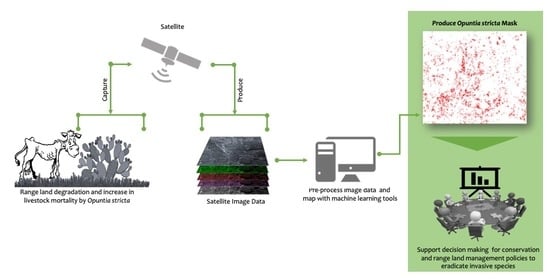
Mapping Opuntia stricta in the Arid and Semi-Arid Environment of Kenya Using Sentinel-2 Imagery and Ensemble Machine Learning Classifiers
Abstract
1. Introduction
2. Materials and Methods
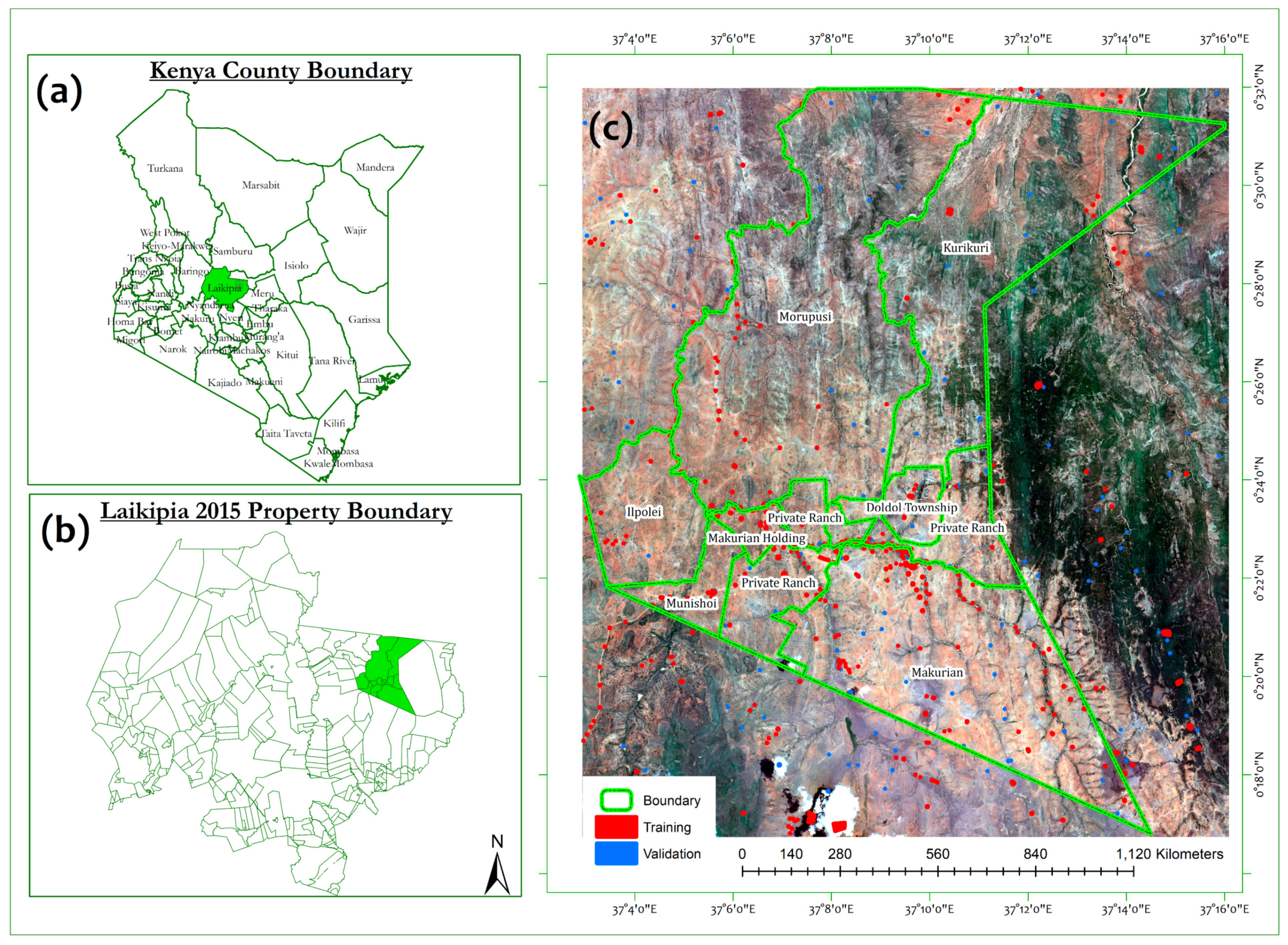
2.1. Description of the Study Area
2.2. Field Data
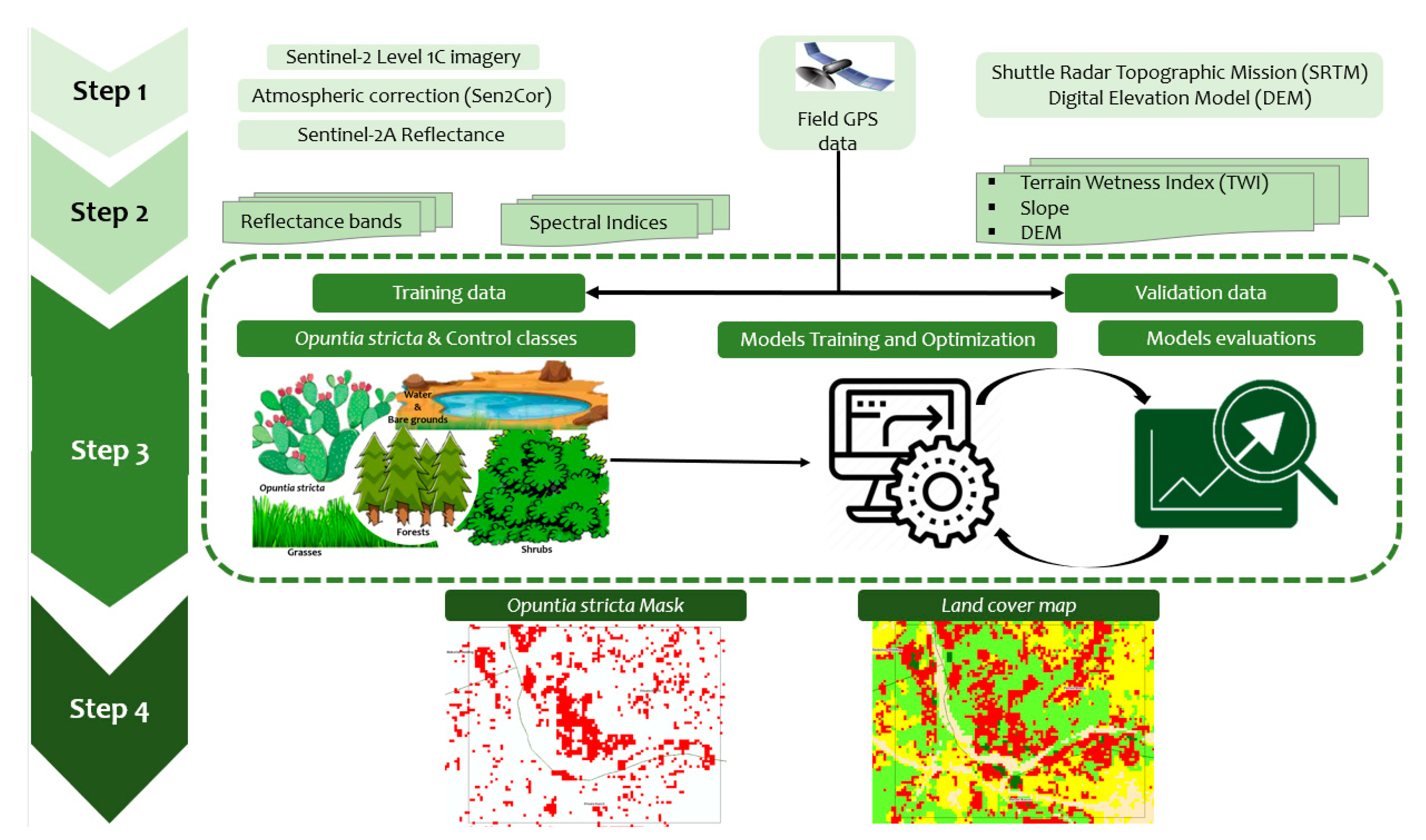
2.3. Satellite Image Analysis and Classification
2.3.1. Satellite Image Acquisition and Pre-Processing
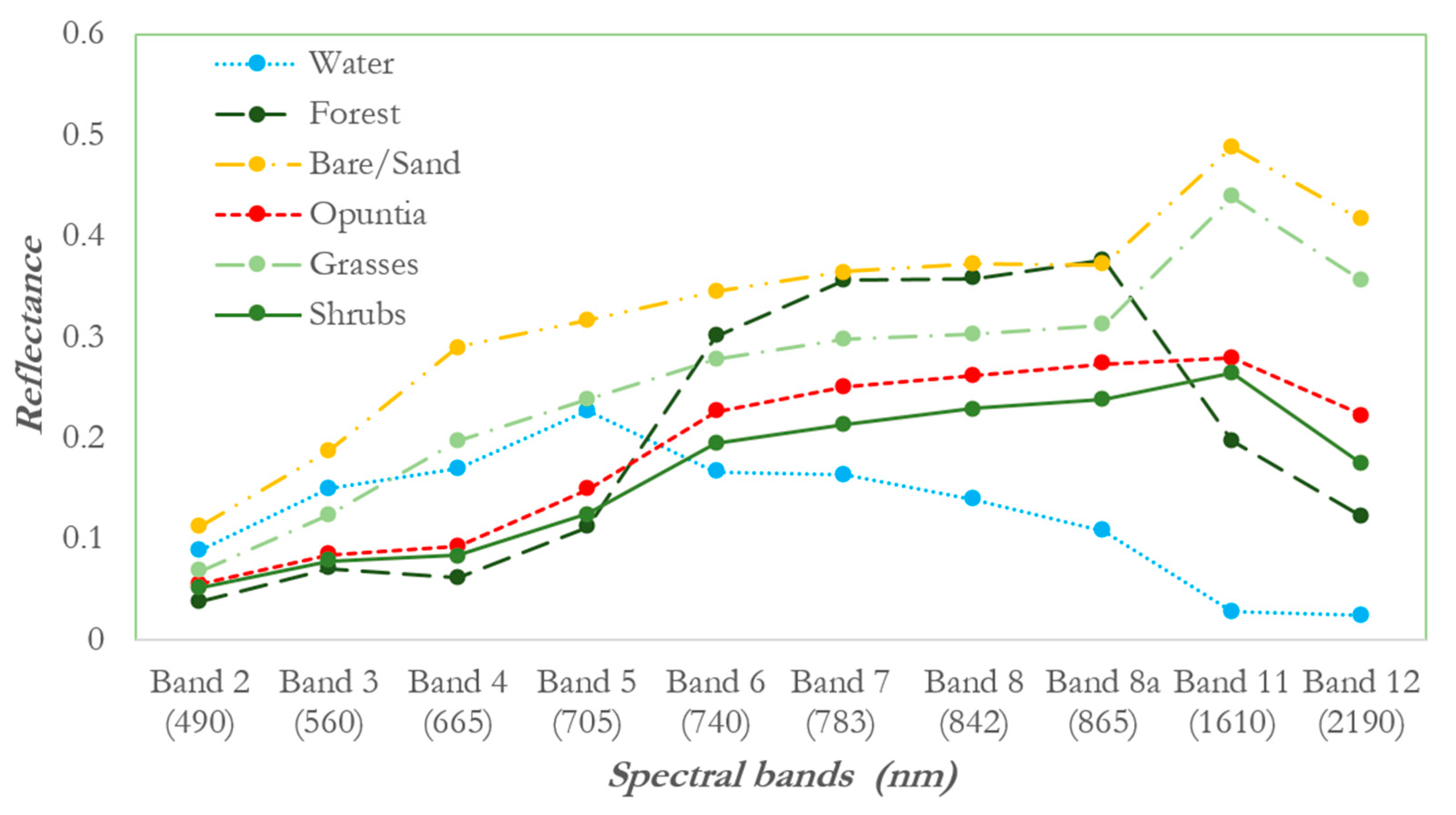
2.3.2. Spectral Separability
2.3.3. Extracting Predictor Variables for Classification
2.3.4. Classification Algorithms and Accuracy Assessment
3. Results
3.1. Analysis of Spectral Separability of Opuntia Stricta and Other Control Classes
3.2. Image Classification
4. Discussion
4.1. Model Evaluation and Spatial Coverage
4.2. Policy Implication, Limitation and Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hungate, B.A.; Barbier, E.B.; Ando, A.W.; Marks, S.P.; Reich, P.B.; van Gestel, N.; Cardinale, B.J. The economic value of grassland species for carbon storage. Sci. Adv. 2017, 3, e1601880. [Google Scholar] [CrossRef] [PubMed]
- Dass, P.; Houlton, B.Z.; Wang, Y.; Warlind, D. Grasslands may be more reliable carbon sinks than forests in California. Environ. Res. Lett. 2018, 13, 074027. [Google Scholar] [CrossRef]
- Huguenin-Elie, O.; Delaby, L.; Klumpp, K. The Role of Grasslands in Biogeochemical Cycles and Biodiversity Conservation. 2018, pp. 3–29. Available online: https://shop.bdspublishing.com/checkout/Store/bds/Detail/Product/3-190-9781786762009-001 (accessed on 16 January 2020).
- Wu, G.; Liu, Y.; Cui, Z.; Liu, Y.; Shi, Z.; Yin, R.; Kardol, P. Trade-off between vegetation type, soil erosion control and surface water in global semi-arid regions: A meta-analysis. J. Appl. Ecol. 2020, 57, 875–885. [Google Scholar] [CrossRef]
- Liu, Y.F.; Liu, Y.; Shi, Z.H.; López-Vicente, M.; Wu, G.L. Effectiveness of re-vegetated forest and grassland on soil erosion control in the semi-arid Loess Plateau. Catena 2020, 195, 104787. [Google Scholar] [CrossRef]
- Phelps, L.N.; Kaplan, J.O. Land use for animal production in global change studies: Defining and characterizing a framework. Glob. Chang. Biol. 2017, 23, 4457–4471. [Google Scholar] [CrossRef]
- Ludwig, L.; Isele, J.; Rahmann, G.; Idel, A.; Hülsebusch, C. A pilot study into biomass yield and composition under increased stocking rates and increased stocking densities on a Namibian organic beef cattle and sheep farm. Org. Agric. 2019, 9, 249–261. [Google Scholar] [CrossRef]
- Suttie, J.M.; Reynolds, S.G.; Batello, C. Grasslands of the World; Plant Production and Protection Series; Food & Agriculture Organization: Rome, Italy, 2005; Volume 34. [Google Scholar]
- Panunzi, E. Are Grasslands under Threat? Brief Analysis of FAO Statistical Data on Pasture and Fodder Crops; Rome UN Food Agric Organisation: Rome, Italy, 2008. [Google Scholar]
- FAO. Nutrition and Livestock–Technical Guidance to Harness the Potential of Livestock for Improved Nutrition of Vulnerable Populations in Programme Planning; FAO: Rome, Italy, 2020. [Google Scholar]
- Alexandratos, N.; Bruinsma, J. World Agriculture towards 2030/2050: The 2012 Revision; FAO: Rome, Italy, 2012. [Google Scholar]
- Nicholson, S.E. A detailed look at the recent drought situation in the Greater Horn of Africa. J. Arid Environ. 2014, 103, 71–79. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0140196313002322 (accessed on 10 March 2021). [CrossRef]
- Njoka, J.T.; Yanda, P.; Maganga, F.; Liwenga, E.; Kateka, A.; Henku, A.; Mabhuye, E.; Malik, N.; Bavo, C. Kenya: Country Situation Assessment. Pathways to Resilience in Semi-arid Economies (PRISE); UoN: Nairobi, Kenya, 2016. [Google Scholar]
- Mganga, K.Z.; Musimba, N.K.R.; Nyariki, D.M. Combining Sustainable Land Management Technologies to Combat Land Degradation and Improve Rural Livelihoods in Semi-arid Lands in Kenya. Environ. Manag. 2015, 56, 1538–1548. Available online: http://link.springer.com/10.1007/s00267-015-0579-9 (accessed on 10 March 2021). [CrossRef]
- Witt, A. Guide to the Naturalized and Invasive Plants of Laikipia [Internet]; Witt, A., Ed.; CABI: Wallingford, UK, 2017; Available online: http://www.cabi.org/cabebooks/ebook/20173158960 (accessed on 10 March 2021).
- Shiferaw, H.; Teketay, D.; Nemomissa, S.; Assefa, F. Some biological characteristics that foster the invasion of Prosopis juliflora (Sw.) DC at Middle Awash Rift Valley Area, north-eastern Ethiopia. J. Arid Environ. 2004, 58, 135–154. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0140196303001460 (accessed on 10 March 2021). [CrossRef]
- Kimothi, M.M.; Dasari, A. Methodology to map the spread of an invasive plant (Lantana camara L.) in forest ecosystems using Indian remote sensing satellite data. Int. J. Remote Sens. 2010, 31, 3273–3289. [Google Scholar] [CrossRef]
- Kaur, R.; Gonzáles, W.L.; Llambi, L.D.; Soriano, P.J.; Callaway, R.M.; Rout, M.E.; Gallaher, T.J. Community Impacts of Prosopis juliflora Invasion: Biogeographic and Congeneric Comparisons. PLoS ONE 2012, 7, e44966. [Google Scholar] [CrossRef] [PubMed]
- Githae, E. Status of Opuntia invasions in the arid and semi-arid lands of Kenya. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2018, 13, 1–7. Available online: http://www.cabi.org/cabreviews/review/20183117948 (accessed on 10 March 2021).
- Witt, A.; Beale, T.; van Wilgen, B.W. An assessment of the distribution and potential ecological impacts of invasive alien plant species in eastern Africa. Trans. R. Soc. S. Afr. 2018, 73, 217–236. [Google Scholar] [CrossRef]
- Andreas, J.; Buono, N.; Di Lello, S.; Gomarasca, M.; Heine, C.; Mason, S.; Nori, M.; Saavedra, K.V.T.R.; Jenet, A.; Buono, N.; et al. The Path to Greener Pastures: Pastoralism, the Backbone of the World’s Drylands; Vétérinaires Sans Frontières International: Brussels, Belgium, 2016. [Google Scholar]
- Schoof, N.; Luick, R. Pastures and pastoralism. In Ecology; Oxford University Press: Oxford, UK, 2018; Available online: http://oxfordbibliographiesonline.com/view/document/obo-9780199830060/obo-9780199830060-0207.xml (accessed on 10 March 2021).
- Shackleton, R.T.; Witt, A.B.R.; Aool, W.; Pratt, C.F. Distribution of the invasive alien weed, Lantana camara, and its ecological and livelihood impacts in eastern Africa. Afr. J. Range Forage Sci. 2017, 34, 1–11. [Google Scholar] [CrossRef]
- Muturi, G.M.; Poorter, L.; Mohren, G.M.J.; Kigomo, B.N. Ecological impact of Prosopis species invasion in Turkwel riverine forest, Kenya. J. Arid Environ. 2013, 92, 89–97. Available online: http://linkinghub.elsevier.com/retrieve/pii/S0140196313000153 (accessed on 10 March 2021). [CrossRef]
- Bekele, K.; Haji, J.; Legesse, B.; Schaffner, U. Economic impacts of Prosopis spp. invasions on dryland ecosystem services in Ethiopia and Kenya: Evidence from choice experimental data. J. Arid Environ. 2018, 158, 9–18. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0140196318304245 (accessed on 10 March 2021). [CrossRef]
- Laikipia County Government. Laikipia County Statistical Abstract 2019 Laikipia. 2019. Available online: https://laikipia.go.ke (accessed on 29 April 2020).
- Ouko, E.; Omondi, S.; Mugo, R.; Wahome, A.; Kasera, K.; Nkurunziza, E.; Wambua, M. Modeling Invasive Plant Species in Kenya’s Northern Rangelands. Front. Environ. Sci. 2020, 8, 69. [Google Scholar] [CrossRef]
- Shackleton, R.T.; Witt, A.B.R.R.; Piroris, F.M.; van Wilgen, B.W. Distribution and socio-ecological impacts of the invasive alien cactus Opuntia stricta in eastern Africa. Biol. Invasions 2017, 19, 2427–2441. [Google Scholar] [CrossRef]
- Witt, A.B.R.R.; Kiambi, S.; Beale, T.; Van Wilgen, B.W. A preliminary assessment of the extent and potential impacts of alien plant invasions in the Serengeti-Mara ecosystem, East Africa. Koedoe 2017, 59, 1–16. [Google Scholar] [CrossRef]
- CABI. Invasive Species Compendium 2019 [cited 6 April 2020]. Available online: https://www.cabi.org/isc/datasheet/37728 (accessed on 6 April 2020).
- Feugang, J.M.; Konarski, P.; Zou, D.; Stintzing, F.C.; Zou, C. Nutritional and medicinal use of Cactus pear (Opuntia spp.) cladodes and fruits. Front. Biosci. 2006, 11, 2574–2589. [Google Scholar] [CrossRef]
- Novoa, A.; Le Roux, J.J.; Robertson, M.P.; Wilson, J.R.U.; Richardson, D.M. Introduced and invasive cactus species: A global review. AoB Plants 2015, 7. [Google Scholar] [CrossRef]
- Strum, S.C.; Stirling, G.; Mutunga, S.K. The perfect storm: Land use change promotes Opuntia stricta’s invasion of pastoral rangelands in Kenya. J. Arid Environ. 2015, 118, 37–47. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0140196315000452 (accessed on 10 March 2021). [CrossRef]
- Padrón, B.; Nogales, M.; Traveset, A.; Vilà, M.; Martínez-Abraín, A.; Padilla, D.P.; Marrero, P. Integration of invasive Opuntia spp. by native and alien seed dispersers in the Mediterranean area and the Canary Islands. Biol. Invasions 2011, 13, 831–844. [Google Scholar] [CrossRef]
- Nefzaoui, A.; Inglese, P.; Belay, T. Improved utilization of cactus pear for food, feed, soil and water conservation and other products in Africa. In Proceedings of the International Workshop, Mekelle, Ethiopia, 19–21 October 2010. [Google Scholar]
- Kunyanga, C.N.; Vellingiri, V.; Imungi, K.J. Nutritional quality, phytochemical composition and health protective effects of an under-utilized prickly cactus fruit (Opuntia stricta Haw) collected from Kenya. Afr. J. Food Agric. Nutr. Dev. 2014, 14, 9561–9577. [Google Scholar]
- Maema, L.P.; Potgieter, M.; Mahlo, S.M. Invasive alien plant species used for the treatment of various diseases in Limpopo Province, South Africa. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 223–231. Available online: http://journals.sfu.ca/africanem/index.php/ajtcam/article/view/4039/pdf (accessed on 10 March 2021). [CrossRef]
- Shackleton, R.T.; Shackleton, C.M.; Kull, C.A. The role of invasive alien species in shaping local livelihoods and human well-being: A review. J. Environ. Manag. 2019, 229, 145–157. [Google Scholar] [CrossRef]
- Beinart, W.; Wotshela, L. Prickly pear in the Eastern Cape since the 1950s-perspectives from interviews. Kronos J. Cape Hist. 2003, 28, 191–209. [Google Scholar]
- Beinart, W. Prickly Pear: A Social History of a Plant in the Eastern Cape; NYU Press: New York, NY, USA, 2011. [Google Scholar]
- Witt, A.B.R.; Nunda, W.; Makale, F.; Reynolds, K. A preliminary analysis of the costs and benefits of the biological control agent Dactylopius opuntiae on Opuntia stricta in Laikipia County, Kenya. BioControl 2020, 65, 515–523. [Google Scholar] [CrossRef]
- Ismail, R.; Mutanga, O.; Peerbhay, K. The identification and remote detection of alien invasive plants in commercial forests: An Overview. S. Afr. J. Geomat. 2016, 5, 49. Available online: http://www.ajol.info/index.php/sajg/article/view/131503 (accessed on 10 March 2021). [CrossRef]
- Matongera, T.N.; Mutanga, O.; Dube, T.; Sibanda, M. Detection and mapping the spatial distribution of bracken fern weeds using the Landsat 8 OLI new generation sensor. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 93–103. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0303243416302045 (accessed on 10 March 2021). [CrossRef]
- Pu, R. Detecting and Mapping Invasive Plant Species by Using Hyperspectral Data. Hyperspectr. Remote Sens. Veg. 2016, 447. [Google Scholar] [CrossRef]
- Skowronek, S.; Ewald, M.; Isermann, M.; Van De Kerchove, R.; Lenoir, J.; Aerts, R.; Feilhauer, H. Mapping an invasive bryophyte species using hyperspectral remote sensing data. Biol. Invasions 2017, 19, 239–254. [Google Scholar] [CrossRef]
- Joshi, C.M.; Skidmore, A.K.; van Andel, J.; de Leeuw, J.; van Duren, I.C. Mapping Cryptic Invaders and Invasibility of Tropical Forest Ecosystems: Chromolaena odorata in Nepal. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, May 2006. [Google Scholar]
- Goetz, A.F.H. Three decades of hyperspectral remote sensing of the Earth: A personal view. Remote Sens. Environ. 2009, 113, S5–S16. Available online: https://linkinghub.elsevier.com/retrieve/pii/S003442570900073X (accessed on 10 March 2021). [CrossRef]
- Shiferaw, H.; Bewket, W.; Eckert, S. Performances of machine learning algorithms for mapping fractional cover of an invasive plant species in a dryland ecosystem. Ecol. Evol. 2019, 9, 2562–2574. [Google Scholar] [CrossRef]
- Rajah, P.; Odindi, J.; Mutanga, O.; Kiala, Z. The utility of Sentinel-2 Vegetation Indices (VIs) and Sentinel-1 Synthetic Aperture Radar (SAR) for invasive alien species detection and mapping. Nat. Conserv. 2019, 35, 41–61. Available online: https://natureconservation.pensoft.net/article/29588/ (accessed on 10 March 2021). [CrossRef]
- Vaz, A.S.; Alcaraz-Segura, D.; Campos, J.C.J.C.; Vicente, J.R.; Honrado, J.P.J.P. Managing plant invasions through the lens of remote sensing: A review of progress and the way forward. Sci. Total Environ. 2018, 642, 1328–1339. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0048969718322101 (accessed on 10 March 2021). [CrossRef]
- (GoK) G of K. Vision 2030 Development Strategy for Northern Kenya and Other Arid Lands. Final Report, Nairobi, Kenya. 2012. Available online: https://www.ndma.go.ke/index.php/resource-center/policy-documents/send/44-policy-documents/4300-vision-2030-development-strategy-for-asals (accessed on 19 March 2020).
- NDMA. Annual Report and Financial Statements for Financial Year Ending, 30 June 2018, Nairobi. 2018. Available online: https://www.ndma.go.ke/ (accessed on 19 March 2020).
- Ojwang, G.O.; Agatsiva, J.; Situma, C. Analysis of Climate Change and Variability Risks in the Smallholder Sector: Case Studies of the Laikipia and Narok Districts Representing Major Agro-Ecological Zones in Kenya, Rome. 2010. Available online: http://www.fao.org/3/i1785e/i1785e00.pdf (accessed on 12 February 2020).
- KNBS. 2019 Kenya Population and Housing Cencus, Nairobi. 2019. Available online: https://www.knbs.or.ke/ (accessed on 19 March 2020).
- Evans, L.A.; Adams, W.M. Fencing elephants: The hidden politics of wildlife fencing in Laikipia, Kenya. Land Use Policy 2016, 51, 215–228. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0264837715003518 (accessed on 10 March 2021). [CrossRef]
- Klopp, J.M. Pilfering the public: The problem of land grabbing in contemporary Kenya. Afr. Today 2000, 47, 7–26. [Google Scholar] [CrossRef]
- Mkutu, K. Pastoralism and Conflict in the Horn of Africa; Saferworld Organisation: London, UK, 2001. [Google Scholar]
- Letai, J. Land Deals in Kenya: The Genesis of Land Deals in Kenya and Its Implication on Pastoral Livelihoods: A Case Study of Laikipia District, 2011, Nairobi. 2011. Available online: https://landportal.org/sites/default/files/land_deals_in_kenya-initial_report_for_laikipia_district2.pdf (accessed on 19 March 2020).
- Idris, A. Taking the camel through the eye of a needle: Enhancing pastoral resilience through education policy in Kenya. Resil. Interdiscip. Perspect. Sci. Humanit. 2011, 2, 25–38. [Google Scholar]
- Williams, A. The Abandoned Lands of Laikipia Land Use Options Study, Laikipia. 2013. Available online: https://laikipia.org/laikipia-county/ (accessed on 19 March 2020).
- Bond, J. Conflict, Development and Security at the Agro–Pastoral–Wildlife Nexus: A Case of Laikipia County, Kenya. J. Dev. Stud. 2014, 50, 991–1008. [Google Scholar] [CrossRef]
- NEMA. State of Environment Report Laikipia County. 2013. Available online: https://laikipia.org/wp-content/uploads/2020/03/STATE-OF-ENVIRONMENT-REPORT.docx (accessed on 19 March 2020).
- Gatti, A.; Bertolini, A. Sentinel-2 Products Specification Document. 2013. Available online: https://sentinel.esa.int/documents/247904/685211/Sentinel-2-Products-Specification-Document (accessed on 23 February 2020).
- Main-Knorn, M.; Pflug, B.; Louis, J.; Debaecker, V.; Müller-Wilm, U.; Gascon, F. Sen2Cor for Sentinel-2. In Image and Signal Processing for Remote Sensing XXIII; Bruzzone, L., Bovolo, F., Benediktsson, J.A., Eds.; SPIE: Belingham, WA, USA, 2017; p. 3. [Google Scholar] [CrossRef]
- SNAP. SNAP—ESA Sentinel Application Platform v7.0. 2020. Available online: http://step.esa.int/main/toolboxes/sentinel-2-toolbox/ (accessed on 11 August 2020).
- Adam, E.; Mureriwa, N.; Newete, S. Mapping Prosopis glandulosa (mesquite) in the semi-arid environment of South Africa using high-resolution WorldView-2 imagery and machine learning classifiers. J. Arid Environ. 2017, 145, 43–51. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0140196317301052 (accessed on 10 March 2021). [CrossRef]
- Kailath, T. The Divergence and Bhattacharyya Distance Measures in Signal Selection. IEEE Trans. Commun. 1967, 15, 52–60. Available online: http://ieeexplore.ieee.org/document/1089532/ (accessed on 10 March 2021). [CrossRef]
- Jensen, J.R. Introductory Digital Image Processing: A Remote Sensing Perspective; Prentice-Hall Inc.: Hoboken, NJ, USA, 1996. [Google Scholar]
- Trigg, S.; Flasse, S. An evaluation of different bi-spectral spaces for discriminating burned shrub-savannah. Int. J. Remote Sens. 2001, 22, 2641–2647. [Google Scholar] [CrossRef]
- Chemura, A.; Mutanga, O. Developing detailed age-specific thematic maps for coffee (Coffea arabica L.) in heterogeneous agricultural landscapes using random forests applied on Landsat 8 multispectral sensor. Geocarto Int. 2017, 32, 759–776. [Google Scholar] [CrossRef]
- Yang, C.; Li, Q.; Wu, G.; Chen, J. A Highly efficient method for training sample selection in remote sensing classification. In Proceedings of the 2018 26th International Conference on Geoinformatics IEEE, Kunming, China, 28–30 June 2018; pp. 1–5. Available online: https://ieeexplore.ieee.org/document/8557085/ (accessed on 10 March 2021).
- Harris Geospatial. ENVI Image Analysis Software; Harris Geospatial: Broomfield, CO, USA, 2011; Available online: https://www.harrisgeospatial.com/ (accessed on 10 March 2021).
- Jafari, R.; Bashari, H.; Tarkesh, M. Discriminating and monitoring rangeland condition classes with MODIS NDVI and EVI indices in Iranian arid and semi-arid lands. Arid Land Res. Manag. 2017, 31, 94–110. [Google Scholar] [CrossRef]
- Zuo, L.; Liu, R.; Liu, Y.; Shang, R. Effect of Mathematical Expression of Vegetation Indices on the Estimation of Phenology Trends from Satellite Data. Chin. Geogr. Sci. 2019, 29, 756–767. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant Remote Sensing Vegetation Indices: A Review of Developments and Applications. J. Sens. 2017, 2017, 1–17. Available online: https://www.hindawi.com/journals/js/2017/1353691/ (accessed on 10 March 2021). [CrossRef]
- Wessel, M.; Brandmeier, M.; Tiede, D. Evaluation of Different Machine Learning Algorithms for Scalable Classification of Tree Types and Tree Species Based on Sentinel-2 Data. Remote Sens. 2018, 10, 1419. Available online: https://www.mdpi.com/2072-4292/10/9/1419 (accessed on 10 March 2021). [CrossRef]
- Svinurai, W.; Hassen, A.; Tesfamariam, E.; Ramoelo, A. Performance of ratio-based, soil-adjusted and atmospherically corrected multispectral vegetation indices in predicting herbaceous aboveground biomass in a Colophospermum mopane tree-shrub savanna. Grass Forage Sci. 2018, 73, 727–739. [Google Scholar] [CrossRef]
- Mudereri, B.T.; Dube, T.; Adel-Rahman, E.M.; Niassy, S.; Kimathi, E.; Khan, Z.; Landmann, T. A comparative analysis of PlanetScope and Sentinel-2 space-borne sensors in mapping Striga weed using Guided Regularised Random Forest classification ensemble. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2/W13, 701–708. Available online: https://www.int-arch-photogramm-remote-sens-spatial-inf-sci.net/XLII-2-W13/701/2019/ (accessed on 10 March 2021).
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Richardson, A.J.; Wiegand, C.L. Distinguishing vegetation from soil background information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Rouse, J.W., Jr.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with Erts; NASA SP-351, 3rd ERTS-1 Symposium; NASA: Washington, DC, USA, 1974.
- Crippen, R. Calculating the vegetation index faster. Remote Sens. Environ. 1990, 34, 71–73. Available online: https://linkinghub.elsevier.com/retrieve/pii/003442579090085Z (accessed on 10 March 2021). [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. Available online: http://ieeexplore.ieee.org/document/134076/ (accessed on 10 March 2021). [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. Available online: https://linkinghub.elsevier.com/retrieve/pii/0034425794901341 (accessed on 10 March 2021). [CrossRef]
- Zevenbergen, L.W.; Thorne, C.R. Quantitative analysis of land surface topography. Earth Surf. Process. Landf. 1987, 12, 47–56. [Google Scholar] [CrossRef]
- Mattivi, P.; Franci, F.; Lambertini, A.; Bitelli, G. TWI computation: A comparison of different open source GISs. Open Geospatial Data Softw. Stand. 2019, 4, 6. [Google Scholar] [CrossRef]
- Yang, C.; Wu, G.; Ding, K.; Shi, T.; Li, Q.; Wang, J. Improving Land Use/Land Cover Classification by Integrating Pixel Unmixing and Decision Tree Methods. Remote Sens. 2017, 9, 1222. Available online: https://www.mdpi.com/2072-4292/9/12/1222 (accessed on 10 March 2021). [CrossRef]
- Hurskainen, P.; Adhikari, H.; Siljander, M.; Pellikka, P.K.E.; Hemp, A. Auxiliary datasets improve accuracy of object-based land use/land cover classification in heterogeneous savanna landscapes. Remote Sens. Environ. 2019, 233, 111354. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0034425719303736 (accessed on 10 March 2021). [CrossRef]
- Mwaniki, W.M.; Möller, S.M. Knowledge based multi-source, time series classification: A case study of central region of Kenya. Appl. Geogr. 2015, 60, 58–68. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0143622815000715 (accessed on 10 March 2021). [CrossRef]
- Franklin, J. Predictive vegetation mapping: Geographic modelling of biospatial patterns in relation to environmental gradients. Prog. Phys. Geogr. Earth Environ. 1995, 19, 474–499. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System. 2009. Available online: http://qgis.org (accessed on 8 January 2020).
- Chen, T.; Guestrin, C. XGBoost. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Abdi, A.M. Land cover and land use classification performance of machine learning algorithms in a boreal landscape using Sentinel-2 data. GISci. Remote Sens. 2020, 57, 1–20. [Google Scholar] [CrossRef]
- Man, C.D.; Nguyen, T.T.; Bui, H.Q.; Lasko, K.; Nguyen, T.N.T. Improvement of land-cover classification over frequently cloud-covered areas using Landsat 8 time-series composites and an ensemble of supervised classifiers. Int. J. Remote Sens. 2018, 39, 1243–1255. [Google Scholar] [CrossRef]
- Freeman, E.A.; Moisen, G.G.; Coulston, J.W.; Wilson, B.T. Random forests and stochastic gradient boosting for predicting tree canopy cover: Comparing tuning processes and model performance. Can. J. For. Res. 2016, 46, 323–339. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Bradter, U.; O’Connell, J.; Kunin, W.E.; Boffey, C.W.H.; Ellis, R.J.; Benton, T.G. Classifying grass-dominated habitats from remotely sensed data: The influence of spectral resolution, acquisition time and the vegetation classification system on accuracy and thematic resolution. Sci. Total Environ. 2020, 711, 134584. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0048969719345759 (accessed on 10 March 2021). [CrossRef]
- Hunter, F.D.L.; Mitchard, E.T.A.; Tyrrell, P.; Russell, S. Inter-Seasonal Time Series Imagery Enhances Classification Accuracy of Grazing Resource and Land Degradation Maps in a Savanna Ecosystem. Remote Sens. 2020, 12, 198. Available online: https://www.mdpi.com/2072-4292/12/1/198 (accessed on 10 March 2021). [CrossRef]
- Strobl, C.; Malley, J.; Tutz, G. An introduction to recursive partitioning: Rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol. Methods 2009, 14, 323–348. [Google Scholar] [CrossRef]
- Bergstra, J.S.; Bardenet, R.; Bengio, Y.; Kégl, B. Algorithms for hyper-parameter optimization. In Proceedings of the 25th Annual Conference on Advances in Neural Information Processing Systems, Granada, Spain, 6–12 December 2011; pp. 2546–2554. [Google Scholar]
- Luo, G. A review of automatic selection methods for machine learning algorithms and hyper-parameter values. Netw. Model. Anal. Health Inform. Bioinform. 2016, 5, 18. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Duchesnay, E. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. Available online: https://www.cambridge.org/core/product/identifier/S0376892997000088/type/journal_article (accessed on 10 March 2021). [CrossRef]
- Olofsson, P.; Foody, G.M.; Stehman, S.V.; Woodcock, C.E. Making better use of accuracy data in land change studies: Estimating accuracy and area and quantifying uncertainty using stratified estimation. Remote Sens. Environ. 2013, 129, 122–131. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0034425712004191 (accessed on 10 March 2021). [CrossRef]
- Lillesand, T.; Kiefer, R.W.; Chipman, J. Remote Sensing and Image Interpretation; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Pontius, R., Jr. Comparison of Categorical Maps. Photogramm. Eng. Remote Sens. 2000, 66, 1011–1016. Available online: http://www2.clarku.edu/~rpontius/ (accessed on 10 March 2021).
- Pontius, R.G.; Millones, M. Death to Kappa: Birth of quantity disagreement and allocation disagreement for accuracy assessment. Int. J. Remote Sens. 2011, 32, 4407–4429. [Google Scholar] [CrossRef]
- Bradley, B.A.; Blumenthal, D.M.; Wilcove, D.S.; Ziska, L.H. Predicting plant invasions in an era of global change. Trends Ecol. Evol. 2010, 25, 310–318. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0169534709003693 (accessed on 10 March 2021). [CrossRef]
- Bogan, S.A.; Antonarakis, A.S.; Moorcroft, P.R. Imaging spectrometry-derived estimates of regional ecosystem composition for the Sierra Nevada, California. Remote Sens. Environ. 2019, 228, 14–30. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0034425719301221 (accessed on 10 March 2021). [CrossRef]
- Shoko, C.; Mutanga, O. Examining the strength of the newly-launched Sentinel 2 MSI sensor in detecting and discriminating subtle differences between C3 and C4 grass species. ISPRS J. Photogramm. Remote Sens. 2017, 129, 32–40. [Google Scholar] [CrossRef]
- Githae, E. Prickly Pear Cactus Invasion: A Major Threat to Biodiversity and Food Security in the Drylands of Kenya; Swedish University of Agricultural Sciences: Uppsala, Sweden, 2018; Available online: https://www.kilimo.go.ke/wp-content/uploads/2018/12/Eunice-Githae-Policy-Brief.pdf (accessed on 10 March 2021).
- (GoK) G of K. National Wildlife Strategy 2030 [Internet], Nairobi, Kenya. 2018. Available online: https://www.tourism.go.ke/wp-content/uploads/2018/06/NWS2030-FINAL-JUNE-12-2018-1.pdf (accessed on 19 March 2020).
- Ndungu, L.; Oware, M.; Omondi, S.; Wahome, A.; Mugo, R.; Adams, E. Application of MODIS NDVI for Monitoring Kenyan Rangelands Through a Web Based Decision Support Tool. Front. Environ. Sci. 2019, 7, 187. [Google Scholar] [CrossRef]
- Witt, A.B.R.; Nunda, W.; Beale, T.; Kriticos, D.J. A preliminary assessment of the presence and distribution of invasive and potentially invasive alien plant species in Laikipia County, Kenya, a biodiversity hotspot. KOEDOE Afr. Prot. Area Conserv. Sci. 2020, 62, 1–10. Available online: http://www.koedoe.co.za/index.php/KOEDOE/article/view/1605 (accessed on 10 March 2021).
- Yin, Q.; Hong, W.; Zhang, F.; Pottier, E. Optimal Combination of Polarimetric Features for Vegetation Classification in PolSAR Image. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 3919–3931. Available online: https://ieeexplore.ieee.org/document/8901433/ (accessed on 10 March 2021). [CrossRef]
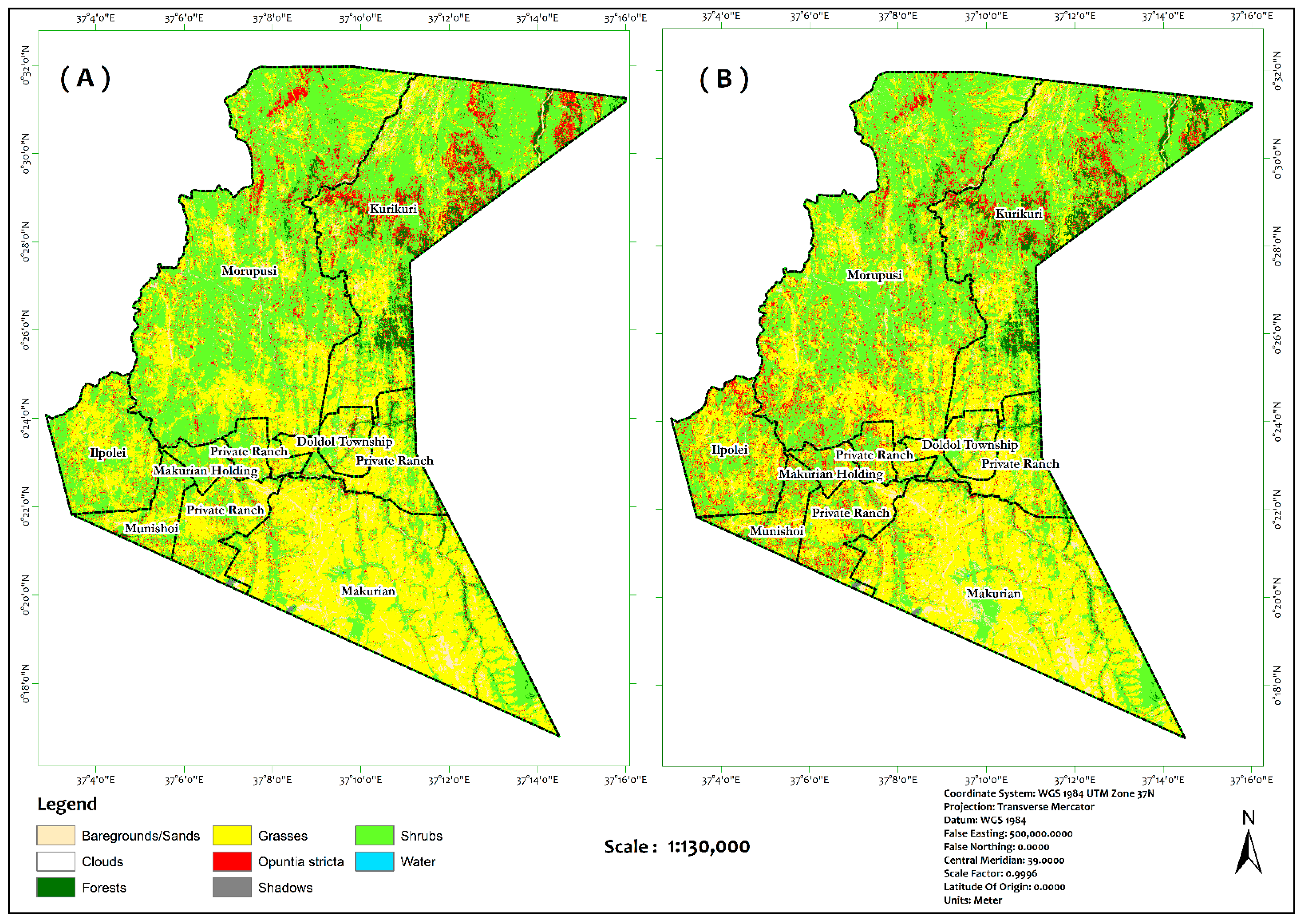

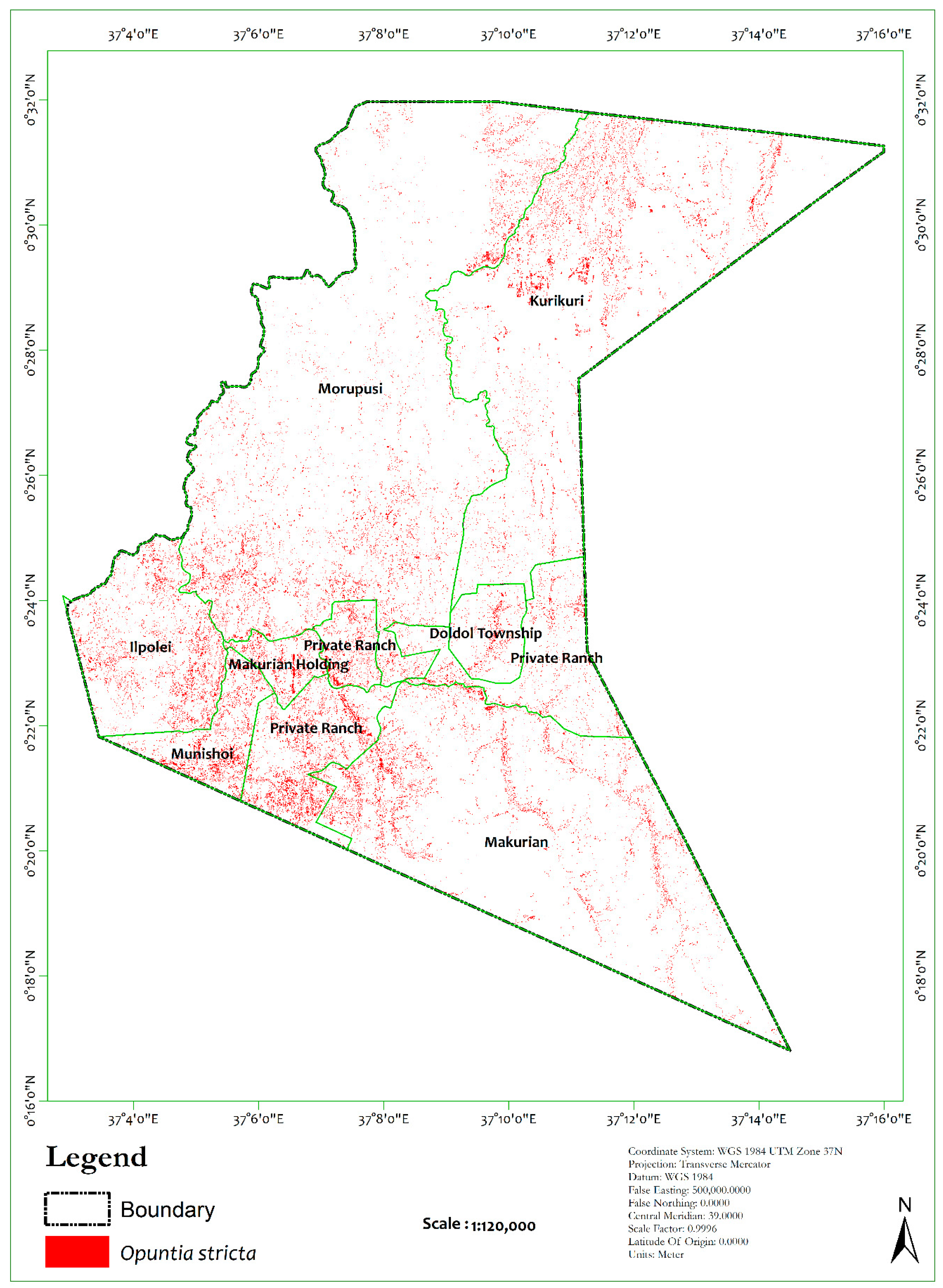
| Classes | Class ID | Description | Samples | |
|---|---|---|---|---|
| Training | Validation | |||
| Bare ground | 0 | Dry exposed soils | 134 | 18 |
| Clouds | 1 | White material | 2 | 2 |
| Forests | 2 | High-density woody vegetation | 41 | 20 |
| Grasses | 3 | Open and high-density low vegetation | 75 | 38 |
| Opuntia stricta | 4 | Target cactus vegetation | 144 | 63 |
| Shadows | 5 | Black material | 1 | 1 |
| Shrubs | 6 | Low lying vegetation | 77 | 20 |
| Water | 7 | Open water | 6 | 3 |
| Band | Spectral Region | Spatial Resolution (m) | Central Wavelength (nm) |
|---|---|---|---|
| 2 | Blue | 10 | 492.4 |
| 3 | Green | 10 | 559.8 |
| 4 | Red | 10 | 664.6 |
| 5 | Red edge | 20 | 704.1 |
| 6 | Red edge | 20 | 740.5 |
| 7 | Red edge | 20 | 782.8 |
| 8 | Near- infrared | 10 | 832.8 |
| 8A | Near -infrared | 20 | 864.7 |
| 11 | Shortwave Infrared | 20 | 1613.7 |
| 12 | Shortwave Infrared | 20 | 2202.4 |
| Model | Hyper-Parameter Value | Definition |
|---|---|---|
| XGB | max_depth = 3, learning_rate = 0.1, n_estimators = 100 | maximum depth of a tree to which changes makes the model complex |
| learning rate step size shrinkage used in the updates hence preventing the overfitting | ||
| maximum number of iterations to the training | ||
| RF | max_depth = 5, n_estimators = 500 | maximum number of levels for each decision tree |
| tree numbers in the forest |
| Opuntia stricta | Shrubs | Grasses | Bare Grounds | Forests | Water | |
|---|---|---|---|---|---|---|
| Opuntia stricta | - | 1.59 | 1.83 | 1.99 | 1.99 | 2.00 |
| Shrubs | 1.27 | - | 1.89 | 1.99 | 1.99 | 2.00 |
| Grasses | 1.66 | 1.83 | - | 1.68 | 2.00 | 2.00 |
| Bare grounds | 1.92 | 1.95 | 1.49 | - | 2.00 | 2.00 |
| Forests | 1.92 | 1.94 | 1.99 | 1.99 | - | 2.00 |
| Water | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | - |
| Land Cover | Bare Ground | Cloud | Forest | Grass | Opuntia | Shadows | Shrubs | Water | User Accuracy | |
|---|---|---|---|---|---|---|---|---|---|---|
| Classification Data | Bare | 24 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 88.89% |
| Cloud | 0 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 100.00% | |
| Forest | 0 | 0 | 23 | 0 | 1 | 0 | 2 | 0 | 88.46% | |
| Grass | 1 | 0 | 0 | 33 | 9 | 0 | 6 | 0 | 67.35% | |
| Opuntia | 0 | 0 | 0 | 2 | 35 | 0 | 3 | 0 | 87.50% | |
| Shadows | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 100.00% | |
| Shrubs | 1 | 0 | 2 | 0 | 28 | 0 | 31 | 4 | 46.97% | |
| Water | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 100.00% | |
| Weights | 27 | 45 | 26 | 49 | 40 | 29 | 66 | 32 | ||
| Producer Accuracy | 92.31% | 100.00% | 92.00% | 86.84% | 47.95% | 100.00% | 73.81% | 88.89% | ||
| Overall Accuracy | 0.802 | |||||||||
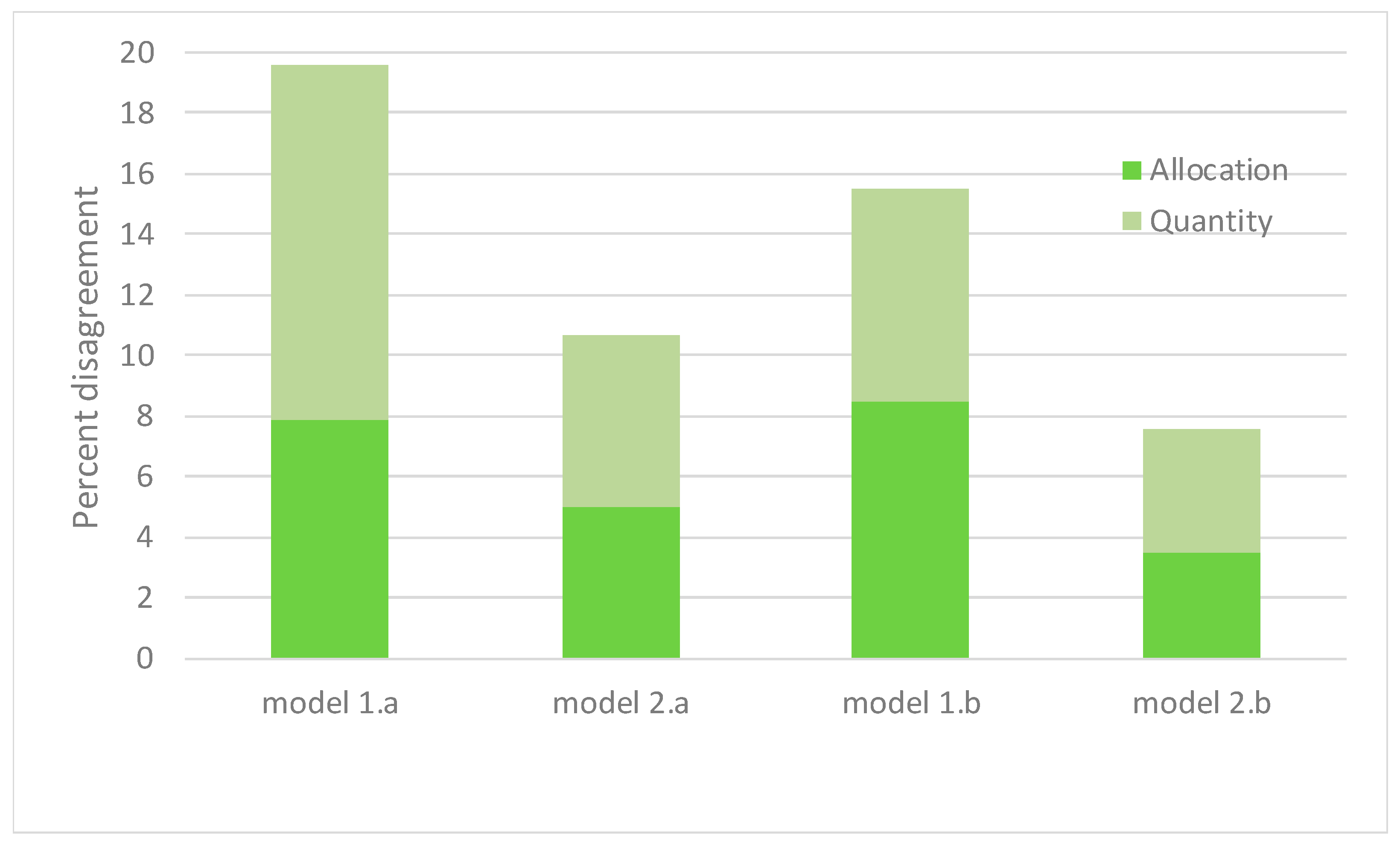
| Allocation Disaggrement | 0.079 | |||||||||
| Quantity Disaggrement | 0.117 | |||||||||
| Kappa | 0.77 | |||||||||
| Reference Data | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Land Cover | Bare Ground | Cloud | Forest | Grass | Opuntia | Shadows | Shrubs | Water | User Accuracy | |
| Classification Data | Bare | 23 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 92.00% |
| Cloud | 0 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 100.00% | |
| Forest | 0 | 0 | 20 | 0 | 1 | 0 | 0 | 0 | 95.24% | |
| Grass | 3 | 0 | 0 | 34 | 11 | 0 | 6 | 0 | 62.96% | |
| Opuntia | 0 | 0 | 1 | 2 | 57 | 0 | 0 | 0 | 95.00% | |
| Shadows | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 100.00% | |
| Shrubs | 0 | 0 | 4 | 0 | 4 | 0 | 36 | 0 | 81.82% | |
| Water | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 100.00% | |
| Weights | 25 | 45 | 21 | 54 | 60 | 29 | 44 | 36 | ||
| Producer Accuracy | 88.46% | 100.00% | 80.00% | 89.47% | 78.08% | 100.00% | 85.71% | 100.00% | ||
| Overall Accuracy | 0.891 | |||||||||
| Allocation Disaggrement | 0.050 | |||||||||
| Quantity Disaggrement | 0.057 | |||||||||
| +Kappa | 0.87 | |||||||||
| Reference Data | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Land Covers | Bare ground | Cloud | Forest | Grass | Opuntia | Shadows | Shrubs | Water | User Accuracy | |
| Classification Data | Bareground | 26 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 83.87% |
| Cloud | 0 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 100.00% | |
| Forest | 0 | 0 | 21 | 0 | 0 | 0 | 3 | 0 | 87.50% | |
| Grass | 0 | 0 | 0 | 30 | 6 | 0 | 6 | 0 | 71.43% | |
| Opuntia | 0 | 0 | 0 | 1 | 48 | 0 | 3 | 0 | 92.31% | |
| Shadows | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 100.00% | |
| Shrubs | 0 | 0 | 4 | 2 | 19 | 0 | 30 | 0 | 54.55% | |
| Water | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 100.00% | |
| Weights | 31 | 45 | 24 | 42 | 52 | 29 | 55 | 36 | ||
| Producer Accuracy | 100.00% | 100.00% | 84.00% | 78.95% | 65.75% | 100.00% | 71.43% | 100.00% | ||
| Overall Accuracy | 0.843 | |||||||||
| Allocation Disaggrement | 0.085 | |||||||||
| Quantity Disaggrement | 0.070 | |||||||||
| Kappa | 0.82 | |||||||||
| Reference Data | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Land Cover | Bare ground | Cloud | Forest | Grass | Opuntia | Shadows | Shrubs | Water | User Accuracy | |
| Classification Data | Bare | 25 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 86.21% |
| Cloud | 0 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 100.00% | |
| Forest | 0 | 0 | 23 | 0 | 0 | 0 | 1 | 0 | 95.83% | |
| Grass | 1 | 0 | 0 | 34 | 9 | 0 | 3 | 0 | 72.34% | |
| Opuntia | 0 | 0 | 1 | 0 | 60 | 0 | 0 | 0 | 98.36% | |
| Shadows | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 100.00% | |
| Shrubs | 0 | 0 | 1 | 0 | 4 | 0 | 38 | 0 | 88.37% | |
| Water | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 100.00% | |
| Weights | 29 | 45 | 24 | 47 | 61 | 29 | 43 | 36 | ||
| Producer Accuracy | 96.15% | 100.00% | 92.00% | 89.47% | 82.19% | 100.00% | 90.48% | 100.00% | ||
| Overall Accuracy | 0.923 | |||||||||
| Allocation Disaggrement | 0.035 | |||||||||
| Quantity Disaggrement | 0.041 | |||||||||
| Kappa | 0.91 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muthoka, J.M.; Salakpi, E.E.; Ouko, E.; Yi, Z.-F.; Antonarakis, A.S.; Rowhani, P. Mapping Opuntia stricta in the Arid and Semi-Arid Environment of Kenya Using Sentinel-2 Imagery and Ensemble Machine Learning Classifiers. Remote Sens. 2021, 13, 1494. https://doi.org/10.3390/rs13081494
Muthoka JM, Salakpi EE, Ouko E, Yi Z-F, Antonarakis AS, Rowhani P. Mapping Opuntia stricta in the Arid and Semi-Arid Environment of Kenya Using Sentinel-2 Imagery and Ensemble Machine Learning Classifiers. Remote Sensing. 2021; 13(8):1494. https://doi.org/10.3390/rs13081494
Chicago/Turabian StyleMuthoka, James M., Edward E. Salakpi, Edward Ouko, Zhuang-Fang Yi, Alexander S. Antonarakis, and Pedram Rowhani. 2021. "Mapping Opuntia stricta in the Arid and Semi-Arid Environment of Kenya Using Sentinel-2 Imagery and Ensemble Machine Learning Classifiers" Remote Sensing 13, no. 8: 1494. https://doi.org/10.3390/rs13081494
APA StyleMuthoka, J. M., Salakpi, E. E., Ouko, E., Yi, Z.-F., Antonarakis, A. S., & Rowhani, P. (2021). Mapping Opuntia stricta in the Arid and Semi-Arid Environment of Kenya Using Sentinel-2 Imagery and Ensemble Machine Learning Classifiers. Remote Sensing, 13(8), 1494. https://doi.org/10.3390/rs13081494