Multiple Endmember Spectral Mixture Analysis (MESMA) Applied to the Study of Habitat Diversity in the Fine-Grained Landscapes of the Cantabrian Mountains
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Data Sources and Preparatory Steps
2.2.1. Landsat Imagery
- Band 1: ultra-blue, 0.43–0.45-µm wavelength
- Band 2: blue, 0.45–0.51-µm wavelength
- Band 3: green, 0.53–0.59-µm wavelength
- Band 4: red, 0.64–0.67-µm wavelength
- Band 5: near-infrared, 0.85–0.88-µm wavelength
- Band 6: shortwave infrared 1, 1.57–1.65-µm wavelength
- Band 7: shortwave infrared 2, 2.11–2.29-µm wavelength
2.2.2. Reference Data
2.3. MESMA Procedure
2.3.1. Spectral Library: Candidate and Optimal Endmembers
2.3.2. Spectral Unmixing: Obtention of Fraction Images and Shade Normalization
2.4. Accuracy Assessment of MESMA Fraction Images
2.5. Diversity Analysis
3. Results
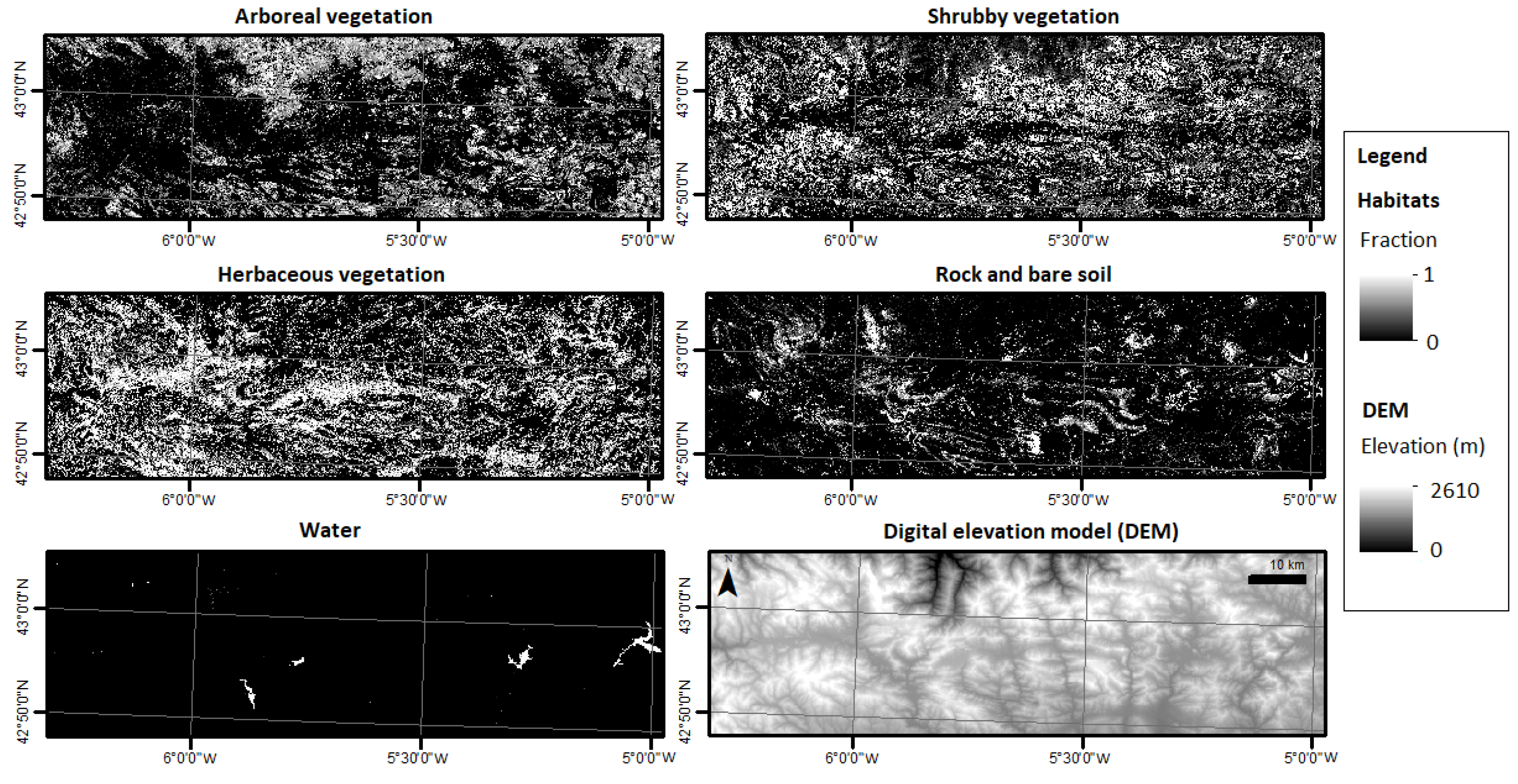
3.1. MESMA Results and Accuracy Assessment
3.1.1. Optimal Endmembers
3.1.2. Spectral Unmixing
3.1.3. Accuracy Assessment of MESMA Fraction Images
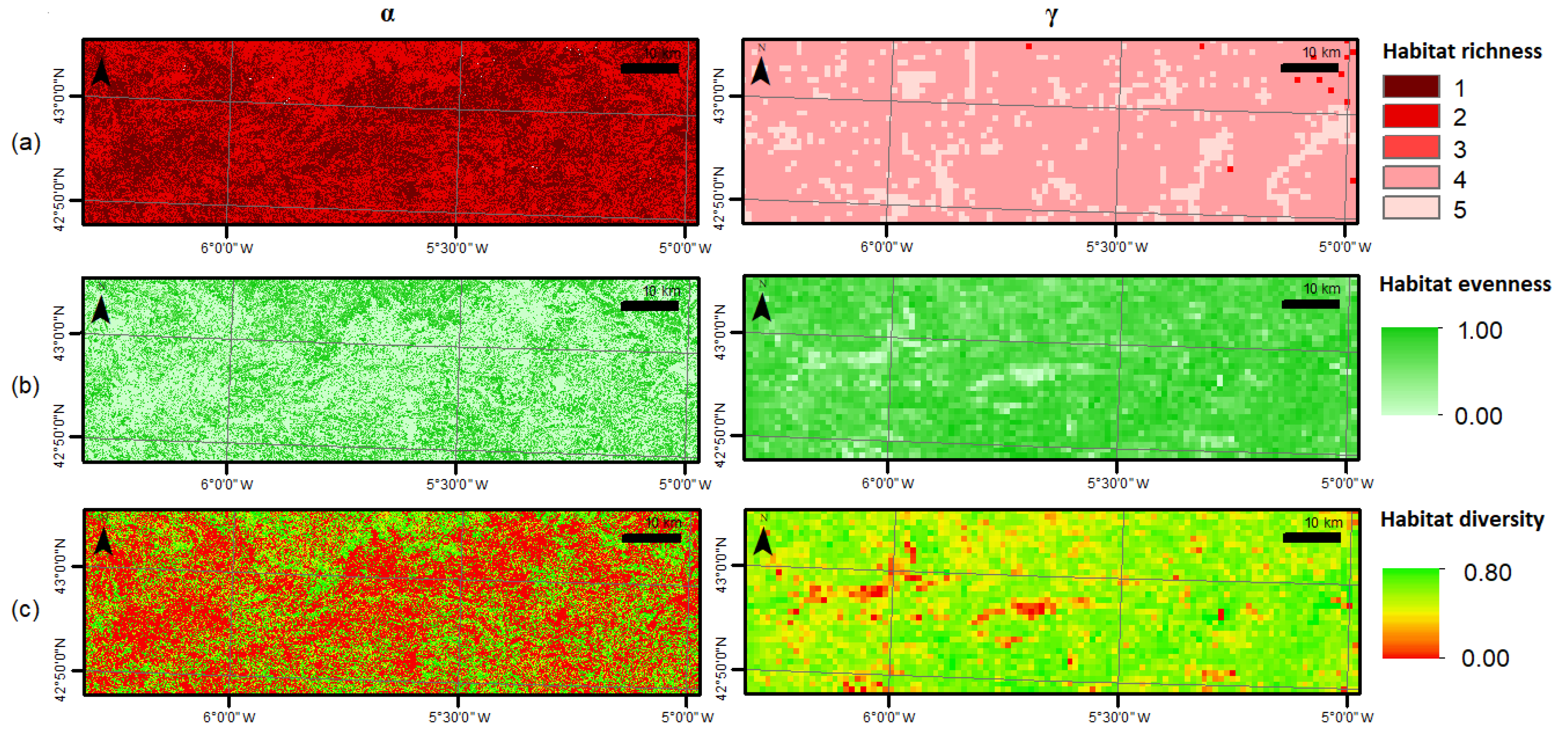
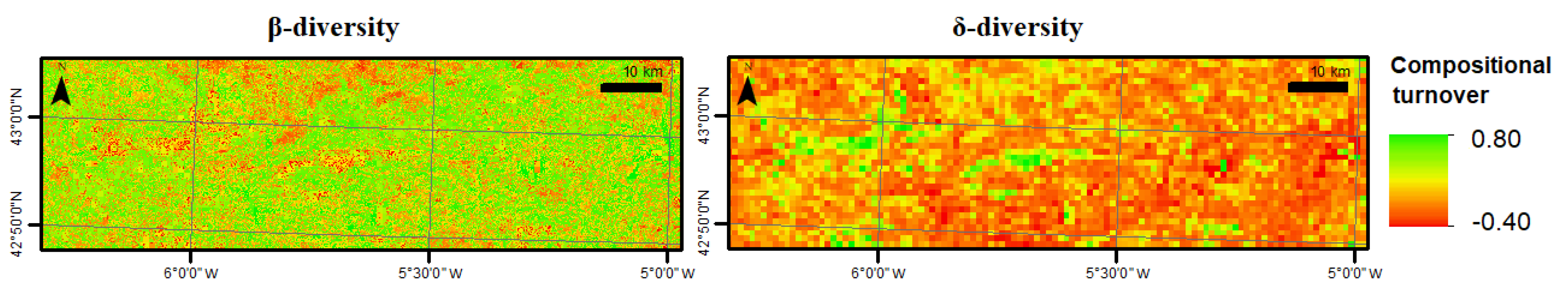
3.2. Habitat Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Pixels (n) | Pixels (%) | |
|---|---|---|
| Class models | ||
| Arboreal vegetation | 293,772 | 7.16 |
| Shrubby vegetation | 725,083 | 17.68 |
| Herbaceous vegetation | 892,727 | 21.77 |
| Rock (and bare soil) | 191,955 | 4.68 |
| Water | 20,409 | 0.50 |
| Arboreal and herbaceous | 438,983 | 10.70 |
| Arboreal, herbaceous and rock | 867 | 0.02 |
| Arboreal, herbaceous and water | 1 | 0.00 |
| Arboreal and rock | 136,413 | 3.33 |
| Arboreal, rock and water | 99 | 0.00 |
| Arboreal and shrubby | 769,141 | 18.76 |
| Arboreal and water | 454 | 0.01 |
| Herbaceous and rock | 331,387 | 8.08 |
| Herbaceous, rock and shrubby | 21 | 0.00 |
| Herbaceous, rock and water | 10 | 0.00 |
| Herbaceous and shrubby | 103,727 | 2.53 |
| Herbaceous and water | 244 | 0.01 |
| Rock and shrubby | 194,126 | 4.73 |
| Rock and water | 1394 | 0.03 |
| Shrubby and water | 125 | 0.00 |
| Total | 4,100,938 | 100 |
References
- Poiani, K.A.; Richter, B.D.; Anderson, M.G.; Richter, H.E. Biodiversity Conservation at Multiple Scales: Functional Sites, Landscapes, and Networks. BioScience 2000, 50, 133–146. [Google Scholar] [CrossRef]
- Léveque, C.; Mounlou, J.-C. Biodiversity; Wiley: Cornwall, UK, 2003; pp. 13–38. [Google Scholar]
- MacArthur, R.H.; MacArthur, J.W. On bird species diversity. Ecology 1961, 42, 594–598. [Google Scholar] [CrossRef]
- Tews, J.; Brose, U.; Grimm, V.; Tielborger, K.; Wichmann, M.C.; Schwager, M.; Jeltsch, F. Animal species diversity driven by habitat heterogeneity/diversity: The importance of keystone structures. J. Biogeogr. 2004, 31, 79–92. [Google Scholar] [CrossRef]
- Pacheco, R.; Vasconcelos, H.L. Habitat diversity enhances ant diversity in a naturally heterogeneous Brazilian landscape. Biodivers. Conserv. 2012, 21, 797–809. [Google Scholar] [CrossRef]
- Currat, M.; Ray, N.; Excoffier, L. SPLATCHE: A program to simulate genetic diversity taking into account environmental heterogeneity. Mol. Ecol. Notes 2004, 4, 139–142. [Google Scholar] [CrossRef]
- Vanbergen, A.J.; Watt, A.D.; Mitchell, R.; Truscott, A.-M.; Palmer, S.C.F.; Ivits, E.; Eggleton, P.; Jones, T.H.; Sousa, J.P. Scale-specific correlations between habitat heterogeneity and soil fauna diversity along a landscape structure gradient. Oecologia 2007, 153, 713–725. [Google Scholar] [CrossRef]
- García-Llamas, P.; Calvo, L.; De la Cruz, M.; Suárez-Seoane, S. Landscape heterogeneity as a surrogate of biodiversity in mountain systems: What is the most appropriate spatial analytical unit? Ecol. Indic. 2018, 85, 285–294. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1949. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Margalef, D.R. Information theory in ecology. Gen. Syst. 1958, 3, 36–71. [Google Scholar]
- Smith, B.; Wilson, J.B. A Consumer’s Guide to Evenness Indices. Oikos 1996, 76, 70–82. [Google Scholar] [CrossRef]
- Rocchini, D.; Delucchi, L.; Bacaro, G.; Cavallini, P.; Feilhauer, H.; Foody, G.M.; He, K.S.; Nagendra, H.; Porta, C.; Ricotta, C.; et al. Calculating landscape diversity with information-theory based indices: A GRASS GIS solution. Ecol. Inf. 2013, 17, 82–93. [Google Scholar] [CrossRef]
- Whittaker, R.J.; Willis, K.J.; Field, R. Scale and species richness: Towards a general, hierarchical theory of species diversity. J. Biogeogr. 2001, 28, 453–470. [Google Scholar] [CrossRef]
- McGarigal, K.; Wan, H.Y.; Zeller, K.A.; Timm, B.C.; Cushman, S.A. Multi-scale habitat selection modeling: A review and outlook. Landsc. Ecol. 2016, 31, 1161–1175. [Google Scholar] [CrossRef]
- Fox, B.J.; Fox, M.D. Factors determining mammal species richness on habitat islands and isolates: Habitat diversity, disturbance, species interactions and guild assembly rules. Glob. Ecol. Biogeogr. 2000, 9, 19–37. [Google Scholar] [CrossRef]
- Kallimanis, A.S.; Mazaris, A.D.; Tzanopoulos, J.; Halley, J.M.; Pantis, J.D.; Sgardelis, S.P. How does habitat diversity affect the species–area relationship? Glob. Ecol. Biogeogr. 2008, 17, 532–538. [Google Scholar] [CrossRef]
- Whittaker, R.H. Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 1960, 30, 279–338. [Google Scholar] [CrossRef]
- Shmida, A.; Wilson, M.V. Biological determinants of species diversity. J. Biogeogr. 1985, 12, 1–20. [Google Scholar] [CrossRef]
- Weber, D.; Schaepman-Strub, G.; Ecker, K. Predicting habitat quality of protected dry grasslands using Landsat NDVI phenology. Ecol. Indic. 2018, 91, 447–460. [Google Scholar] [CrossRef]
- Erinjery, J.; Singh, M.; Kent, R. Mapping and assessment of vegetation types in the tropical rainforests of the Western Ghats using multispectral Sentinel-2 and SAR Sentinel-1 satellite imagery. Remote Sens. Environ. 2018, 216, 345–354. [Google Scholar] [CrossRef]
- Clerici, N.; Weissteiner, C.J.; Gerard, F. Exploring the Use of MODIS NDVI-Based Phenology Indicators for Classifying Forest General Habitat Categories. Remote Sens. 2012, 4, 1781–1803. [Google Scholar] [CrossRef]
- Valerio, F.; Ferreira, E.; Godinho, S.; Pita, R.; Mira, A.; Fernandes, N.; Santos, S.M. Predicting Microhabitat Suitability for an Endangered Small Mammal Using Sentinel-2 Data. Remote Sens. 2020, 12, 562. [Google Scholar] [CrossRef]
- Schulz, C.; Koch, R.; Cierjacks, A.; Kleinschmit, B. Land change and loss of landscape diversity at the Caatinga phytogeographical domain—Analysis of pattern-process relationships with MODIS land cover products (2001–2012). J. Arid Environ. 2017, 136, 54–74. [Google Scholar] [CrossRef]
- Pan, Y.; Hu, T.; Zhu, X.; Zhang, J.; Wang, X. Mapping Cropland Distributions Using a Hard and Soft Classification Model. IEEE Trans. Geosci. Remote Sens. 2012, 50, 4301–4312. [Google Scholar] [CrossRef]
- Nichol, J.E.; Wong, M.S.; Corlett, R.; Nichol, D.W. Assessing avian habitat fragmentation in urban areas of Hong Kong (Kowloon) at high spatial resolution using spectral unmixing. Landsc. Urban Plan. 2010, 95, 54–60. [Google Scholar] [CrossRef]
- Somers, B.; Asner, G.P.; Tits, L.; Coppin, P. Endmember variability in Spectral Mixture Analysis: A review. Remote Sens. Environ. 2011, 115, 1603–1616. [Google Scholar] [CrossRef]
- Roberts, D.A.; Gardner, M.; Church, R.; Ustin, S.; Scheer, G.; Green, R.O. Mapping Chaparral in the Santa Monica Mountains Using Multiple Endmember Spectral Mixture Models. Remote Sens. Environ. 1998, 65, 267–279. [Google Scholar] [CrossRef]
- Quintano, C.; Fernández-Manso, A.; Roberts, D. Burn severity mapping from Landsat MESMA fraction images and Land Surface Temperature. Remote Sens. Environ. 2017, 190, 83–95. [Google Scholar] [CrossRef]
- Fernandez-Guisuraga, J.M.; Calvo, L.; Suárez-Seoane, S. Comparison of pixel unmixing models in the evaluation of post-fire forest resilience based on temporal series of satellite imagery at moderate and very high spatial resolution. ISPRS J. Photogramm. Remote Sens. 2020, 164, 217–228. [Google Scholar] [CrossRef]
- Fernández-Manso, A.; Quintano, C.; Roberts, D. Evaluation of potential of multiple endmember spectral mixture analysis (MESMA) for surface coal mining affected area mapping in different world forest ecosystems. Remote Sens. Environ. 2012, 127, 181–193. [Google Scholar] [CrossRef]
- Schaaf, A.N.; Dennison, P.E.; Fryer, G.K.; Roth, K.L.; Roberts, D.A. Mapping Plant Functional Types at Multiple Spatial Resolutions Using Imaging Spectrometer Data. GISci. Remote Sens. 2011, 48, 324–344. [Google Scholar] [CrossRef]
- Quintano, C.; Fernández-Manso, A.; Calvo, L.; Roberts, D.A. Vegetation and Soil Fire Damage Analysis Based on Species Distribution Modeling Trained with Multispectral Satellite Data. Remote Sens. 2019, 11, 1832. [Google Scholar] [CrossRef]
- Dashti, H.; Poley, A.; Glenn, N.F.; Ilangakoon, N.; Spaete, L.; Roberts, D.; Enterkine, J.; Flores, A.; Ustin, S.; Mitchell, J. Regional Scale Dryland Vegetation Classification with an Integrated Lidar-Hyperspectral Approach. Remote Sens. 2019, 11, 2141. [Google Scholar] [CrossRef]
- Lyu, X.; Li, X.; Dang, D.; Dou, H.; Xuan, X.; Liu, S.; Li, M.; Gong, J. A new method for grassland degradation monitoring by vegetation species composition using hyperspectral remote sensing. Ecol. Indic. 2020, 114, 106310. [Google Scholar] [CrossRef]
- Powell, R.L.; Roberts, D.A.; Dennison, P.E.; Hess, L.L. Sub-pixel mapping of urban land cover using multiple endmember spectral mixture analysis: Manaus, Brazil. Remote Sens. Environ. 2007, 106, 253–267. [Google Scholar] [CrossRef]
- Degerickx, J.; Okujeni, A.; Iordache, M.-D.; Hermy, M.; Van der Linden, S.; Somers, B. A Novel Spectral Library Pruning Technique for Spectral Unmixing of Urban Land Cover. Remote Sens. 2017, 9, 565. [Google Scholar] [CrossRef]
- Degerickx, J.; Roberts, D.A.; Somers, B. Enhancing the performance of Multiple Endmember Spectral Mixture Analysis (MESMA) for urban land cover mapping using airborne lidar data and band selection. Remote Sens. Environ. 2019, 221, 260–273. [Google Scholar] [CrossRef]
- Miller, D.L.; Alonzo, M.; Roberts, D.A.; Tague, C.; McFadden, J.P. Drought Response of Urban Trees and Turfgrass using Airborne Imaging Spectroscopy. Remote Sens. Environ. 2020, 240, 111646. [Google Scholar] [CrossRef]
- Ninyerola, M.; Pons, X.; Roure, J.M. Atlas Climático Digital de la Península Ibérica. Metodología y Aplicaciones en Bioclimatología y Geobotánica; Universidad Autónoma de Barcelona: Barcelona, Spain, 2005; Available online: http://www.opengis.uab.es/wms/iberia/mms/index.htm (accessed on 1 December 2020).
- Morán-Ordóñez, A.; Suárez-Seoane, S.; Calvo, L.; de Luis, E. Using predictive models as a spatially explicit support tool for managing cultural landscapes. Appl. Geogr. 2011, 31, 839–848. [Google Scholar] [CrossRef]
- García, D.; Quevedo, M.; Obeso, J.R.; Abajo, A. Fragmentation patterns and protection of montane forest in the Cantabrian range (NW Spain). For. Ecol. Manag. 2005, 208, 29–43. [Google Scholar] [CrossRef]
- Blanco-Fontao, B.; Quevedo, M.; Obeso, J.R. Abandonment of traditional uses in mountain areas: Typological thinking versus hard data in the Cantabrian Mountains (NW Spain). Biodivers. Conserv. 2011, 20, 1133–1140. [Google Scholar] [CrossRef]
- Morán-Ordóñez, A.; Suárez-Seoane, S.; Marcos, E.; de Luis, E.; Calvo, L. The heathland economy in South-West Europe: Cantabrian Mountain (Spain). In Economy and Ecology of Heathlands; Diemont, A., Heijman, W.J.M., Spiel, H., Webb, N.R., Eds.; KNNV Publishing: Zeist, The Netherlands, 2013; pp. 93–104. [Google Scholar] [CrossRef]
- Council Directive 92/43/EEC of 21 May 1992 on the Conservation of Natural Habitats and of Wild Fauna and Flora. Off. J. Eur. Commun. 1992, 206, 7–50. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:31992L0043&from=EN (accessed on 1 December 2020).
- Álvarez-Martínez, J.M.; Suárez-Seoane, S.; De Luis Calabuig, E. Modelling the risk of land cover change from environmental and socio-economic drivers in heterogeneous and changing landscapes: The role of uncertainty. Landsc. Urban Plan. 2011, 101, 108–119. [Google Scholar] [CrossRef]
- García-Llamas, P.; Geijzendorffer, I.R.; García-Nieto, A.P.; Calvo, L.; Suárez-Seoane, S. Impact of land cover change on ecosystem service supply in mountain systems: A case study in the Cantabrian Mountains (NW of Spain). Reg. Environ. Chang. 2019, 19, 529–542. [Google Scholar] [CrossRef]
- US. Geological Survey and EROS Data Center. EarthExplorer; U.S. Dept. of the Interior, U.S. Geological Survey: Reston, VA, USA, 2020. Available online: https://earthexplorer.usgs.gov/ (accessed on 1 December 2020).
- USGS. Landsat 8 Collection 1 (C1) Land Surface Reflectance Code (LaSRC) Product Guide. Version 3.0; Department of the Interior U.S. Geological Survey: Sioux Falls, SD, USA, 2020.
- Instituto Geográfico Nacional y el Centro Nacional de Información Geográfica. Plan Nacional de Ortofotografía Aérea; Ministerio de Transportes, Movilidad y Agenda Urbana: Madrid, Spain, 2020. Available online: https://pnoa.ign.es/ (accessed on 1 December 2020).
- Huck, J.; Turtle, F. Polygon Divider (Version 0.6). 2020. Available online: https://github.com/jonnyhuck/RFCL-PolygonDivider (accessed on 1 December 2020).
- QGIS Development Team. QGIS Geographic Information System; Open Source Geospatial Foundation Project. (Version 3.14.16). 2020. Available online: http://qgis.osgeo.org (accessed on 1 December 2020).
- Quintano, C.; Fernández-Manso, A.; Roberts, D.A. Multiple Endmember Spectral Mixture Analysis (MESMA) to map burn severity levels from Landsat images in Mediterranean countries. Remote Sens. Environ. 2013, 136, 76–88. [Google Scholar] [CrossRef]
- Crabbé, A.H.; Jakimow, B.; Somers, B.; Roberts, D.A.; Halligan, K.; Dennison, P.; Dudley, K. Spectral Library QGIS Plugin (Version 1.0.9). 2020. Available online: https://bitbucket.org/kul-reseco/spectral-libraries (accessed on 1 December 2020).
- Tane, Z.; Roberts, D.; Veraverbeke, S.; Casas, A.; Ramirez, C.; Ustin, S. Evaluating Endmember and Band Selection Techniques for Multiple Endmember Spectral Mixture Analysis using Post-Fire Imaging Spectroscopy. Remote Sens. 2018, 10, 389. [Google Scholar] [CrossRef]
- Roberts, D.A.; Dennison, P.E.; Gardner, M.; Hetzel, Y.; Ustin, S.L.; Lee, C. Evaluation of the potential of hyperion for fire danger assessment by comparison to the airborne visible/infrared imaging spectrometer. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1297–1310. [Google Scholar] [CrossRef]
- Roberts, D.A.; Quattrochi, D.A.; Hulley, G.C.; Hook, S.J.; Green, R.O. Synergies between VSWIR and TIR data for the urban environment: An evaluation of the potential for the hyperspectral infrared imager (HyspIRI) decadal survey mission. Remote Sens. Environ. 2012, 117, 83–101. [Google Scholar] [CrossRef]
- Crabbé, A.H.; Somers, B.; Roberts, D.A.; Halligan, K.; Dennison, P.; Dudley, K. MESMA QGIS Plugin (Version 1.0.7). 2020. Available online: https://bitbucket.org/kul-reseco/mesma (accessed on 1 December 2020).
- Jurgiel, B. Point Sampling Tool (Version 0.5.3). 2020. Available online: http://github.com/borysiasty/pointsamplingtool (accessed on 1 December 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 1 December 2020).
- Brewer, W.L.; Lippitt, C.L.; Lippitt, C.D.; Litvak, M.E. Assessing drought-induced change in a piñon-juniper woodland with Landsat: A multiple endmember spectral mixture analysis approach. Int. J. Remote Sens. 2017, 38, 4156–4176. [Google Scholar] [CrossRef]
- Roth, K.L.; Dennison, P.E.; Roberts, D.A. Comparing endmember selection techniques for accurate mapping of plant species and land cover using imaging spectrometer data. Remote Sens. Environ. 2012, 127, 139–152. [Google Scholar] [CrossRef]
- Roth, K.L.; Roberts, D.A.; Dennison, P.E.; Alonzo, M.; Peterson, S.H.; Beland, M. Differentiating plant species within and across diverse ecosystems with imaging spectroscopy. Remote Sens. Environ. 2015, 167, 135–151. [Google Scholar] [CrossRef]
- Rashed, T.; Weeks, J.; Roberts, D.; Rogan, J.; Powell, R. Measuring the physical compositions of urban morphology using multiple endmember spectral mixture models. Photogramm. Eng. Remote Sens. 2003, 69, 1011–1020. [Google Scholar] [CrossRef]
- McWire, K.; Minor, T.; Fenstermaker, L. Hyperspectral Mixture Modeling for Quantifying Sparse Vegetation Cover in Arid Environments. Remote Sens. Environ. 2000, 72, 360–374. [Google Scholar] [CrossRef]
- Chen, X.; Li, L. A Comparison of Spectral Mixture Analysis Methods for Urban Landscape Using LANDSAT ETM+ Data: Los Angeles, CA; The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences: Beijing, China, 2008. [Google Scholar]
- Somers, B.; Delalieux, S.; Verstraeten, W.W.; Verbessel, J.; Lhermitte, S.; Coppin, P. Magnitude- and Shape-Related Feature Integration in Hyperspectral Mixture Analysis to Monitor Weeds in Citrus Orchards. IEEE Trans. Geosci. Remote Sens. 2009, 47, 3630–3642. [Google Scholar] [CrossRef]
- Zhang, J.; Rivard, B.; Sanchez-Azofeifa, A. Derivative spectral unmixing of hyperspectral data applied to mixtures of lichen and rock. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1934–1940. [Google Scholar] [CrossRef]
- Tits, L.; Keersmaecker, W.D.; Somers, B.; Asner, G.P.; Farifteh, J.; Coppin, P. Hyperspectral shape-based unmixing to improve intra- and interclass variability for forest and agro-ecosystem monitoring. ISPRS J. Photogramm. Remote Sens. 2012, 74, 163–174. [Google Scholar] [CrossRef]
- Fernández-García, V.; Quintano, C.; Taboada, A.; Marcos, E.; Calvo, L.; Fernández-Manso, A. Remote Sensing Applied to the Study of Fire Regime Attributes and Their Influence on Post-Fire Greenness Recovery in Pine Ecosystems. Remote Sens. 2018, 10, 733. [Google Scholar] [CrossRef]
- Dudley, K.L.; Dennison, P.E.; Roth, K.L.; Roberts, D.A.; Coates, A.R. A multi-temporal spectral library approach for mapping vegetation species across spatial and temporal phenological gradients. Remote Sens. Environ. 2015, 167, 121–134. [Google Scholar] [CrossRef]
- Mitraka, Z.; Del Frate, F.; Carbone, F. Nonlinear Spectral Unmixing of Landsat Imagery for Urban Surface Cover Mapping. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3340–3350. [Google Scholar] [CrossRef]
- Yu, J.; Chen, D.; Lin, Y.; Ye, S. Comparison of linear and nonlinear spectral unmixing approaches: A case study with multispectral TM imagery. Int. J. Remote Sens. 2017, 38, 773–795. [Google Scholar] [CrossRef]




| Pixels (n) | Pixels (%) | Cover (Mean ± SD) | |
|---|---|---|---|
| Classes | |||
| Arboreal vegetation | 1,639,730 | 39.98 | 23.96 ± 33.59 |
| Shrubby vegetation | 1,792,223 | 43.70 | 31.49 ± 39.86 |
| Herbaceous vegetation | 1,767,967 | 43.11 | 33.93 ± 42.50 |
| Rock and bare soil | 856,272 | 20.88 | 10.05 ± 24.51 |
| Water | 22,736 | 0.55 | 0.53 ± 7.16 |
| Class models | |||
| One-habitat type model | 2,123,946 | 51.79 | |
| Two-habitat type model | 1,975,994 | 48.18 | |
| Three-habitat type model | 998 | 0.02 | |
| Total | 4,100,938 | 100 |
| Diversity Metric | Spatial Scale (km2) | Value (Mean ± SD) |
|---|---|---|
| α-richness | 0.0009 | 1.48 ± 0.50 |
| γ-richness | 1 | 4.15 ± 0.36 |
| ε-richness | 3630 | 5.00 ± 0.00 |
| α-evenness * | 0.0009 | 0.40 ± 0.43 * |
| γ-evenness | 1 | 0.80 ± 0.12 |
| ε-evenness | 3630 | 0.90 ± 0.00 |
| α-diversity | 0.0009 | 0.20 ± 0.22 |
| γ-diversity | 1 | 0.60 ± 0.09 |
| ε-diversity | 3630 | 0.72 ± 0.00 |
| β-diversity | 0.0009 | 0.40 ± 0.23 |
| δ-diversity | 1 | 0.11 ± 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-García, V.; Marcos, E.; Fernández-Guisuraga, J.M.; Fernández-Manso, A.; Quintano, C.; Suárez-Seoane, S.; Calvo, L. Multiple Endmember Spectral Mixture Analysis (MESMA) Applied to the Study of Habitat Diversity in the Fine-Grained Landscapes of the Cantabrian Mountains. Remote Sens. 2021, 13, 979. https://doi.org/10.3390/rs13050979
Fernández-García V, Marcos E, Fernández-Guisuraga JM, Fernández-Manso A, Quintano C, Suárez-Seoane S, Calvo L. Multiple Endmember Spectral Mixture Analysis (MESMA) Applied to the Study of Habitat Diversity in the Fine-Grained Landscapes of the Cantabrian Mountains. Remote Sensing. 2021; 13(5):979. https://doi.org/10.3390/rs13050979
Chicago/Turabian StyleFernández-García, Víctor, Elena Marcos, José Manuel Fernández-Guisuraga, Alfonso Fernández-Manso, Carmen Quintano, Susana Suárez-Seoane, and Leonor Calvo. 2021. "Multiple Endmember Spectral Mixture Analysis (MESMA) Applied to the Study of Habitat Diversity in the Fine-Grained Landscapes of the Cantabrian Mountains" Remote Sensing 13, no. 5: 979. https://doi.org/10.3390/rs13050979
APA StyleFernández-García, V., Marcos, E., Fernández-Guisuraga, J. M., Fernández-Manso, A., Quintano, C., Suárez-Seoane, S., & Calvo, L. (2021). Multiple Endmember Spectral Mixture Analysis (MESMA) Applied to the Study of Habitat Diversity in the Fine-Grained Landscapes of the Cantabrian Mountains. Remote Sensing, 13(5), 979. https://doi.org/10.3390/rs13050979












