Abstract
Modern plant phenotyping requires tools that are robust to noise and missing data, while being able to efficiently process large numbers of plants. Here, we studied the skeletonization of plant architectures from 3D point clouds, which is critical for many downstream tasks, including analyses of plant shape, morphology, and branching angles. Specifically, we developed an algorithm to improve skeletonization at branch points (forks) by leveraging the geometric properties of cylinders around branch points. We tested this algorithm on a diverse set of high-resolution 3D point clouds of tomato and tobacco plants, grown in five environments and across multiple developmental timepoints. Compared to existing methods for 3D skeletonization, our method efficiently and more accurately estimated branching angles even in areas with noisy, missing, or non-uniformly sampled data. Our method is also applicable to inorganic datasets, such as scans of industrial pipes or urban scenes containing networks of complex cylindrical shapes.
1. Introduction
The skeleton of a 3D object is a thinned 1D representation (often in the form of a graph) that captures its basic geometry and shape. Skeletons have broad applications in computer vision, such as for object matching, surface reconstruction, feature tracking, and computer animation [1]. Accordingly, there are many approaches for extracting skeleton graphs from 3D meshes or point clouds [2,3,4,5,6,7,8,9].
More recently, skeletonization has become critical to modern plant phenotyping [10,11,12]. The advent of high-throughput 3D imaging platforms, such as light detection and ranging (LiDAR), has generated large datasets of point clouds of plant shoot architectures [13,14]. Fast and accurate determination of branch angles is important for many downstream tasks, including the study of plant growth and development [15], measuring light interception by leaves [16], and for inferring genotype-to-phenotype relationships [17], both in the lab and in the field [18,19]. In addition, assessing and quantifying changes in plant morphology is often used for measuring agricultural yields [20], analyzing stress responses and plant-environment interactions [21], and facilitating functional genomics studies [22]. Plant skeletonization is also important in forestry [23], ecology [24], urban planning [25], and engineering [26]. All of these applications require methods for accurately skeletonizing shapes from 3D data.
Compared to man-made structures, skeletonizing natural, organic structures has many challenges, including dealing with the curvature of branches (i.e., no straight lines), the tapering of branches (i.e., unequal radii over the length of the branch), the non-uniformity of branch lengths, and other nuisances, such as trichomes (the fine hairs on plant stems), which disrupt fitting with simple primitives. These issues lie alongside the usual difficulties associated with processing point clouds, such as missing data, non-uniform point density, and registration errors [27,28,29]. As a result, while advancements in data acquisition have made it possible to rapidly scan plants, methods for analyzing them in real-time remain a bottleneck.
Here, we describe a fast skeletonization method that can generate accurate branch angles from noisy 3D point clouds of shoot architectures. Our method builds upon our previous work [30] by analyzing the geometry of normal vectors around branch points, projecting these vectors onto a Gaussian sphere, and then robustly clustering them into unique individual branches that better align with the underlying geometry.
As plant shapes vary across species and are highly plastic to environmental changes, we tested our method on a dataset of two species (tomato and tobacco) grown under five conditions (ambient light, shade, high-heat, high-light, and drought), and scanned at multiple developmental time points, to assess the ability of our method to handle natural variability. Compared to three existing skeletonization methods (Laplacian-based contraction [3], -medial skeletonization [7], and PypeTree [5]), the branch angles we generated were, on average, 1.71 to 2.69 times (in absolute error) and 1.61 to 2.48 times (in percent error) more accurate. Finally, our method runs in seconds, making it amenable to real-time phenotyping applications.
Related Work
Several methods for extracting skeleton graphs from 3D data are designed to function on closed polygonal meshes. However, it is not straightforward to reliably produce watertight shapes from point clouds due to challenges posed by self-occlusion and light reflection by the target surface [2,29]. Surface reconstruction is often unreliable, since the errors introduced can lead to inaccurate skeletons [3]. Therefore, we focused on approaches that can be applied directly to unorganized 3D point clouds.
Algorithms for skeleton extraction from unorganized 3D point clouds broadly fall into four categories: topological thinning, distance field-based, general field-based, or geometric [1]. Some approaches, such as topological thinning and distance fields, typically work on voxelized data. However, these methods are sensitive to the discretization resolution used, and they can have poor results when the point cloud is noisy, non-uniform, or incomplete. In practice, some methods combine elements from multiple categories, and thus the distinctions between categories are not strict.
Topological thinning or contraction algorithms start at the surface of an object and shrink it to a 1D skeletal representation [1]. This is done by iteratively removing voxels from the shape’s boundary while preserving the basic topology. As each voxel can be evaluated locally, thinning algorithms are generally computationally efficient. However, they are susceptible to the excessive removal of branch endpoints, which can lead to truncated skeletons [1]. Au et al. [4] developed a mesh contraction algorithm for non-volumetrized data that uses the Laplacian operator to smoothly collapse an object into a skeletal shape. Subsequently, Cao et al. [3] adapted this Laplacian thinning approach to work directly on raw point clouds. However, this method requires careful tuning of contraction parameters to remain faithful to the object’s topology and is sensitive to the sampling density. As the Laplacian operator requires the calculation of higher-order derivatives, it is also computationally expensive and not guaranteed to be numerically stable.
Distance field-based methods apply the distance transform, which identifies the shortest distance from each computed interior point to the boundary of the object [31]. Any locally-centered voxels are putative members of the shape’s skeleton, which are subsequently pruned by thinning or clustering. The remaining voxels are re-connected to produce a final skeleton graph. Although these methods are efficient, they are sensitive to boundary noise and suffer from poor centeredness due to errors at the re-connecting step [1]. Huang et al. [7] applied a spatially-localized version of the -median to local neighborhoods of points, and then applied contraction and re-centering to generate a final skeleton. However, this method is sensitive to sampling density and uniformity. Some methods for trees first apply the -median [32] or other distance-based measures [33] to extract a coarse skeleton, which is then used to guide optimization to recover missing data from the point cloud. This new information is then used to refine the original skeleton.
General field-based approaches apply functions other than the distance transform to point sets, such as potential fields or Newtonian repulsion [1]. For example, Sharf et al. [8] performed surface reconstruction by growing a deformable blob that captures the point cloud’s shape and then extracts a skeleton by finding its centerline. In general, these methods tend to be robust to noise at object boundaries. However, if the data is incomplete (e.g., if there are holes or sparsely sampled areas), the blob can leak outside of the underlying shape. In addition, some of these methods require calculating higher-order derivatives and, thus, may be inefficient for large point clouds.
Geometric methods encompass approaches that work directly on raw point cloud data. For example, Voronoi tesselation can produce an approximation of a shape’s medial surface [34,35], which can then be pruned into a skeleton. However, this process is highly sensitive to noise [1]. Alternatively, Reeb graphs are more robust to outliers but can be sensitive to the object’s orientation [1]. PypeTree [5], which builds off the method by Verroust and Lazarus [6], is designed specifically for point clouds of plant architectures. It calculates geodesic distances from the plant’s root to the rest of the point cloud. Skeleton points are then extracted from the centroids of the points in each level set. However, if a level set spans a fork, points that belong to multiple branches will not be differentiated. Therefore, PypeTree can produce geometric errors and poor angle measurements at forks. ROSA [2] uses normal information and rotational symmetry to extract skeletons from point clouds that have an underlying cylindrical shape. ROSA performs well on incomplete point clouds; however, it is generally less robust to noise and outliers, requiring preprocessing and denoising before it can be applied to raw data [2].
2. Methods
2.1. Overview
As input, we are given a point cloud of m points, where each three-dimensional point describes a location on the surface of the plant. Each has an associated normal vector representing its 3D orientation; these vectors form a set of normals . In our dataset, normal information was provided by the 3D scanner.
Our method can be divided into two parts (Figure 1): computation of the initial skeleton, and refinement of the skeleton at branch points (forks). The first part is done using the approach by Ziamtsov and Navlakha [30], which builds off of PypeTree [5]. We refer to the improved version as PypeTree*, to distinguish it from the original method [5].
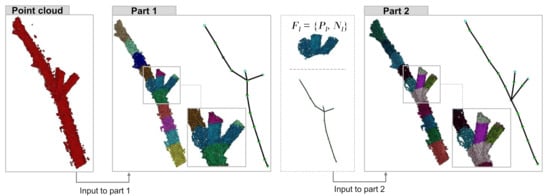
Figure 1.
Overview. As input, we are provided a 3D point cloud of a plant architecture. A partial point cloud of the region around a branch point (fork) is shown. In Part 1, we compute its skeleton. Each color represents points and normal vectors that belong to a node in the graph. In Part 2, the geometry and branch angles around a fork are refined to better match the underlying plant shape.
Briefly, PypeTree* divides the points P into level sets, where each level l consists of points that lie within a similar geodesic distance from the root of the tree. For each level, a node of the skeleton graph is created at the centroid of that level’s points. For example, Figure 1 (Part 1) shows nodes (i.e., levels) in different colors, with their associated points. Forks are detected by running a connected components algorithm on ; if consists of, say, two disconnected components—points on the left and right of the fork, each with a similar geodesic distance to the root—then the level is split into two, and two nodes are created, one at the centroid of each component. The output is a skeleton graph , where the nodes over all l; each level has an associated set of points and normal vectors ; and the edges E connect nodes across successive levels.
This approach is fairly fast on large point clouds; however, it often suffers from geometric errors at forks. Even if there is a fork at level l, in practice, often consists of only one component, and it is not until level where the two branches of the fork are sufficiently separated from each other that their corresponding points no longer belong to the same component. In terms of the skeleton, this means that generates a single node in the graph, and generates two nodes, each connected to the centroid of . This severely compromises the angle estimation at the fork, since the branch point is detected late. Furthermore, the location of the centroid can be biased (due to averaging of points that belong to different branches), leading to inaccurately-placed nodes within forks, and again, incorrect angles.
Thus, the second part of our method refines the skeleton geometry around each fork, using as input the points and normal vectors associated with each fork (Figure 1 (Part 2)). If normals are unavailable, existing methods for normal estimation [36,37] can be applied before using our method.
2.2. Gaussian Sphere Mapping
The points (Figure 2A) and normal vectors for each fork node l are processed as follows. For simplicity, we drop the subscript l from the notation below, since the same logic is applied to each fork node/level l. We do not require that forks consist of exactly two children (branches).
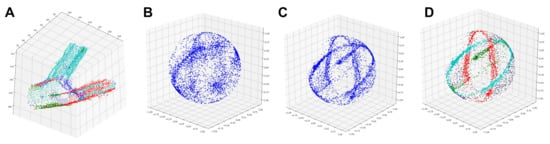
Figure 2.
Gaussian spheres. (A) Points that correspond to an example fork node. (B) Mapping of normal vectors to a Gaussian sphere. (C) Mapping of smoothed normals to a Gaussian sphere. (D) Correspondence between colored rings and original branches in panel A.
First, for each normal vector associated with a point in the level, we compute its local neighborhood of points j where , where r defines the radius of the neighborhood. We then compute a smoothed version of the normal vector by averaging the normal vectors around its local neighborhood:
This reduces the normal vector noise around the fork.
Second, we map each to a Gaussian sphere. Every normal is a unit vector, and thus if we imagine it as a position, it will lay on the surface of a unit sphere. Mapping all normals onto a unit sphere will produce a Gaussian sphere. Comparing Figure 2B (unsmoothed) to Figure 2C (smoothed) shows how smoothening tightens the spread of normal vectors and generates a clear pattern of rings. All normal vectors that map onto the same ring belong to the same cylinder, which represents a branch of the fork. Assuming all branches have an approximately cylindrical shape, the goal is to identify the normal vectors that belong to different cylinders/branches. The number of rings to identify is set to the number of children of the fork node in the skeleton graph from Part 1.
For example, the fork in Figure 2A has three cylinders (a mother branch and two daughter branches), which matches the three rings we observe in Figure 2C; these rings are color-coded in Figure 2D to match their original branches.
Finding rings in a Gaussian sphere can be challenging when there is missing data, such as for the mother branch (green) in Figure 2A. As a result, the ring of green points is incomplete in Figure 2C,D. Next, we discuss how to extract rings that correspond to different cylinders/branches from a Gaussian sphere.
2.3. Sampling of Cylinder Axes
We extract rings from a Gaussian sphere (Figure 3A) by investigating the cross product between normal vectors:
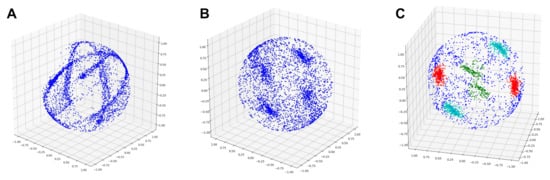
Figure 3.
Sampling of cylinder axes. (A) Mapping of smoothed normals to a Gaussian sphere, for an example fork. (B) Cylinder axes identified using Pipe-run (random sampling). (C) Cylinder axes identified using our sampling approach. All rings are represented by well-separated clusters, including the ring with missing data.
If the cross product is taken between two normal vectors that belong to the same ring, then is a perpendicular vector that points along the axis of the cylinder/branch. If we continue taking cross products between normal vectors that belong to the same ring and mapping them onto the Gaussian sphere, clear clusters will form on the Gaussian sphere. However, the problem is that we do not know a priori which normal vectors belong to the same ring.
Previously, a method called Pipe-run [26] was proposed to extract pipes from noisy point clouds using Gaussian spheres. Pipe-run draws random pairs of normal vectors and computes their cross products, but this method has several shortcomings. First, it assumes that most of the randomly drawn normal vectors come from the same ring, which works well when the input contains a single cylinder. However, in our case, there are multiple rings, each with a different orientation. Second, each cylinder may not have the same number of points, since branches could vary in thickness and size. Thus, sampling an equal number of points per cylinder could produce spurious axes. Third, cylinders with missing data would be poorly represented by random sampling. Indeed, Figure 3B shows that Pipe-run sampling does not effectively cluster the three cylinders. Other methods, such as RANSAC [38], are also highly susceptible to noise and missing data.
Our sampling approach is based on two key observations. First, pairs of normal vectors that belong to the same ring likely have corresponding points that lie close to each other in space. Therefore, we sample pairs of points that lie within a local neighborhood, defined by a radius r. Second, pairs of normal vectors that belong to the same ring will lie on a circle with approximately the same radius. To compute the approximate radius between a point and every point in its neighborhood, we used a method called the radius-based surface descriptor (RSD) [39]:
The RSD takes two points and and their normal vectors as input and computes the approximate radius of an assumed circle, if the points were lying on the circle. For example, if the points represented a perfect cylinder, would be constant over all pairs. In reality, the data is noisy and the approximated radii tend to vary.
Given these observations, our sampling method is as follows. Given a point , we compute its approximate radius with all points that lie within a distance r from . We then sort all the approximate radii and choose the point associated with the median radius. Finally, we compute the cross product between the smoothed normal vectors of points and . This results in a single sample per point, and a robust cylinder axis supported by ’s neighborhood.
Figure 3C shows the results of our sampling method, which reduces noise. There are six clusters corresponding to the three rings, because the sign of the cross product depends on the order of operation; thus, we generally expect two opposite vectors for each ring.
2.4. Clustering Cylinder Axes
The previous step generates a set consisting of an for each point i. Our next goal is to cluster the values in such that each cluster corresponds to a different cylinder axis. We performed unsupervised clustering using the mean-shift algorithm [40] because it deals well with outliers [26]. The algorithm identifies clusters as locations in space with high sampling density. Formally, the algorithm iterates as follows:
where is the current point of highest density, is the previous point, and is the radial basis kernel function (RBF). Initially, is selected randomly from . After every iteration, is re-projected back to the surface of the sphere. The process stops when the distance between the current and previous point is very small (e.g., <). The number of clusters is known from the skeleton graph (i.e., the number of branches at the fork).
Once the process converges, we obtain a vector , which represents the cylinder axis of one branch i. Before searching for the next axis, we perform two steps to find additional points that belong to the same axis. First, we find all normal vectors that are within 10 degrees of being perpendicular to . The resulting normal vectors and their corresponding points constitute a ring/cylinder. Second, we remove from set all that are within 10 degrees of . We then repeat the clustering procedure on a new random point to find the next cylinder axis.
2.4.1. A Modified Clustering Algorithm
We modified this algorithm in two ways for better performance for our application. First, the RBF computes , but as mentioned above, the sign of the cross-product can be inverted based on the order of the points. Thus, vectors that lie on opposite sides belong to the same axis. To account for this, we make the following adjustment:
This allows for more precise computation of the direction of higher density.
Second, we reduce (a kernel parameter) as the number of iterations increases. A large prevents the algorithm from getting stuck at a local high density location initially; then, as the algorithm settles, we reduce to hone in on the highest density location more precisely. This is akin to reducing the learning rate over time during gradient descent optimization. Specifically, when the distance between current and previous step falls below a threshold , we reduce by an order of magnitude. If the distance becomes larger than , then we continue iterating with the new . Otherwise (if the distance remains below ), we say that the algorithm has converged. This dynamic adjustment of becomes more important in forks with higher numbers of branches.
2.4.2. Dealing with Ambiguous Points
One final issue remains: each ring (cylinder) of the Gaussian sphere intersects with at least one other ring in two places, and it is unclear to which ring the points at these intersections should be assigned. We call such points ambiguous. In other words, when we extract normal vectors that are with 10 degrees of being perpendicular to cylinder axis , there are multiple axes that satisfy this rule. The more branches a fork has, the more intersections there are and the more ambiguity there will be. Figure 4A shows another example fork, and Figure 4B highlights its ambiguous points in red.
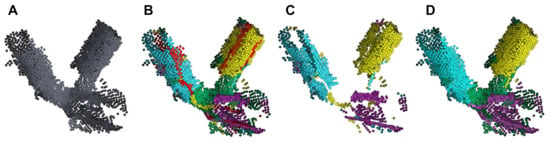
Figure 4.
Clustering of ambiguous points. (A) Initial fork points. (B) Results after initial clustering. Cyan, yellow, purple: non-ambiguous points (i.e., points assigned to a single cylinder axis). Red: ambiguous points. Green: noise (i.e., points not assigned to any ring). (C) Non-ambiguous points only. (D) Final clustering after resolving the assignment of ambiguous points to a single axis/branch. The cylinder accuracy is improved.
To resolve ambiguous points into their correct branches, we perform the following steps. First, we compute the centroid over all non-ambiguous points in ring i. Non-ambiguous points are those that were clustered to only one cylinder axis. These points are shown in Figure 4B (cyan, yellow, and purple points). Points that were not within 10 degrees of being perpendicular to any are marked as noise and shown in green. Second, viewing each i as a plane with corresponding axis and point , we project each ambiguous point onto each plane i. We then compute the Manhattan distance between each projected point and each . Ambiguous points are assigned to the ring (axis) with the shortest Manhattan distance.
Figure 4D shows the final clustering after resolving ambiguous points. Resolution of ambiguous points not only improves cylinder accuracy but could also be useful for estimating branch volumes.
2.5. Final Skeleton Graph
The clustering step produces an axis for each cylinder ring found in the Gaussian sphere. Each of these axes represents a direction of a splitting branch. To generate the final skeleton graph, we need to identify the new position of the branch point, and then connect the new branches to this branch point.
The identified axes () are 3D vectors that will generally not intersect. We find points of intersection between all pairs of axis vectors by using the skew lines intersection algorithm. The idea is to find the shortest line segments that connect each pair of initial axes. For each pair, the point of intersection is defined as the midpoint of the line segment.
For example, for axes i and , we turned the cylinder axes and centroids into 3D line equations:
and solved the linear system of equations for u where
Inserting and back into the linear equations yields the two closest points on those lines, which form a line segment. The center of this line segment is the intersection point of and . The average of the intersection points, across all pairs of lines, is the new position of the fork node. Lastly, we create a new node for each branch axis vector i at its centroid and insert these nodes in the old skeleton graph. We then add an edge between the intersection point and the new nodes for each branch.
For example, in Figure 1 (Part 2), we show how a single fork node is divided into three nodes, corresponding to the mother branch and two daughter branches. The graph structure outside the fork node remains the same. This procedure enhances the geometry of the skeleton graph at fork areas, thus, making angle measurements more accurate as we evaluate next.
2.6. Plant Data
We tested our method on point clouds generated from 3D scans of tomato (Solanum lycopersicum cv m82D) and tobacco (Nicotiana benthamiana) plants [10,30,41]. To capture a wide range of shoot architectures and branch angles, plants were grown under several conditions: an ambient light control, shade, high-temperature, high-light, and drought. These conditions promote diverse phenotypes and are representative of realistic growth environments.
We used the FaroArm EDGE Model 14000, which is a non-invasive laser scanner that provides micron-level resolution, with an error ± 25 m. Smaller plants had about 10,000 points (with a height of ≈30 cm), whereas the largest plants had about 100,000 points (height of up to ≈80 cm), excluding leaf/lamina points. The average distance between points was about 0.125 cm. Not all LiDAR sensors acquire data at the same density; in our conclusions, we describe how our method’s parameters could be adapted to point clouds with variable densities.
Each plant was scanned at developmental timepoints 5, 12, and 20 days post-germination. We analyzed 31 forks (18 tomato and 13 tobacco) from 12 plants (8 tomato and 4 tobacco), comprising a total of 76 angles; many forks split two ways, but some split ways.
Prior to applying our method, three pre-processing steps were applied to the raw scanned point cloud (Figure 5). First, the object of interest was selected within the point cloud, and all background objects were manually removed. For example, in our scans, we removed the pot in which the plant grew from the point cloud (Figure 5A→B).
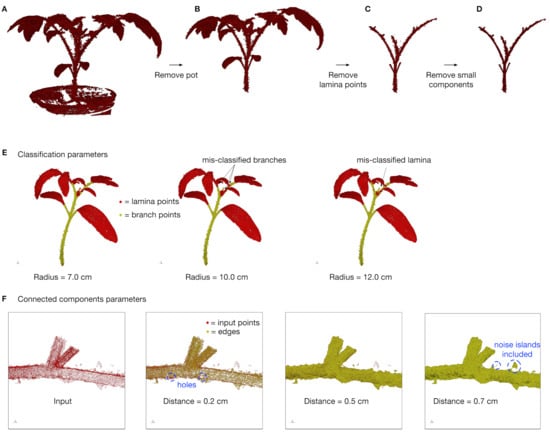
Figure 5.
Pre-processing steps. (A) The raw input point cloud after scanning. (B) The point cloud after manually removing the pot. (C) The point cloud after removing lamina points with deep learning classification [30]. (D) The final set of branch points after removing small connected components. (E) The effect of the radius parameter on classification performance. The radius parameter is used to compute fast point feature histograms [42], which are used as feature vectors for classification. (F) The effect of the distance parameter on the connected components algorithm. The distance parameter is used to identify and remove isolated islands of points prior to applying our skeletonization algorithm.
Second, points that correspond to the lamina/leaves were removed from the point cloud. To do this, we applied a classification algorithm [30] to automatically identify branch and leaf points. This algorithm used a deep learning model, where, for each point, we computed a feature vector using fast point feature histograms (FPFH [42]) with a radius parameter. The radius parameter was designed to capture neighborhood points around a point of interest to compute a descriptive, local feature vector for the point. The geometry around a lamina point is often flat or plane-like, whereas the geometry around a branch point often has a cylindrical shape. Thus, the radius should be large enough to capture these differences, but not so large that it jeopardizes them (particularly since there are many more lamina points than branch points); in addition, the radius should be adjusted based on the point cloud density (typically, lower density → larger radius). For example, Figure 5E shows examples of different radii that induce different errors, including mis-classifying branch points as lamina points (Figure 5E, Radius = 10.0 cm) and mis-classifying lamina points as branch points (Figure 5E, Radius = 12.0 cm). We used a radius of 5.0 cm, which was selected after performing a grid search and selecting the radius yielding the highest classification performance. In Ziamtsov and Navlakha (2019), we performed a full analysis of how different radii affect both classification performance and time complexity. The deep learning network architecture consisted of an input layer with 33 nodes (FPFH features), three hidden layers with 33, 66, and 33 nodes per layer, respectively, and an output layer with two nodes (lamina or branch). We applied the model to each point, and all lamina points were removed (Figure 5B→C).
Third, we ran a connected components algorithm on the identified branch points, where two points are considered neighbors if they are within a distance of 0.5 cm from each other. We then discarded points from all components except the largest component (Figure 5C→D). This removed small “noise islands” that were detached from the plant. Very small distances led to the removal of points in the middle of branches (see blue circles in Figure 5F, Distance = 0.2 cm), which caused holes in the branches. These small (non-noise) islands are expected when points are non-uniformly distributed. On the other hand, large distances caused actual noise islands to be included within the branching (see blue circles in Figure 5F, Distance = 0.7 cm), which can lead to an incorrect skeleton and branch angles. Thus, the value of the distance parameter depends on the density of the point cloud and should be selected so that branch points maintain a solid, cylindrical shape without capturing peripheral noise. Once the branch points were identified, our method was then applied directly.
3. Results
We compared the accuracy and run-time of our method to three popular approaches for skeleton extraction from 3D point clouds: PypeTree* [5,30], -medial skeletonization [7], and Laplacian-based contraction [3]. PypeTree* [5,30] is a geometric method specifically tailored for plant biology applications. The -medial skeleton [7] is a distance field-based algorithm designed to operate on general point clouds. Laplacian-based contraction [3] is a thinning-based algorithm and is the successor to the well-known ROSA method [2]. Each of these methods represents a different family of skeleton extraction algorithms (see related work for details). For each method, we used a grid search to identify the best set of parameters yielding the most visually accurate skeletons. We used the publicly available code and implementation for each method.
We measured the angles produced at each fork and compared them to ground-truth angles, which were measured manually via cylindrical fitting in third party point cloud visualization software. To test our method’s robustness, we selected a range of plants with varying sizes, amounts of missing data, and levels of registration noise. Figure 6 shows a subset of 7 of the 12 plants evaluated, consisting of tomato plants spanning four growth conditions.
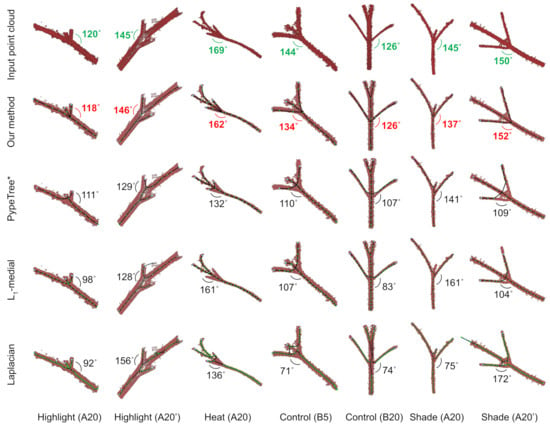
Figure 6.
Branch angle prediction for four methods. Seven forks are shown (columns), each from tomato plants across different conditions. The name “Highlight (A20)” means the fork comes from a plant grown in highlight conditions scanned on developmental day 20. Plants are given names “A” or “B” for replicates. The apostrophe symbol denotes a different fork that belongs to the same plant. Each row shows the skeletons and estimated angles for each of the four methods; the top row shows the ground-truth angle, computed manually. Only one angle is highlighted in each fork.
3.1. Accuracy of Branch Angles
Our method consistently generated accurate skeletons at forks and remained more robust to noise and missing data compared to other methods. Figure 6 shows example forks from tomato plants (columns) and the ground-truth angle calculation for each fork (top row), followed by the skeletons and estimated angles for each of the four methods (subsequent rows). For example, on the Highlight (A20) plant, the ground-truth angle was 120. The skeleton extracted by our method estimated the angle to be 118, whereas the estimates of the other methods were much worse: 111(PypeTree*), 98 (-medial), and 92 (Laplacian).
These seven examples are illustrated because they represent a range of challenges faced by skeletonization algorithms. For example, Highlight (A20), Control (B5), and Shade (A20’) all show noisy forks with hairs (trichomes) on the stem and with registration errors; Highlight (A20) and Highlight (A20’) show examples of missing data at a fork; and Shade (A20) and Heat (A20) show “webbed” forks.
Each method struggled with overcoming these challenges. For example, -medial often missed small branches due to stochastic sampling (Heat A20, missed two branches). PypeTree* mainly struggled with averaging points from different branches at forks, which tended to pull the branch point away from the main axis, in turn reducing the quality of angles (e.g., Shade (A20’) and Heat (A20)). Laplacian was the least resilient to noise (e.g., Shade (A20’), Control (B5)), and closely packed branches often produced loops, spurious branches, and erroneous skeleton shapes (Shade (A20’), Control (B5)). All three methods performed the worst on webbed forks.
For our method, for the three plants with noisy forks and registration errors, the estimated angles had an average percent error of 4.9%; for the two plants with missing data at the fork, our method had an error of 3.5%; and for the two plants with webbed forks, our method had an error of 5.0%. Thus, while our method performed better than the other methods in overcoming these challenges, it was most sensitive to registration errors and noise in the form of webbed forks.
Over all 18 tomato forks analyzed (from 8 plants in 5 conditions), in terms of absolute errors, our method was on average 1.96-times more accurate than PypeTree*; 2.25-times more accurate than -medial; and 3.15-times more accurate than Laplacian. In terms of percent errors, our method was on average 1.90-times more accurate than PypeTree*; 2.21-times more accurate than -medial; and 2.92-times more accurate than Laplacian.
Importantly, our method demonstrated similar gains when tested on forks from tobacco plants. Over 13 forks from four tobacco plants across two conditions, in terms of absolute errors, our method was on average 1.39-times more accurate than PypeTree*; 2.13-times more accurate than ; and 2.16-times more accurate than Laplacian. In terms of percent errors, our method was on average 1.26-times more accurate than PypeTree*; 1.99-times more accurate than -medial; and 1.99-times more accurate than Laplacian. Thus, our method was able to generalize well across two species.
The results from all 31 forks are quantified in Figure 7, demonstrating our method’s improved estimation of branch angles in terms of absolute errors (Figure 7A) and percent errors (Figure 7B).
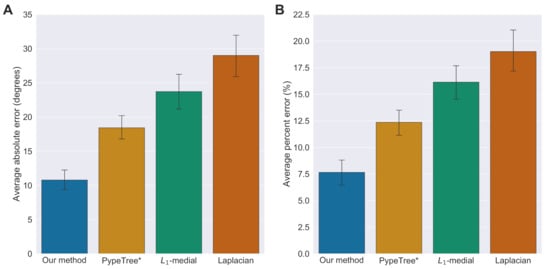
Figure 7.
Accuracy of predicted angles for all methods. (A) Average absolute errors for each method, defined as the absolute value of the ground-truth angle minus the predicted angle. Error bars represent the standard error of the mean. (B) Average percent errors for each method, defined as the ratio of the absolute error to the ground-truth angle. Error bars represent the standard error of the mean.
3.2. Run-Time Performance
For each fork, we compared the amount of time each method took to run. Measurements were made on a Windows 10 Pro 64-bit machine with an Intel i7-2600 CPU @ 3.4 GHz, and 16 GB RAM. As the -medial method is stochastic (i.e., it requires selection of random seeds during initialization), we averaged its run-time over four runs.
Table 1 shows that PypeTree* and our method are the two fastest algorithms. On average, our method is only 0.17-times slower than PypeTree*, suggesting that the error correction process we applied on top of the original PypeTree algorithm only leads to a small loss in timing efficiency. The other two methods (-medial and Laplacian) are significantly slower. Specifically, our method is 2.81-times faster than -medial and 41-times faster than Laplacian. -medial was fairly competitive on larger plants with more cloud points, but showed significantly slower times on smaller plants compared to PypeTree* and our method.

Table 1.
Run-time comparison for all methods. All measurements shown are in seconds.
In summary, our method can compute branch angles for large plants in seconds and generally produces more accurate branch angles than existing methods.
4. Conclusions
In this work, we presented a fast method that improved the extraction of skeleton graphs from point clouds of plant shoot architectures. We tested our method on high-resolution 3D point cloud data from two species of plants (tomato and tobacco) and showed significant improvement in the accuracy of angle estimates compared to three competing approaches. Our method has the potential to be deployed in large-scale phenotyping applications with noisy point clouds, and is designed to be used with minimal pre-processing. For example, our method can also be applied to point clouds generated from technologies other than LiDAR, such as multi-view photogrammetry [43]. In general, such technologies face challenges not typically faced by laser scanning, including white balance matching, feature matching across images for registration, and errors associated with approximating the camera position and camera sensor noise. However, once these challenges are addressed and normal vectors are estimated, our method can be used after the appropriate pre-processing steps are applied. All of our algorithms are available and implemented in P3D, our open-source plant phenotyping toolkit [44].
There are three main challenges to accurate skeletonization: noise, variable point density, and missing data. To review, our method deals with noise in individual normal vectors (e.g., due to trichomes) by averaging normal directions within a local neighborhood. To handle point clouds with different densities, our method includes a radius parameter that delineates the size of the neighborhood; for lower densities, the radius may need to be increased. If different parts of the point cloud have different densities, then the radius parameter may need to adapt to the local density of the point cloud. Finally, for handling missing data, our method clusters normal vectors that belong to the same ring on the Gaussian sphere; this allows us to cluster branches even if they have holes.
Our method (Branch-Pipe) includes five main parameters: (1) the radius of the neighborhood for averaging normal vectors; (2) the radius of the neighborhood for sampling cylinder axes; (3) the RBF kernel parameter in the clustering step; (4) the stopping condition in the clustering step; and (5) the degree cutoff, where normals are assigned to the same ring if they are within a certain range. We offer the following guidance for how to set these parameters. The first four parameters depend on the density of the point cloud with sparser point clouds requiring higher values of these parameters. For example, the first two parameters (the two radii) should be set to capture just enough points to establish a good local representation. In practice, we used the same value for both radii. The fifth parameter depends on the noise level of the normal vectors; we observed through an empirical experiment that normal vectors of a cylindrical branch isolated from our data set had about 10 degrees of error.
There are several lessons that we learned from applying these methods to noisy plant architectures. First, accurate angle estimation benefits from considering normal vector directions at forks. Spawning branches have small angles and can be very close to each other, which causes points to cluster together; using normal vectors to distinguish each branch thus becomes critical. Second, while highly parameterized methods (e.g., Laplacian) provide more flexibility, in practice, it was often difficult to tune the parameters to work across a range of different fork types. Third, non-deterministic methods that require random down-sampling (e.g., -medial) increase efficiency; however, we found that the underlying skeletons could be susceptible to noise in the sampling process, often leading to broken skeletons and non-trivial angle variation across runs. Fourth, taking advantage of some plant-specific geometric characteristics—such as the assumption that branches are cylindrical, the absence of loops in the skeleton, and the tapering of branch radii—can lead to improvements in the overall skeleton quality and performance.
There are several avenues for future work. First, the identification of forks requires that each level set (colored points in Figure 1) is sampled with enough points from each branch. PypeTree* generates each level based on distances from the root; however, sometimes this approach places a border between two adjacent levels that lies right in the middle of a fork. While uncommon, this can compromise the quality of predicted angles. Thus, designing automated methods that improve the identification of levels such that the borders of adjacent levels do not cut through a fork remains an open challenge. Second, the angle quality can be improved by better calculating the positions of nodes within levels. Currently, our method defines a node in the skeleton graph to be the centroid of the points in the level. Developing a method that considers normals when calculating the centroid could improve not only the angle predictions but also the quality of the entire skeleton graph. Third, when we insert new nodes into the skeleton after clustering, we assume that all branches emit from the same node; while this is certainly true in many cases, there are times when branches can be offset relative to each other. Adding a cleverer node-merging mechanism would further improve the quality of the skeletons. Finally, while we designed our method to be species-agnostic, we only tested it on two Solanaceous species: tomato (Solanum lycopersicum cv m82D) and tobacco (Nicotiana benthamiana). Evaluating it on broader datasets with more diverse phenotypes could identify additional challenges.
Author Contributions
Conceptualization, I.Z. and S.N.; methodology, I.Z., K.F. and S.N.; software, I.Z.; validation, I.Z., K.F.; writing—original draft preparation, I.Z., K.F. and S.N.; writing—review and editing, I.Z., K.F. and S.N.; funding acquisition, S.N. All authors have read and agreed to the published version of the manuscript.
Funding
S.N. was supported by the Pew Charitable Trusts, the National Science Foundation under award CAREER DBI-1846554, and funding from the Simons Center for Quantitative Biology at Cold Spring Harbor Laboratory.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data and code executable are available at: https://github.com/iziamtso/P3D, accessed on 14 September 2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cornea, N.D.; Silver, D.; Min, P. Curve-Skeleton Properties, Applications, and Algorithms. IEEE Trans. Vis. Comput. Graph. 2007, 13, 530–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagliasacchi, A.; Zhang, H.; Cohen-Or, D. Curve Skeleton Extraction from Incomplete Point Cloud. In ACM SIGGRAPH 2009 Papers; Association for Computing Machinery: New York, NY, USA, 2009; pp. 1–9. [Google Scholar]
- Cao, J.; Tagliasacchi, A.; Olson, M.; Zhang, H.; Su, Z. Point Cloud Skeletons via Laplacian Based Contraction. In Proceedings of the 2010 Shape Modeling International Conference, Aix-en-Provence, France, 21–23 June 2010; pp. 187–197. [Google Scholar] [CrossRef] [Green Version]
- Au, O.K.C.; Tai, C.L.; Chu, H.K.; Cohen-Or, D.; Lee, T.Y. Skeleton Extraction by Mesh Contraction. ACM Trans. Graph. 2008, 27, 1–10. [Google Scholar] [CrossRef]
- Delagrange, S.; Jauvin, C.; Rochon, P. PypeTree: A Tool for Reconstructing Tree Perennial Tissues from Point Clouds. Sensors 2014, 14, 4271–4289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verroust, A.; Lazarus, F. Extracting Skeletal Curves from 3D Scattered Data. Vis. Comput. 2000, 16, 15–25. [Google Scholar] [CrossRef]
- Huang, H.; Wu, S.; Cohen-Or, D.; Gong, M.; Zhang, H.; Li, G.; Chen, B. L1-Medial Skeleton of Point Cloud. ACM Trans. Graph. 2013, 32, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sharf, A.; Lewiner, T.; Shamir, A.; Kobbelt, L. On-the-fly Curve-skeleton Computation for 3D Shapes. In Computer Graphics Forum; Blackwell Publishing Ltd.: Oxford, UK, 2007; Volume 26, pp. 323–328. [Google Scholar]
- Ai, M.; Yao, Y.; Hu, Q.; Wang, Y.; Wang, W. An Automatic Tree Skeleton Extraction Approach Based on Multi-View Slicing Using Terrestrial LiDAR Scans Data. Remote Sens. 2020, 12, 3824. [Google Scholar] [CrossRef]
- Conn, A.; Pedmale, U.V.; Chory, J.; Navlakha, S. High-Resolution Laser Scanning Reveals Plant Architectures That Reflect Universal Network Design Principles. Cell Syst. 2017, 5, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucksch, A.; Atta-Boateng, A.; Azihou, A.F.; Battogtokh, D.; Baumgartner, A.; Binder, B.M.; Braybrook, S.A.; Chang, C.; Coneva, V.; DeWitt, T.J.; et al. Morphological plant modeling: Unleashing geometric and topological potential within the plant sciences. Front. Plant Sci. 2017, 8, 900. [Google Scholar] [CrossRef] [PubMed]
- Prusinkiewicz, P.; Lindenmayer, A. The Algorithmic Beauty of Plants; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Perez-Sanz, F.; Navarro, P.J.; Egea-Cortines, M. Plant Phenomics: An Overview of Image Acquisition Technologies and Image Data Analysis Algorithms. GigaScience 2017, 6, gix092. [Google Scholar] [CrossRef] [Green Version]
- Pieruschka, R.; Schurr, U. Plant phenotyping: Past, present, and future. Plant Phenomics 2019, 2019, 7507131. [Google Scholar] [CrossRef]
- Li, M.; Frank, M.H.; Coneva, V.; Mio, W.; Chitwood, D.H.; Topp, C.N. The Persistent Homology Mathematical Framework Provides Enhanced Genotype-to-Phenotype Associations for Plant Morphology. Plant Physiol. 2018, 177, 1382–1395. [Google Scholar] [CrossRef] [Green Version]
- Gaetan, L.; Serge, C.; Annie, E.; Didier, C.; Frederic, B. Characterization of Whole Plant Leaf Area Properties Using Laser Scanner Point Clouds. In Proceedings of the 2012 IEEE 4th International Symposium on Plant Growth Modeling, Simulation, Visualization and Applications, Shanghai, China, 31 October–3 November 2012; pp. 250–253. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Bassel, G.W. Linking Genes to Shape in Plants Using Morphometrics. Annu. Rev. Genet. 2020, 54, 417–437. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Snider, J.L.; Chee, P.W. In-Field High Throughput Phenotyping and Cotton Plant Growth Analysis Using LiDAR. Front. Plant Sci. 2018, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Andújar, D.; Rueda-Ayala, V.; Moreno, H.; Rosell-Polo, J.; Escolá, A.; Valero, C.; Gerhards, R.; Fernández-Quintanilla, C.; Dorado, J.; Griepentrog, H.W. Discriminating Crop, Weeds and Soil Surface with a Terrestrial LIDAR Sensor. Sensors 2013, 13, 14662–14675. [Google Scholar] [CrossRef] [Green Version]
- Mathan, J.; Bhattacharya, J.; Ranjan, A. Enhancing Crop Yield by Optimizing Plant Developmental Features. Development 2016, 143, 3283–3294. [Google Scholar] [CrossRef] [Green Version]
- Giovannetti, M.; Małolepszy, A.; Göschl, C.; Busch, W. Large-Scale Phenotyping of Root Traits in the Model Legume Lotus Japonicus. In Plant Genomics; Springer: New York, NY, USA, 2017; Volume 1610, pp. 155–167. [Google Scholar] [CrossRef]
- Yang, W.; Duan, L.; Chen, G.; Xiong, L.; Liu, Q. Plant phenomics and high-throughput phenotyping: Accelerating rice functional genomics using multidisciplinary technologies. Curr. Opin. Plant Biol. 2013, 16, 180–187. [Google Scholar] [CrossRef]
- Hackenberg, J.; Spiecker, H.; Calders, K.; Disney, M.; Raumonen, P. SimpleTree—An efficient open source tool to build tree models from TLS clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Morsdorf, F.; Meier, E.; Kötz, B.; Itten, K.I.; Dobbertin, M.; Allgöwer, B. LIDAR-based geometric reconstruction of boreal type forest stands at single tree level for forest and wildland fire management. Remote Sens. Environ. 2004, 92, 353–362. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H.; Dai, M.; Ma, W.; Quan, L. Data-driven synthetic modeling of trees. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1214–1226. [Google Scholar] [CrossRef]
- Qiu, R.; Zhou, Q.Y.; Neumann, U. Pipe-Run Extraction and Reconstruction from Point Clouds. In Computer Vision–ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer International Publishing: Cham, Switzerland, 2014; Volume 8691, pp. 17–30. [Google Scholar] [CrossRef] [Green Version]
- Han, X.F.; Jin, J.S.; Wang, M.J.; Jiang, W.; Gao, L.; Xiao, L. A review of algorithms for filtering the 3D point cloud. Signal Process. Image Commun. 2017, 57, 103–112. [Google Scholar] [CrossRef]
- Pauly, M.; Mitra, N.J.; Guibas, L.J. Uncertainty and Variability in Point Cloud Surface Data. In Proceedings of the Eurographics Symposium on Point-Based Graphics, SPBG’04. Zurich, Switzerland, 2–4 June 2004; Eurographics Association: Goslar, Germany, 2004; pp. 77–84. [Google Scholar]
- Xia, S.; Chen, D.; Wang, R.; Li, J.; Zhang, X. Geometric primitives in LiDAR point clouds: A review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 685–707. [Google Scholar] [CrossRef]
- Ziamtsov, I.; Navlakha, S. Machine Learning Approaches to Improve Three Basic Plant Phenotyping Tasks Using Three-Dimensional Point Clouds. Plant Physiol. 2019, 181, 1425–1440. [Google Scholar] [CrossRef] [Green Version]
- Hassouna, M.S.; Farag, A.A. Robust centerline extraction framework using level sets. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 458–465. [Google Scholar]
- Mei, J.; Zhang, L.; Wu, S.; Wang, Z.; Zhang, L. 3D tree modeling from incomplete point clouds via optimization and L 1-MST. Int. J. Geogr. Inf. Sci. 2017, 31, 999–1021. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L.; Fang, T.; Mathiopoulos, P.T.; Qu, H.; Chen, D.; Wang, Y. A structure-aware global optimization method for reconstructing 3-D tree models from terrestrial laser scanning data. IEEE Trans. Geosci. Remote Sens. 2014, 52, 5653–5669. [Google Scholar] [CrossRef]
- Ogniewicz, R.; Ilg, M. Voronoi Skeletons: Theory and Applications. In Proceedings of the 1992 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Champaign, IL, USA, 15–18 June 1992; pp. 63–69. [Google Scholar] [CrossRef]
- Dey, T.K.; Sun, J. Defining and computing curve-skeletons with medial geodesic function. In Proceedings of the Symposium on Geometry Processing, Cagliari, Italy, 26–28 June 2006; Volume 6, pp. 143–152. [Google Scholar]
- Mitra, N.J.; Nguyen, A. Estimating surface normals in noisy point cloud data. In Proceedings of the Nineteenth Annual Symposium on Computational Geometry, San Diego, CA, USA, 8–10 June 2003; pp. 322–328. [Google Scholar]
- Zhang, J.; Cao, J.; Liu, X.; Wang, J.; Liu, J.; Shi, X. Point cloud normal estimation via low-rank subspace clustering. Comput. Graph. 2013, 37, 697–706. [Google Scholar] [CrossRef]
- Fischler, M.A.; Bolles, R.C. Random Sample Consensus: A Paradigm for Model Fitting with Applications to Image Analysis and Automated Cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Marton, Z.C.; Pangercic, D.; Blodow, N.; Kleinehellefort, J.; Beetz, M. General 3D modelling of novel objects from a single view. In Proceedings of the 2010 IEEE/RSJ International Conference on Intelligent Robots and Systems, Taipei, Taiwan, 18–22 October 2010; pp. 3700–3705. [Google Scholar] [CrossRef] [Green Version]
- Comaniciu, D.; Meer, P. Mean Shift: A Robust Approach Toward Feature Space Analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef] [Green Version]
- Conn, A.; Pedmale, U.V.; Chory, J.; Stevens, C.F.; Navlakha, S. A Statistical Description of Plant Shoot Architecture. Curr. Biol. 2017, 27, 2078–2088.e3. [Google Scholar] [CrossRef] [Green Version]
- Rusu, R.B.; Blodow, N.; Beetz, M. Fast Point Feature Histograms (FPFH) for 3D registration. In Proceedings of the 2009 IEEE International Conference on Robotics and Automation, Kobe, Japan, 12–17 May 2009; pp. 3212–3217. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Slaughter, D.C.; Max, N.; Maloof, J.N.; Sinha, N. Structured light-based 3D reconstruction system for plants. Sensors 2015, 15, 18587–18612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziamtsov, I.; Navlakha, S. Plant 3D (P3D): A Plant Phenotyping Toolkit for 3D Point Clouds. Bioinformatics 2020, 36, 3949–3950. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).