning. Trans. Chin. Soc. Agric. Mach. 2019, 50, 9. [Google Scholar]
- Löw, F.; Michel, U.; Dech, S.; Conrad, C. Impact of Feature Selection on the Accuracy and Spatial Uncertainty of Per-Field Crop Classification Using Support Vector Machines. ISPRS J. Photogramm. Remote Sens. 2013, 85, 102–119. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. In The Handbook of Brain Theory and Neural Networks; MIT Press: Cambridge, MA, USA, 1995; Volume 3361. [Google Scholar]
- Karim, F.; Majumdar, S.; Darabi, H.; Harford, S. Multivariate LSTM-FCNs for Time Series Classification. Neural Netw. 2019, 116, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An Assessment of the Effectiveness of a Random Forest Classifier for Land-Cover Classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Tuanmu, M.N.; Viña, A.; Bearer, S.; Xu, W.; Ouyang, Z.; Zhang, H.; Liu, J. Mapping Understory Vegetation Using Phenological Characteristics Derived from Remotely Sensed Data. Remote Sens. Environ. 2010, 114, 1833–1844. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Z.; Li, Z.; Chen, Y.; Zhang, L.; Cao, J.; Tao, F. Identifying the Spatiotemporal Changes of Annual Harvesting Areas for Three Staple Crops in China by Integrating Multi-Data Sources. Environ. Res. Lett. 2020, 15, 074003. [Google Scholar] [CrossRef]
- Smola, A.J.; Schölkopf, B. A Tutorial on Support Vector Regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Altman, N. Classification and Regression Trees. Nat. Methods 2017, 14, 757–758. [Google Scholar] [CrossRef]
- Jin, S.; Yang, L.; Zhu, Z.; Homer, C. A Land Cover Change Detection and Classification Protocol for Updating Alaska NLCD 2001 to 2011. Remote Sens. Environ. 2017, 195, 44–55. [Google Scholar] [CrossRef]
- Tomek, I. A Generalization of the K-NNRule. IEEE Trans. Syst. Man Cybern. 1976, SMC-6, 121–126. [Google Scholar] [CrossRef]
- Menahem, E.; Rokach, L.; Elovici, Y. Troika—An Improved Stacking Schema for Classification Tasks. Inf. Sci. 2009, 179, 4097–4122. [Google Scholar] [CrossRef] [Green Version]
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Phys. Lett. B 2014, 299, 345–350. [Google Scholar] [CrossRef]
- Gadiraju, K.K.; Ramachandra, B.; Chen, Z.; Vatsavai, R.R. Multimodal Deep Learning Based Crop Classification Using Multispectral and Multitemporal Satellite Imagery. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Virtual Event, 6–10 July 2020; pp. 3234–3242. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness and Correlation. arXiv 2020, arXiv:2010.16061. Available online: https://arxiv.org/abs/2010.16061 (accessed on 1 March 2021).
- Massey, R.; Sankey, T.T.; Congalton, R.G.; Yadav, K.; Thenkabail, P.S.; Ozdogan, M.; Meador, A.J.S. MODISphenology-derived, multi-year distribution of conterminous US crop types. Remote Sens. Environ. 2017, 198, 490–503. [Google Scholar] [CrossRef]
- Xun, L.; Wang, P.; Li, L.; Wang, L.; Kong, Q. Identifying crop planting areas using Fourier-transformed feature of time series MODIS leaf area index and sparse-representation-based classification in the North China Plain. Int. J. Remote Sens. 2019, 40, 2034–2052. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.B.; Tan, M.H.; Xin, L.J. Remote sensing monitoring of winter wheat sowing area changes in the North China Plain from 2001 to 2011. J. Agric. Eng. 2015, 31, 190–199. [Google Scholar]
- Yao, Y.; Rosasco, L.; Caponnetto, A. On Early Stopping in Gradient Descent Learning. Constr. Approx. 2007, 26, 289–315. [Google Scholar] [CrossRef]

| Abbreviations Used in This Study | Description | Data Set | Test Set | ||
|---|---|---|---|---|---|
| Number of Pixels | Areal Percentage/% | Number of Pixels | Areal Percentage/% | ||
| WT | Wheat | 69,117 | 30.04 | 1429 | 29.69 |
| CN | Corn | 16,334 | 7.10 | 351 | 7.29 |
| WC | Wheat–Corn | 92,791 | 40.34 | 1956 | 40.64 |
| ER | Early rice | 50,533 | 21.97 | 1051 | 21.84 |
| EL | Early rice–Late rice | 1261 | 0.55 | 26 | 0.54 |
| Total | 230,036 | 100 | 4813 | 100 | |
| Classifier | Parameter | Description |
|---|---|---|
| SVM | C | C is the penalty coefficient used to control the loss function. |
| gamma | gamma denotes the kernel function coefficient. | |
| RF | n_estimators | n_estimators shows the ability and complexity of RF to learn from data. |
| min_samples_split | min_samples_split expresses the minimum number of samples needed to split internal nodes. | |
| min_samples_leaf | min_samples_leaf indicates the minimum sample tree on the leaf nodes. | |
| max_features | max_features mean the number of features to be considered when finding the optimal splitting point. | |
| KNN | n_neighbors | n_neighbors show the number of neighboring samples. |
| leaf_size | leaf_size represents the size of the sphere tree or kd tree. |
| Predicted | |||
|---|---|---|---|
| Positive | Negative | ||
| Actual | Positive | True positives (TP) | False negatives (FN) |
| Negative | False positives (FP) | True negatives (TN) | |
| Hyper-Parameter | Candidate Values | Data | Selected Values for Input Sets |
|---|---|---|---|
| N_neighbors Leaf_size | 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 200, 300, 500, 800, 1200 10, 20, 30, 60, 100 | EVI NDVI NaE | 50 100 100 100 10 30 |
| EVI | NDVI | NaE | |
| Overall Accuracy | 67.98% | 72.31% | 76.20% |
| F1-score | 0.6724 | 0.7159 | 0.7616 |
| kappa coefficient | 0.5317 | 0.5941 | 0.6536 |
| Classifier | Hyper-Parameter | Candidate Values | Selected Values for Input Sets |
|---|---|---|---|
| SVM | C | 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 200, 300, 500, 800, 1200 | 10 |
| gamma | 0.1, 1, 2, 10, ‘auto’ | 0.1 | |
| RF | N_estimators | 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000 | 800 |
| Min_samples_split | 2, 5, 10, 15, 20, 100 | 2 | |
| Min_samples_leaf | 1, 2, 3, 4, 5, 10 | 1 | |
| Max_features | ‘log2′, ’sqrt’, ’auto’ | ‘sqrt’ |
| Reference Classes | Predict Classes | ||||
|---|---|---|---|---|---|
| WT | CN | WC | ER | EL | |
| WT | 61 | −4 | −37 | −20 | 0 |
| CN | 1 | 4 | −3 | −2 | 0 |
| WC | −7 | 6 | 8 | −7 | 0 |
| ER | −11 | −1 | −19 | 29 | 2 |
| EL | 1 | 0 | 0 | −4 | 3 |
| Crop Type | Stacking | Conv1D | LSTM | Reference Map |
|---|---|---|---|---|
| Areal Percentage/% | ||||
| WT | 30.01 | 24.56 | 21.35 | 29.69 |
| CN | 6.97 | 6.17 | 6.29 | 7.29 |
| WC | 40.31 | 46.54 | 49.25 | 40.64 |
| ER | 22.18 | 22.28 | 22.64 | 21.84 |
| EL | 0.53 | 0.45 | 0.47 | 0.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhang, J.; Xun, L.; Wang, J.; Wu, Z.; Henchiri, M.; Zhang, S.; Zhang, S.; Bai, Y.; Yang, S.; et al. Evaluating the Effectiveness of Machine Learning and Deep Learning Models Combined Time-Series Satellite Data for Multiple Crop Types Classification over a Large-Scale Region. Remote Sens. 2022, 14, 2341. https://doi.org/10.3390/rs14102341
Wang X, Zhang J, Xun L, Wang J, Wu Z, Henchiri M, Zhang S, Zhang S, Bai Y, Yang S, et al. Evaluating the Effectiveness of Machine Learning and Deep Learning Models Combined Time-Series Satellite Data for Multiple Crop Types Classification over a Large-Scale Region. Remote Sensing. 2022; 14(10):2341. https://doi.org/10.3390/rs14102341
Chicago/Turabian StyleWang, Xue, Jiahua Zhang, Lan Xun, Jingwen Wang, Zhenjiang Wu, Malak Henchiri, Shichao Zhang, Sha Zhang, Yun Bai, Shanshan Yang, and et al. 2022. "Evaluating the Effectiveness of Machine Learning and Deep Learning Models Combined Time-Series Satellite Data for Multiple Crop Types Classification over a Large-Scale Region" Remote Sensing 14, no. 10: 2341. https://doi.org/10.3390/rs14102341
APA StyleWang, X., Zhang, J., Xun, L., Wang, J., Wu, Z., Henchiri, M., Zhang, S., Zhang, S., Bai, Y., Yang, S., Li, S., & Yu, X. (2022). Evaluating the Effectiveness of Machine Learning and Deep Learning Models Combined Time-Series Satellite Data for Multiple Crop Types Classification over a Large-Scale Region. Remote Sensing, 14(10), 2341. https://doi.org/10.3390/rs14102341







